Abstract
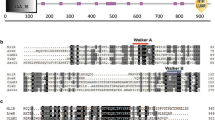
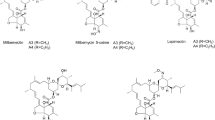
Milbemycins A3/A4 are important 16-membered macrolides which have been commercialized and widely used as pesticide and veterinary medicine. However, similar to other milbemycin producers, the production of milbemycins A3/A4 in Streptomyces bingchenggensis is usually accompanied with undesired by-products such as C5-O-methylmilbemycins B2/B3 (α-class) and β1/β2 (β-class) together with nanchangmycin. In order to obtain high yield milbemycins A3/A4-producing strains that produce milbemycins A3/A4 as main components, milD, a putative C5-O-methyltransferase gene of S. bingchenggensis, was biofunctionally investigated by heterologous expression in Escherichia coli. Enzymatic analysis indicated that MilD can catalyze both α-class (A3/A4) and β-class milbemycins (β11) into C5-O-methylmilbemycins B2/B3 and β1, respectively, suggesting little effect of furan ring formed between C6 and C8a on the C5-O-methylation catalyzed by MilD. Deletion of milD gene resulted in the elimination of C5-O-methylmilbemycins B2/B3 and β1/β2 together with an increased yield of milbemycins A3/A4 in disruption strain BCJ13. Further disruption of the gene nanLD encoding loading module of polyketide synthase responsible for the biosynthesis of nanchangmycin led to strain BCJ36 that abolished the production of nanchangmycin. Importantly, mutant strain BCJ36 (∆milD∆nanLD) produced milbemycins A3/A4 as main secondary metabolites with a yield of 2312 ± 47 μg/ml, which was approximately 74 % higher than that of the initial strain S. bingchenggensis BC-109-6 (1326 ± 37 μg/ml).






Similar content being viewed by others
References
Baltz RH (2011) Strain improvement in actinomycetes in the postgenomic era. J Ind Microbiol Biotechnol 38:657–666
Chen Y, Smanski MJ, Shen B (2010) Improvement of secondary metabolite production in Streptomyces by manipulating pathway regulation. Appl Microbiol Biotechnol 86:19–25
Clark EDB, Schwarz E, Rudolph, R (1999) Inhibition of aggregation side reactions during in vitro protein folding. In: Ronald W (ed) Methods in enzymology. Academic, pp 217–236
Cropp A, Chen S, Liu H, Zhang W, Reynolds KA (2001) Genetic approaches for controlling ratios of related polyketide products in fermentation processes. J Ind Microbiol Biotechnol 27:368–377
Gao H, Zhuo Y, Ashforth E, Zhang L (2010) Engineering of a genome-reduced host: practical application of synthetic biology in the overproduction of desired secondary metabolites. Protein Cell 1:621–626
Gomez-Escrlbano JP, Bibb MJ (2011) Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol 4:207–215
He Y, Sun Y, Liu T, Zhou X, Bai L, Deng Z (2010) Cloning of separate meilingmycin biosynthesis gene clusters by use of acyltransferase–ketoreductase didomain PCR amplification. Appl Environ Microbiol 76:3283–3292
Hong YS, Lee JH, Kim HS, Kim KW, Lee JJ (2001) Targeted gene disruption of the avermectin B O-methyltransferase gene in Streptomyces avermitilis. Biotechnol Lett 23:1765–1770
Ibrahim RK, Bruneau A, Bantignies B (1998) Plant O-methyltransferases: molecular analysis, common signature and classification. Plant Mol Biol 36(1):1–10
Ikeda H, Ōmura S (1997) Avermectin biosynthesis. Chem Rev 97:2591–2609
Ikeda H, Nonomiya T, Usami M, Ohta T, Ōmura S (1999) Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc Natl Acad Sci U S A 96:9509–9514
Jung M, Saito A, Buescher G, Maurer M (2002) Milbemycin oxime. In: Vercruysse J, Rew RS (eds) Macrocyclic lactones in anti-parasitic therapy. CAB International, pp 51–74
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical streptomyces genetics. John Innes Foundation Norwich, UK
Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H (2010) Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad U S A 107:2646–2651
Lee HN, Kim HJ, Kim P, Lee HS, Kim ES (2012) Minimal polyketide pathway expression in an actinorhodin cluster-deleted and regulation-stimulated Streptomyces coelicolor. J Ind Microbiol Biotechnol 39:805–811
Lomovskaya N, Doi-Katayama Y, Filippini S, Nastro C, Fonstein L, Gallo M, Colombo AL, Hutchinso CR (1998) The Streptomyces peucetius dpsY and dnrX genes govern early and late steps of daunorubicin and doxorubicin biosynthesis. J Bacteriol 180:2379–2386
Lomovskaya N, Otten SL, Doi-Katayama Y, Fonstein L, Liu XC, Takatsu T, Inventi-Solari A, Filippini S, Torti F, Colombo AL, Hutchinson CR (1999) Doxorubicin overproduction in Streptomyces peucetius: cloning and characterization of the dnrU ketoreductase and dnrV genes and the doxA cytochrome P-450 hydroxylase gene. J Bacteriol 181:305–318
McCall JW (2005) The safety-net story about macrocyclic lactone heartworm preventives: a review, an update, and recommendation. Vet Parasitol 133:197–206
Mo SJ, Lee SK, Jin YY, Oh CH, Suh JW (2013) Application of a combined approach involving classical random mutagensis and metabolic engineering to enhance FK506 production in Streptomyces sp. RM7011. Appl Microbiol Biotechnol 97:3053–3052
Mosher RH, Paradkar AS, Anders C, Barton B, Jensen SE (1999) Genes specific for the biosynthesis of clavam metabolites antipodal to clavulanic acid are clustered with the gene for clavaminate synthase 1 in Streptomyces clavuligerus. Antimicrob Agents Chemother 43:1215–1224
Ni X, Li D, Yang L, Huang T (2011) Construction of kanamycin B overproducing strain by genetic engineering of Streptomyces tenebrarius. Appl Microbiol Biotechnol 89:723–731
Nonaka K, Kumasaka C, Okamoto Y, Maruyama F, Yoshikawa H (1999) Bioconversion of milbemycin-related compounds: biosynthetic pathway of milbemycins. J Antibiot 52:109–116
Nonaka K, Tsukiyama T, Okamoto Y, Sato K, Kumasaka C, Yammoto T, Maruyama F, Yoshikawa H (2000) New milbemycins from Streptomyces hygroscopicus subsp. aureolacrimosus: fermentation, isolation and structure elucidation. J Antibiot 53:694–704
Olano C, Lombó F, Méndez C, Salas JA (2008) Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng 10:281–292
Paradkar AS, Mosher RH, Anders C, Griffin A, Griffin J, Hughes C, Greaves P, Barton B, Jensen SE (2001) Applications of gene replacement technology to Streptomyces clavuligerus strain development for clavulanic acid production. Appl Environ Microbiol 67:2292–2297
Pluschkell U, Horowitz AR, Ishaaya I (1999) Effect of milbemectin on the sweet potato whitefly, Bemisia tabaci. Phytoparasitica 27:183–191
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Scotti C, Hutchinson CR (1996) Enhanced antibiotic production by manipulation of the Streptomyces peucetius dnrH and dnmT genes involved in doxorubicin (adriamycin) biosynthesis. J Bacteriol 178:7316–7321
Smanski MJ, Yu Z, Casper J, Lin S, Peterson RM, Chen Y, Wendt-Pienkowski E, Rajski SR, Shen B (2011) Dedicated ent-kaurene and ent-atiserene synthases for platensimycin and platencin biosynthesis. Proc Natl Acad Sci U S A 108:13498–13503
Sun Y, Zhou X, Liu J, Bao K, Zhang G, Tu G, Kieser T, Deng Z (2002) ‘Streptomyces nanchangensis’, a producer of the insecticidal polyether antibiotic nanchangmycin and the antiparasitic macrolide meilingmycin, contains multiple polyketide gene clusters. Microbiology 148:361–371
Takiguchi Y, Mishima H, Okuda M, Terao M, Aoki A, Fukuda R (1980) Milbemycins, a new family of macrolide antibiotics: fermentation, isolation and physico-chemical properties. J Antibiot 33:1120–1127
Tanaka Y, Komatsu M, Okamoto S, Tokuyama S, Kaji A, Ikeda H, Ochi K (2009) Antibiotic overproduction by rpsL and rsmG mutants of various actinomycetes. Appl Environ Microbiol 75:4919–4922
Wang XJ, Wang JD, Xiang WS, Zhang J (2009a) Three new milbemycin derivatives from Streptomyces bingchenggensis. J Asian Nat Prod Res 11:597–603
Wang XJ, Wang XC, Xiang WS (2009b) Improvement of milbemycin-producing Streptomyces bingchenggensis by rational screening of ultraviolet-and chemically induced mutants. World J Microbiol Biotechnol 25:1051–1056
Wang XJ, Yan YJ, Zhang B, An J, Wang JJ, Tian J, Jiang L, Chen YH, Huang SX, Yin M, Zhang J, Al G, Liu CX, Zhu ZX, Xiang WS (2010) Genome sequence of the milbemycin-producing bacterium Streptomyces bingchenggensis. J Bacteriol 192:4526–4527
Wei X, Yunxiang L, Yinghua Z (2006) Enhancement and selective production of oligomycin through inactivation of avermectin’s starter unit in Streptomyces avermitilis. Biotechnol Lett 28:911–916
Xiang WS, Wang JD, Wang XJ, Zhang J, Wang Z (2007) Further new milbemycin antibiotics from Streptomyces bingchenggensis. J Antibiot 60:608–613
Xiang WS, Wang JD, Fan HM, Wang XJ, Zhang J (2008) New seco-milbemycins from Streptomyces bingchenggensis: fermentation, isolation and structure elucidation. J Antibiot 61:27–32
Xiang WS, Wang JD, Wang XJ, Zhang J (2009a) Bingchamides A and B, two novel cyclic pentapeptides from the Streptomyces bingchenggensis: fermentation, isolation, structure elucidation and biological properties. J Antibiot 62:501–505
Xiang WS, Wang JD, Wang XJ, Zhang J (2009b) A novel macrolide compound from Streptomyces bingchenggensis: fermentation, isolation, structure elucidation and biological properties. J Antibiot 62:229–231
Yadav P, Singh R (2011) A review on anthelmintic drugs and their future scope. Int J Pharm Pharmaceut Sci 3:17–21
Yu Q, Bai LQ, Zhou XF, Deng ZX (2012) Inactivation of the positive LuxR-type oligomycin biosynthesis regulators OlmRI and OlmRII increases avermectin production in Streptomyces avermitilis. Chinese Sci Bull 57:869–876
Zhang X, Chen Z, Zhao J, Song Y, Wen Y, Li J (2004) Deletion analysis of oligo mycin PKS genes (olmA) in Streptomyces avermitilis. Chinese Sci Bull 49:350–354
Zhou M, Jing X, Xie P, Chen W, Wang T, Xia H, Qin Z (2012) Sequential deletion of all the polyketide synthase and nonribosomal peptide synthetase biosynthetic gene clusters and a 900-kb subtelomeric sequence of the linear chromosome of Streptomyces coelicolor. FEMS Microbiol Lett 33:169–179
Acknowledgments
This research was supported by the National Key Technology R&D Program (2010CB126102), the Outstanding Youth Foundation of China (31225024), the National Natural Science Foundation of China (30971937), the Program for New Century Excellent Talents in University (NCET-08-0668, 1154-NCET-002), the Outstanding Youth Foundation of Heilongjiang Province (JC200706), and the Program for New Teachers in University (20092325120007).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Zhang and An contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhang, J., An, J., Wang, JJ. et al. Genetic engineering of Streptomyces bingchenggensis to produce milbemycins A3/A4 as main components and eliminate the biosynthesis of nanchangmycin. Appl Microbiol Biotechnol 97, 10091–10101 (2013). https://doi.org/10.1007/s00253-013-5255-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-5255-5