Abstract
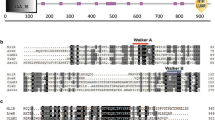
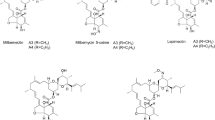
Milbemycins produced by several Streptomyces species are a group of 16-membered macrolides with potent insecticidal and anthelminthic activity. Milbemycin A3/A4, the main components of the milbemycins biosynthetic pathway, and 5-oxomilbemycin A3/A4, the analogs of milbemycin A3/A4 without the reduction of the C-5 keto group, have been developed as acaricides, insecticides, and anthelmintics. However, so far, little is known about the regulation of milbemycins biosynthesis, which has greatly hampered the generation of high producing strains by metabolic engineering. Herein, a TetR family regulator MilR2 (encoded by sbi_00792) was identified being involved in activation of 5-oxomilbemycin A3/A4 biosynthesis in a high 5-oxomilbemycins-producing strain Streptomyces hygroscopicus SIPI-KF. The ΔmilR2 mutant with an in-frame deletion of the MilR2 DNA-binding domain resulted in significantly reduced 5-oxomilbemycin A3/A4 production (approximately 36.9 and 39.7%) at tested two time points, and accordingly introduction of an extra copy of milR2 into SIPI-KF led to enhanced production by 12.6 and 34.4%. We further showed that MilR2 could directly repress the transcription of the gene sbi_00791 encoding a putative hydrolase, which is located divergently from milR2. The precise MilR2-binding site consisting of a 7-nt perfect inverted repeat separated by 10-nt (5′-ACCAACCAGCTGGTAAGGGTTGGT-3′) was defined. In situ mutagenesis of the MilR2-binding site resulted in 19.7 and 13.5% decreases in 5-oxomilbemycin A3/A4 production, which is much lower than the decreased rates of ΔmilR2. Collectively, the results demonstrated that MilR2 serves as an activator for 5-oxomilbemycin A3/A4 production and the function of MilR2 is only partially mediated through its repression on the transcription of sbi_00791.





Similar content being viewed by others
References
Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE (1992) Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116(1):43–49
Burg RW, Miller BM, Baker EE, Birnbaum J, Currie SA, Hartman R, Kong YL, Monaghan RL, Olson G, Putter I, Tunac JB, Wallick H, Stapley EO, Oiwa R, Omura S (1979) Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob Agents Chemother 15(3):361–367
Dun JL, Zhao YW, Zheng GS, Zhu H, Ruan LJ, Wang WF, Ge M, Jiang WH, Lu YH (2015) PapR6, a putative atypical response regulator, functions as a pathway-specific activator of pristinamycin II biosynthesis in Streptomyces pristinaespiralis. J Bacteriol 197(3):441–450
He YL, Sun YH, Liu TG, Zhou XF, Bai LQ, Deng ZX (2010) Cloning of separate meilingmycin biosynthesis gene clusters by use of acyltransferase-ketoreductase didomain PCR amplification. Appl Environ Microbiol 76(10):3283–3292
Ichinose H, Fujimoto Z, Kaneko S (2013) Characterization of an α-L-rhamnosidase from Streptomyces avermitilis. Biosci Biotechnol Biochem 77(1):213–216
Kim MS, Cho WJ, Song MC, Park SW, Kim K, Kim E, Lee N, Nam SJ, Oh KH, Yoon YJ (2017) Engineered biosynthesis of milbemycins in the avermectin high-producing strain Streptomyces avermitilis. Microb Cell Factories 16(1):9
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25(4):402–408
Mishima H, Kurabayashi M, Tamura C, Sato S, Kuwano H, Saito A, Aoki A (1974) Structures of milbemycins α1, α2, α3, α4, α5, α6, α7, α8, α9, α10 and β1. Abstract Papers 18th Symp. Chem Natural Products, symposium papers 18(0):309–316
Mishima H, Kurabayashi M, Tamura C, Sato S, Kuwano H, Saito A, Aoki A (1975) Structures of milbemycin β1, β2, and β3. Tetrahedron Lett 16(10):711–714
Myronovskyi M, Welle E, Fedorenko V, Luzhetskyy A (2011) Glucuronidase as a sensitive and versatile reporter in actinomycetes. Appl Environ Microbiol 77(15):5370–5383
Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R (2005) The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69(2):326–356
Strohl WR (1992) Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res 20(5):961–974
Tiffert Y, Supra P, Wurm R, Wohlleben W, Wagner R, Reuther J (2008) The Streptomyces coelicolor GlnR regulon: identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Mol Microbiol 67(4):861–880
Wang Y, Cen XF, Zhao GP, Wang J (2012) Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J Bacteriol 194(19):5237–5244
Wang XJ, Guo SL, Guo WQ, Xi D, Xiang WS (2009) Role of nsdA in negative regulation of antibiotic production and morphological differentiation in Streptomyces bingchengensis. J Antibiot (Tokyo) 62(6):309–313
Wang R, Mast Y, Wang J, Zhang WW, Zhao GP, Wohlleben W, Lu YH, Jiang WH (2013) Identification of two-component system AfsQ1/Q2 regulon and its cross-regulation with GlnR in Streptomyces coelicolor. Mol Microbiol 87(1):30–48
Wang XJ, Yan YJ, Zhang B, An J, Wang JJ, Tian J, Jiang L, Chen YH, Huang SX, Yin M, Zhang J, Gao AL, Liu CX, Zhu ZX, Xiang WS (2010) Genome sequence of the milbemycin-producing bacterium Streptomyces bingchenggensis. J Bacteriol 192(17):4526–4527
Wang XJ, Zhang B, Yan YJ, An J, Zhang J, Liu CX, Xiang WS (2013) Characterization and analysis of an industrial strain of Streptomyces bingchenggensis by genome sequencing and gene microarray. Genome 56(11):677–689
Wang HY, Zhang J, Zhang YJ, Zhang B, Liu CX, He HR, Wang XJ, Xiang WS (2014) Combined application of plasma mutagenesis and gene engineering leads to 5-oxomilbemycins A3/A4 as main components from Streptomyces bingchenggensis. Appl Microbiol Biotechnol 98(23):9703–9712
Wilkinson CJ, Hughes-Thomas ZA, Martin CJ, Bohm I, Mironenko T, Deacon M, Wheatcroft M, Wirtz G, Staunton J, Leadlay PF (2002) Increasing the efficiency of heterologous promoters in actinomycetes. J Mol Microbiol Biotechnol 4(4):417–426
Xiang WS, Wang JD, Wang XJ, Zhang J (2007) Two new beta-class milbemycins from streptomyces bingchenggensis: fermentation, isolation, structure elucidation and biological properties. J Antibiot (Tokyo) 60(6):351–356
Xiang WS, Wang JD, Wang XJ, Zhang J, Wang Z (2007) Further new milbemycin antibiotics from Streptomyces bingchenggensis. Fermentation, isolation, structural elucidation and biological activities. J Antibiot (Tokyo) 60(10):608–613
Zhang J, An J, Wang JJ, Yan YJ, He HR, Wang XJ, Xiang WS (2013) Genetic engineering of Streptomyces bingchenggensis to produce milbemycins A3/A4 as main components and eliminate the biosynthesis of nanchangmycin. Appl Microbiol Biotechnol 97(23):10091–10101
Zhang Y, He H, Liu H, Wang H, Wang X, Xiang W (2016) Characterization of a pathway-specific activator of milbemycin biosynthesis and improved milbemycin production by its overexpression in Streptomyces bingchenggensis. Microb Cell Factories 15(1):152
Acknowledgements
This work was supported by the National Major Scientific and Technological Special Project for “Significant New Drugs Development” (2014ZX09201001-005-001), the National Natural Science Foundation of China (31770088 and 31570072), and the Derivative Bank of Chinese Biological Resources, CAS (ZSYS-016). We also thank the support from SA-SIBS Scholarship Program.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
All of the authors confirm that ethical principles have been followed in the research as well as in manuscript preparation.
Electronic supplementary material
ESM 1
(PDF 894 kb)
Rights and permissions
About this article
Cite this article
Wei, K., Wu, Y., Li, L. et al. MilR2, a novel TetR family regulator involved in 5-oxomilbemycin A3/A4 biosynthesis in Streptomyces hygroscopicus. Appl Microbiol Biotechnol 102, 8841–8853 (2018). https://doi.org/10.1007/s00253-018-9280-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9280-2