Abstract
Background
Emergence of antimicrobial resistance by Staphylococcus aureus has limited treatment options against its infections. The purpose of this study was to determine the pooled prevalence of resistance to different antimicrobial agents by S. aureus in Ethiopia.
Methods
Web-based search was conducted in the databases of PubMed, Google Scholar, Hinari, Scopus and the Directory of Open Access Journals (DOAJ) to identify potentially eligible published studies. Required data were extracted and entered into Excel spread sheet. Statistical analyses were performed using Stata version 13.0. The metaprop Stata command was used to pool prevalence values. Twenty-one separate meta-analysis were done to estimate the pooled prevalence of the resistance of S. aureus to twenty-one different antimicrobial agents. Heterogeneity among the studies was assessed using the I2 statistic and chi-square test. Publication bias was assessed using Egger’s test. Because of significant heterogeneity amongst the studies, the random effects model was used to pool prevalence values.
Results
The electronic database search yielded 1317 studies among which 45 studies met our inclusion criteria. Our analyses demonstrated very high level of resistance to amoxicillin (77% [95% confidence interval (CI): 68%, 0.85%]), penicillin (76% [95% CI: 67%, 84%]), ampicillin (75% [95% CI: 65%, 85%]), tetracycline (62% [95% CI: 55%, 68%]), methicillin (47% [95% CI: 33%, 61%]), cotrimoxaziole (47% [95% CI: 40%, 55%]), doxycycline (43% [95% CI: 26%, 60%]), and erythromycin (41% [95% CI: 29%, 54%]). Relatively low prevalence of resistance was observed with kanamycin (14% [95% CI: 5%, 25%]) and ciprofloxacin (19% [95% CI: 13%, 26%]). The resistance level to vancomycin is 11% 995% CI: (4%, 20%). High heterogeneity was observed for each of the meta-analysis performed (I2 ranging from 79.36% to 95.93%; all p-values ≤0.01). Eggers’ test did not show a significant publication bias for all antimicrobial agents except for erythromycin and ampicillin.
Conclusions
S. aureus in Ethiopia has gotten notoriously resistant to almost to all of antimicrobial agents in use including, penicillin, cephalosporins, tetracyclines, chloramphenicol, methicillin, vancomycin and sulphonamides. The resistance level to vancomycin is bothersome and requires a due attention. Continued and multidimensional efforts of antimicrobial stewardship program promoting rational use of antibiotics, infection prevention and containment of AMR are urgently needed.
Similar content being viewed by others
Background
Staphylococcus aureus (S. aureus) infection is a major cause of skin, soft tissue, respiratory, bone, joint, and cardiovascular disorders [1]. S. aureus remains a versatile and dangerous pathogen in humans. The frequencies of both community-acquired and hospital-acquired staphylococcal infections have increased steadily. Treatment of these infections has become more difficult because of the emergence of multidrug-resistant strains [2].
Various mechanisms are responsible for S. aureus antimicrobial resistance (AMR). Penicillin is inactivated by β-lactamase. AMR to methicillin confers resistance to all β-lactamase-resistant penicillin’s and cephalosporins which require the presence of the mec gene that encodes penicillin-binding protein [3]. The enterococcal plasmid-bearing gene for resistance to vancomycin has been transferred by conjugation to S. aureus in vitro [4]. Both increased cell-wall synthesis and alterations in the cell wall that prevent vancomycin from reaching sites of cell-wall synthesis have been suggested as mechanisms [4]. Increase in vancomycin use has led to the emergence of two types of glycopeptide-resistant S. aureus. The first one, designated vancomycin intermediate-resistant S. aureus (VISA), is associated with a thickened and poorly cross-linked cell wall is due to continuous exposure to glycopeptide. The second type, vancomycin-resistant S. aureus (VRSA), is due to acquisition from Enterococcus species of the vanA operon resulting in high-level resistance and is a rare phenomenon [5].
In Ethiopia the first published antimicrobial preliminary report on AMR was published by Plorde et al. in 1970 for different microbial agents [6]. Beginning from that time AMR report were made by different antimicrobial surveillances and studies, it showed rapid rise and spread of resistant strains.
Facilitating more appropriate choices of treatment, minimizing the morbidity and mortality due to resistant infections, and preserving the effectiveness of antimicrobials requires summarization and synthesis of the evidence regarding AMR in a country. Appropriately summarized and synthesized evidence is mandatory for updating national treatment guidelines. To our knowledge, no previous meta-analysis or systematic review has been conducted on S. aureus AMR to all antimicrobial commonly in use in Ethiopia. The purpose of this study was, therefore, to determine pooled prevalence of S. aureus resistance to common antimicrobial agents in Ethiopia based on the best available studies.
Methods
Study design
This study did a meta-analysis of prevalence of S. aures resistance to different antimicrobial agents in Ethiopia using the best available studies.
Literature search strategy
Web-based search using PubMed, Google Scholar, Hinari, Scopus and the Directory of Open Access Journals (DOAJ) was conducted in June 2016. Google search was used for unpublished works and government documents. Two of the authors (SD and SF) independently searched for relevant studies to be included in this meta-analysis. The PubMed search was carried out via the EndNote software. Relevant search results from Google scholar, Embase, Scopus and the DOAJ were individually downloaded and manually entered into EndNote. The reference lists of the identified studies were used to identify other relevant studies.
The search was done using various key words: Staphylococcus, antimicrobial resistance, antibiotic resistance, drug resistance, drug susceptibility, antibacterial resistance, Ethiopia. These key terms were used in various combinations using Boolean search technique. We did not limit the search by year or language of publication.
Study selection procedures and criteria
Study selection was performed in two stages independently by two of the authors (SD and SF). First, the titles and abstracts of all retrieved articles were reviewed and then grouped as “eligible for inclusion” if they did address the study question and “ineligible for inclusion” if they did not. Second, articles which were grouped under “eligible for inclusion” were reviewed in full detail for decision.
All available studies and data were included based on the following predefined inclusion criteria. 1) Studies that were original journal articles, short communications, or unpublished works; 2) Studies that did the antimicrobial susceptibility test according to the criteria of the Clinical Laboratory Standards Institute (CLSI) and defined antimicrobial resistance range according to CLSI manual [7], 3) Studies which used human infection sample.
Studies that 1) were duplicates, 2) were based on small number of isolates (1–10), 3) were conducted on non-human samples like on foods, food handlers’ belongings, health workers belongings or health workers carriage and 4) which were based on non-infectious carriage were excluded from this meta-analysis.
Data extraction
Required data were extracted from eligible studies using Excel spreadsheet format prepared for this purpose by AA and SD. The data extracted from eligible studies include name of author(s), year of publication, place where the study was conducted, study design, total number of S. aureus isolate tested in the study, number of resistant S. aureus isolates, and isolate source. If the proportion of drug sensitive isolates (q) was reported, the number of resistant isolates was calculated by multiplying the number of isolates (n) by one minus the proportion of drug sensitive isolates (1-q) and if the proportion of drug resistant isolates was given the number of resistant isolates was found by multiplying the proportion (p) with total number of isolates (n).
Statistical analysis and reporting
Statistical analyses were performed using Stata version 13.0 (Statacorp, LP, college station, TX). The prevalence values from the different studies were pooled using the metaprop command in Stata [8]. We did twenty-one separate meta-analyses to estimate the pooled prevalence of the resistance of S. aureus to twenty one different antimicrobial agents. The number of studies included in each of the meta-analyses ranged from 4 to 39. Heterogeneity amongst the studies was assessed using the I2 statistic. Because of significant heterogeneity amongst the studies the random-effects model (REM) was used to estimate the pooled prevalence and 95% CIs using the DerSimonian and Laird method [9]. The Freeman-Tukey double arcsine transformation was used so that studies reporting proportions near or at 0 and 1 would not be excluded from the meta-analysis. The possible presence of publication bias was checked using Egger’s test [10].
For studies that appeared to report unusually higher prevalence of resistance compared to others, we did sensitivity analysis after dropping the study which we suspected of reporting a higher-than-usual result. If the point estimate of pooled prevalence after dropping a study lies within the 95% CI of the overall pooled estimate for all studies combined, we considered the given study as having non-significant influence on the overall pooled estimate. Otherwise, the study was considered as having significantly influencing the overall estimate.
Results of the current meta-analysis are reported as per the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guideline. The PRISMA checklist was used to ensure inclusion of relevant information (the filled checklist is included as Additional file 1: S1) [11].
Results
Included studies and characteristics
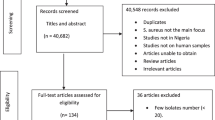
The electronic database search yielded 1317 from PubMed and 17,400 from Google scholar, Hinari, and Google search of which 16,083 articles remained after removing duplicate articles. Title and abstract screening reduced eligible articles to 76 for full text evaluation. After reading the full texts, 31 studies were excluded for various reasons. Thirteen studies were excluded as their report is based on small number of isolates (less than or equal to10) [12,13,14,15,16,17,18,19,20,21,22,23,24], four studies reported crude resistance for all bacterial pathogen isolated [25,26,27,28], eleven did not address our study question [29,30,31,32,33,34,35,36], six studies were based on samples taken from of healthy carriers [37,38,39,40,41,42], one study [43] was part of another study [44], and one study [45] suffered from environmental contamination of the samples during processing. Thus, 45 studies met our inclusion criteria (Fig. 1). Forty-one of the studies were journal articles, three were unpublished works [46,47,48] and one was an official government document from the Drug Administration and Control Authority (DACA) of Ethiopia [49].
S. aureus isolates from a total of 4570 patients were tested for their antimicrobial resistance. The isolates were from ear discharge [50,51,52,53,54,55,56,57], eye discharge [47, 58,59,60], blood [61,62,63,64,65,66,67,68], wound infection [69,70,71,72,73,74], surgical site infection [30, 73, 75,76,77,78], mixed samples [6, 46, 48, 49, 79,80,81,82,83], leprosy ulcer [84, 85], and urine sample [86, 87]. Twenty nine studies used primary data while nineteen studies used records from hospitals or regional laboratories (the characteristics of each included study is summarized Table 1).
Publication bias and heterogeneity
Evidence of high heterogeneity was observed for each of the meta-analyses performed (I2 ranging from 79.36% to 95.93%; all p-values ≤ 0.01). Eggers’ test did not suggest any significant publication bias except for erythromycin and ampicillin (see Additional file 2: S2).
Prevalence of S. aureus resistance to different antimicrobial agents
Summary of the pooled prevalence of S. aureus AMR prevalence for twenty-one different antimicrobial agents and the number of studies included in the meta-analysis for each agent are presented in Table 2. Prevalence of S. aureus resistance for each antimicrobial agent based on pharmacological classification of the agents is given below. As new anti-MRSA agents such as linezolid, daptomycin, tigecycline, telavancin and ceftaroline are rarely available in Ethiopia and no published studies available on resistance to this agents, our results do not cover such agents.
Prevalence of resistance to glycopeptides (vancomycin)
Nineteen studies were included for meta-analysis of vancomycin resistance prevalence. The pooled prevalence for S. aureus resistance to vancomycin in Ethiopia is 11% (95% confidence interval [CI]: 4%, 20%). The forest plot for vancomycin resistance is presented in Fig. 2. The results of sensitivity analysis after exclusion of the two studies that appeared to report outlier prevalence values separately and both together showed non-significant influence of the two studies on the overall estimate. The pooled prevalence of vancomycin resistance when Guta et al. and Desalegn et al. were removed separately was 0.09, (95% CI: 0.03, 0.17). When, both Guta et al. and Desalegn et al. were excluded, vancomycin resistance was 0.07 (95% CI: 0.02, 0.14). All the three pooled values lie within the overall pooled estimate.
Prevalence of resistance to penicillin’s
Here, the pooled prevalence of S. aureus resistance to penicillin G, amoxicillin, ampicillin, and amoxacilin-caluvanic acid was estimated. Resistance to penicillin G was estimated based on 33 studies, to amoxicillin based on 18 studies, to ampicillin based on 27 studies and to amoxacilin-caluvanic acid based on 12 studies. Pooled resistance rates were highest for β-lactamase-sensitive penicillin’s. Resistance to amoxicillin was 77% (95% CI: 68%, 85%), to penicillin G 75% (95% CI: 65%, 85%) and to ampicillin 76% (95% CI: 67%, 84%). Resistance to carbencilin (β-lactam-sensitive antibiotic) was relatively lower than other β-lactam-antibiotics (34% [95% CI: 17%, 54%]).
Relatively lower resistance rate was observed to β-lactamase-resistant penicillin’s: methicillin (47% [95% CI: 33%, 61%]) and amoxicillin-clavulanic acid (30% [95% CI: 19%, 43%]). The forest plots for methicillin and amoxacilin resistance are presented in Figs. 3 and 4, respectively while the forest plots for penicillin G, ampicillin, amoxicillin-clavulanic acid, and carbencillin resistance are presented in Additional file 3: S3, Additional file 4: S4, Additional file 5: S5 and Additional file 6: S6.
Prevalence of resistance to cephalosporins
Prevalence of the resistance of S. aureus to cephalosporins is similar to the prevalence of resistance to β-lactamase-resistant penicillin’s (amoxaclin-clavulanic acid). The prevalence of resistance to cephalothin is 30% (95% CI: 18%, 43%), to ceftriaxone 34% (95% CI: 25%, 43%) and to cefoxitine 27% (95% CI: 6%, 54%). The forest plot for ceftriaxone resistance is presented in Fig. 5 while the forest plots for cephalotine and cefoxitine resistance are presented respectively in Additional file 7: S7 and Additional file 8: S8.
Prevalence of resistance to floroquinolones
Two antimicrobial agents were tested from the floroquinolones: ciprofloxacin and norfloxacilin. Thirty one studies were used to estimate the prevalence of ciprofloxacin resistance and eleven studies were included for the estimation of norfloxacilin resistance. The pooled prevalence of S. aureus resistance to ciprofloxacin was 19% (95% CI: 13%, 26%) and to norfloxacillin 25% (95% CI: 14%, 38%). The forest plot for ciprofloxacin resistance is presented in Fig. 6 while the forest plot for norfloxacilin included as Additional file 9: S9.
Prevalence of resistance to protein synthesis inhibitors
Higher rates of resistance were observed with reversible inhibitors of protein synthesis compared to aminoglycosides (irreversible inhibitors of protein synthesis). Tetracycline showed the highest resistance rate (62% [95% CI: 55%, 68%]) followed by doxycycline 43% (95% CI: 26%, 60%), erythromycin (41% [95% CI: 29%, 54%]), and chloramphenicol (37% [95% CI: 29%, 54%]). Clindamycin and aminoglycosides showed relatively lower level of resistance (Table 2).
The prevalence of resistance to gentamycin is 26% (95% CI: 18%, 34%), to amikacin 23% (95% CI: 7%, 44%) and to kanamycin 14% (95% CI: 5%, 25%). The forest plot for gentamycin resistance is presented in Fig. 7 while the forest plots for erythromycin, chloramphenicol, doxycycline, amikacin, clindamycin, and kanamycin resistance are presented as the Additional file 10: S10, Additional file 11: S11, Additional file 12: S12, Additional file 13: S13, Additional file 14: S14 and Additional file 15: S15.
Prevalence of resistance to antimetabolites
Thirty five studies were included for estimation of pooled prevalence of S. aureus resistance to sulphametaxozole-trimethoprim and found to be 47% (95% CI: 40%, 55%). The forest plot for sulphametaxozole- trimethoprim resistance is presented in Fig. 8.
Comparison of the prevalence of S. aures resistance to different antimicrobial agents
Comparison of the prevalence of S. aures resistance to different antimicrobial agents addressed by this meta-analysis is given in Fig. 9. It is found that the magnitude of S.aureus resistance to the different antimicrobial agents ranges from 11% to vancomycin to 77% to amoxicillin. Accordingly, invitro antimicrobial effectiveness in decreasing order believed to be vancomycin, kanamycin, ciprofloxacilin, amikacin, clindamycin, amoxacilin-clavulanic acid, cephalothin, carbencilin, ceftriaxone, cefoxitine, chloramphenicol, erythromycin, doxycycline, methicillin, cotrimoxazole, tetracycline, ampicillin, pencilin, and amoxacilin.
Discussion
In this meta-analysis, we estimated the pooled prevalence of S. aureus resistance to 21 different antimicrobial agents commonly used in Ethiopia. Generally 45 studies were included for the meta-analysis, however the number of studies included in each meta-analyses ranged from 4 to 39. Overall, the 45 studies provided evidence regarding the level of S. aureus resistance to different antimicrobial agents based on 4530 isolates. It was found that S. aureus resistance to commonly available antimicrobial agents in Ethiopia was alarmingly high ranging from 11% to vancomycin to 77% to amoxicillin.
The pooled estimate of the prevalence of S. aureus resistance particularly to methicillin (MRSA) in Ethiopia is similar to 2014 global surveillance reports of the World Health Organization (WHO) 2014 [88], which showed MRSA prevalence between 33% to 95% in Africa. The pooled prevalence of MRSA in Ethiopia 47% (95% CI: 33%–61%) is within the range of the global WHO report for Africa.
The pooled estimate in study for MRSA is in agreement with the pooled estimate of community acquired-MRSA prevalence in Asia, Europe, and North America which ranges from 23.1% to 47.4% [89]. However, the pooled estimate 47% (95% CI: 33%–61%) MRSA prevalence in Ethiopia is higher than pooled estimate of community acquired MRSA prevalence 30.2% based on 27 retrospective studies and 37.3% based on 5 prospective studies [90]. The higher prevalence in our study may be due to the inclusion of both community acquired and nosocomial infection in the original studies. Nosocomial infection are believed to have higher rate of resistance due to larger exposure rate to antimicrobial agents. Increasing resistance to antimicrobial agents in hospitals is caused by transmission of resistant strains within hospitals by cross colonization of patients via hands of healthcare staff and direct patient to patient contact and subsequent spread [91].
Global pattern of AMR shows variation among different geographic, socioeconomic strata and among studies [49, 88, 92]. Variation may be to differences in time, place, design, and population involved in the study. This may be due to healthcare facilities conditions like implementation and monitoring of infection prevention policies and rational antibiotic usage which varies in different facilities. The most important reason is due to character of the study. Studies are conducted within a specified time and locality. It is reasonable to assume population under study might be infected by the same strains of agent at specified period of time and location. This could be a good reason why heterogeneity tests showed significant variability (p-value ≤0.01) among studies included in this meta-analysis for 2 l antimicrobial agents.
S. aureus acquires resistance by various mechanisms: formation of alternative pathways for sulphonamides [93, 94], production of β-lactamase to β-lactam-sensitive antibiotics, increased efflux to tetracycline [95, 96], presence of acetyltransferase to chloramphenicol, decrease in accumulation to macrolide antibiotics [97], aminoglycoside-modifying enzymes production to aminoglycosides, altered topoisomerase IV and DNA gyrase expression for fluoroquinolones, and expression of mec gene altering penicillin binding protein to β-lactam antibiotics [98]. Since the AMR for β-lactam sensitive β-lactam antibiotics is very high, it can be speculated that most strains of S. aureus found in Ethiopia produce the β-lactamase enzyme. However, there is no molecular study conducted to identify the type of resistant strains and mechanism responsible for resistance in Ethiopia.
Lower rate of resistance was seen with β-lactamase-resistant antibiotics (amoxicillin-clavulanic acid, methicillin, ceftriaxone, cefoxtine, and cephalothin) compared to β-lactamase-sensitive penicillins. Unlike β-lactamase sensitive penicillin’s, resistance to carbeniciln is significantly lower. The lower rate of resistance observed with carbencilin and clindamycin may be due to their infrequent use in Ethiopia [99].
Resistance to methicillin confers resistance to all β-lactamase-resistant penicillins and cephalosporins. This high level of resistance requires the presence of the mec gene that encodes penicillin-binding protein [98]. The implication of high prevalence of MRSA for suspected or verified S. aureus infections such as common skin and wound infections and surgical prophylaxis is that there is a need for better alternatives drugs. Alternative drugs needed to treat or prevent S. aureus infections are more expensive and, because of their adverse effects, monitoring during treatment is advisable which increases the costs even further.
The prevalence of resistance S. aureus to vancomycin 11% 995% CI: (4%, 20%) in this study is bothersome and higher compared to global prevalence estimate [100]. The prevalence of VISA was 2.05% before, 2.63% in 2006–2009, and 7.93% in 2010–2014. Vancomycin resistance is erasing all possible treatment options in Ethiopia for MRSA. The higher prevalence of vancomycin in Ethiopia compared to global estimate may be due to larger and irrational use of antimicrobial agents in Ethiopia,
The prevalence estimates of glycopeptides/vancomycin resistance from Guta et al., and Desalegn et al., were unusually high, however sensitivity analysis showed non-significant influence on the overall pooled prevalence estimate. The prevalence estimates from Guta et al. and Desalegn et al. were unusually high, however sensitivity analysis showed non-significant influence on the overall pooled prevalence estimate. Larger exposure probability to resistant strains due to larger use of vancomycin in hospital settings might have resulted in a relatively higher prevalence of vancomycin resistance in the two studies [73, 77].
In four of the twenty studies (published in 2014 and after) [48, 51, 73, 77], the prevalence of S. aureus resistance to vancomycin is higher than 40%. In contrast, in studies published before 2014 the prevalence of S. aureus resistance to vancomycin in Ethiopia is much lower (0% to 16%). This may indicate a rapid rise and spread of vancomycin resistant S. aureus strains in Ethiopia as the rate of vancomycin use and exposure in Ethiopia increases. This calls for inclusion for effective new anti-MRSA antimicrobial agents for treatment of staphylococcal infections in the national medicine list and effective antimicrobial stewardship programs for prevention and containment of antimicrobial resistance.
Staphylococcal infection in Ethiopia can be better treated by vancomycin, floroquinolones, and aminoglycosides based on the finding of our invitro finding. However, clinical effectiveness study had not yet proved it. Resistance to vancomycin, the only choice for MRSA in Ethiopia, is of a great concern. It is bothersome due to lack of alternative agents in Ethiopia for the treatment of S. aureus infections. Making things worse, alternative new anti-MRSA agents (like linezolid, daptomycin, tigecycline, telavancin, and ceftaroline are rarely available in Ethiopia for treatment of vancomycin resistant S. aureus.
Many factors contribute to AMR. First, lack of infection prevention contributes to recurrent infection then to spread of resistant strains. Second, misuse of antimicrobials from prescription–dispensing-to patient use [101]. In Ethiopia, it is a common practice that antibiotics can be purchased without prescription, which leads to misuse of antibiotics by the public [102]. Third factor could be misuse of antibiotics by health professionals and non-standardized practice [101]. The fourth factor could be poor hospital hygienic conditions [103]. A last contributing factor could be lack of routine antimicrobial susceptibility testing which diverts to empiric therapy [49]. In line to strategies for prevention and containment of S. aureus there is a need for innovative way of halting AMR. Combination therapy and availability of new anti-MRSA agents will play vital role in fighting against AMR to S. aureus.
However, interpretation of the findings of this meta-analysis requires considering the limitations thereof. The limitations arise from the inherent characteristics of the included individual studies. First, this is invitro antimicrobial resistance testing and its direct translation to clinical effectiveness requires caution. Second, many studies involved very limited localities and were done mainly in teaching hospitals in bigger cities where patients with advanced, severe stages, recurrent infections are treated. Hence, the resistance level could have overestimated.
Conclusions
This meta-analysis demonstrates that S. aureus has gotten alarmingly resistant to many of common antimicrobials used in Ethiopia. It is highly resistant to penicillin, cephalosporin, tetracyclines, chloramphenicol, methicillin, sulphonamides, and vancomycin. Resistance to vancomycin is of a great concern and bothersome due to unavailability of treatment options for S. aureus infections in Ethiopia.
Continued and multidimensional efforts of antimicrobial stewardship programme promoting rational use of antimicrobials, infection prevention and containment of AMR are urgently needed. It is deemed necessary to include new anti-MRSA agents in national medicine list to treat resistant strains. Combination therapy, effective in battling AMR in many infectious diseases model, may prove significant advantage in battling resistance to S. aureus. Therapeutic options are urgently needed for patients infected with resistant S. aureus. Further researches focusing on clinical treatment outcome and identifying dynamics promoting resistance, high risk strains and molecular genetic basis of resistance are needed.
Abbreviations
- AMR:
-
Antimicrobial resistance
- MRSA:
-
Methicillin resistant Staphylococcus aureus
- NCCLs:
-
National Committee for clinical Laboratory Standard
- S. aureus :
-
Staphylococcus aureus
- VISA:
-
Vancomycin intermediate Staphylococcus aureus
- VRSA:
-
Vancomycin resistant Staphylococcus aureus
References
Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339(8):520–32.
McGuinness WA, Malachowa N, FR DL. Vancomycin resistance in Staphylococcus Aureus. Yale J Biol Med. 2017;90(2):269–81.
Hiramatsu K. Mechanism of methicillin resistance and genetic background of Staphylococcus Aureus. Nihon Naika Gakkai Zasshi. 1992;81(10):1592–8.
Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC. Methicillin-resistant Staphylococcus Aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–6.
Périchon B, Courvalin P. VanA-type vancomycin-resistant Staphylococcus Aureus. Antimicrob Agents Chemother. 2009;53(11):4580–7.
Plorde JJ, Eng SC, Wright LJ, Debesai A. Antibiotic sensitivities of common bacterial pathogens isolated in Addis Ababa; a preliminary report. Ethiop Med J. 1970;8(3):107–18.
Stephen J, Ronald J, Yvette S, José H, Ivonne D, Robert L, Susan E, Carol A: Manual of antimicrobial susceptibility testing. In. USA; 2005.
Nyaga VN, Arbyn M, Aerts M. Metaprop: a Stata command to perform meta-analysis of binomial data. Archives of Public Health. 2014;72:39.
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88.
Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34.
Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group. Perferred Reporting items for Systematic Review and Meta-analysis:The PRISMA statement. PLoS Med. 2009;6:e1000097.
Abrha A, Abdissa A, Beyene G, Getahun G, Girma T. Bacteraemia among severely malnourished children in jimma university hospital, ethiopia. Ethiopian journal of health sciences. 2011;21(3):175–82.
Alemu A, Moges F, Shiferaw Y, Tafess K, Kassu A, Anagaw B, Agegn A. Bacterial profile and drug susceptibility pattern of urinary tract infection in pregnant women at University of Gondar Teaching Hospital, Northwest Ethiopia. BMC research notes. 2012;5:197.
Beyene G, Tsegaye W. Bacterial uropathogens in urinary tract infection and antibiotic susceptibility pattern in jimma university specialized hospital, southwest ethiopia. Ethiopian journal of health sciences. 2011;21(2):141–6.
Demilie T, Beyene G, Melaku S, Tsegaye W. Urinary bacterial profile and antibiotic susceptibility pattern among pregnant women in north west ethiopia. Ethiopian journal of health sciences. 2012;22(2):121–8.
Tesfahunegn Z, Asrat D, Woldeamanuel Y, Estifanos K. Bacteriology of surgical site and catheter related urinary tract infections among patients admitted in Mekelle hospital, Mekelle, Tigray, Ethiopia. Ethiop Med J. 2009;47(2):117–27.
Assefa Y, Moges F, Endris M, Zereay B, Amare B, Bekele D, Tesfaye S, Mulu A, Belyhun Y. Bacteriological profile and drug susceptibility patterns in dacryocystitis patients attending Gondar University Teaching Hospital, Northwest Ethiopia. BMC Ophthalmol. 2015;15:34.
Gebremariam TT. Bacteriology and risk factors of bacterial keratitis in Jimma, Southwest Ethiopia. Ethiop Med J. 2015;53(4):191–7.
Kibret M, Abera B. Prevalence and antibiogram of bacterial isolates from urinary tract infections at Dessie Health Research Laboratory, Ethiopia. Asian Pac J Trop Biomed. 2014;4(2):164–8.
Tadesse E, Teshome M, Merid Y, Kibret B, Shimelis T. Asymptomatic urinary tract infection among pregnant women attending the antenatal clinic of Hawassa referral hospital, Southern Ethiopia. BMC research notes. 2014;7:155.
Wondimeneh Y, Muluye D, Alemu A, Atinafu A, Yitayew G, Gebrecherkos T, Damtie D, Ferede G. Urinary tract infection among obstetric fistula patients at Gondar University Hospital, northwest Ethiopia. BMC Womens Health. 2014;14:12.
Yeshitela B, Gebre-Selassie S, Feleke Y. Asymptomatic bacteriuria and symptomatic urinary tract infections (UTI) in patients with diabetes mellitus in Tikur Anbessa specialized university hospital, Addis Ababa, Ethiopia. Ethiop Med J. 2012;50(3):239–49.
Gebre-Selassie S. Asymptomatic bacteriuria in pregnancy: epidemiological, clinical and microbiological approach. Ethiop Med J. 1998;36(3):185–92.
Derese B, Kedir H, Teklemariam Z, Weldegebreal F, Balakrishnan S. Bacterial profile of urinary tract infection and antimicrobial susceptibility pattern among pregnant women attending at antenatal Clinic in Dil Chora Referral Hospital, Dire Dawa, eastern Ethiopia. Ther Clin Risk Manag. 2016;12:251–60.
Moges AF, Genetu A, Mengistu G. Antibiotic sensitivities of common bacterial pathogens in urinary tract infections at Gondar hospital, Ethiopia. East Afr Med J. 2002;79(3):140–2.
Muluye D, Wondimeneh Y, Ferede G, Nega T, Adane K, Biadgo B, Tesfa H, Moges F. Bacterial isolates and their antibiotic susceptibility patterns among patients with pus and/or wound discharge at Gondar university hospital. BMC research notes. 2014;7:619.
Melaku S, Gebre-Selassie S, Damtie M, Alamrew K. Hospital acquired infections among surgical, gynaecology and obstetrics patients in Felege-Hiwot referral hospital, Bahir Dar, northwest Ethiopia. Ethiop Med J. 2012;50(2):135–44.
Kebede A, Adamu Y, Bejiga A. Bacteriological study of dacryocystitis among patients attending in Menelik II hospital, Addis Ababa, Ethiopia. Ethiop Med J. 2010;48(1):29–33.
Melaku S, Kibret M, Abera B, Gebre-Sellassie S. Antibiogram of nosocomial urinary tract infections in Felege Hiwot referral hospital, Ethiopia. Afr Health Sci. 2012;12(2):134–9.
Mulu W, Kibru G, Beyene G, Damtie M. Postoperative nosocomial infections and antimicrobial resistance pattern of bacteria isolates among patients admitted at Felege Hiwot referral hospital, Bahirdar, Ethiopia. Ethiopian journal of health sciences. 2012;22(1):7–18.
Seboxa T, Amogne W, Abebe W, Tsegaye T, Azazh A, Hailu W, Fufa K, Grude N, Henriksen TH. High mortality from blood stream infection in Addis Ababa, Ethiopia, is due to antimicrobial resistance. PLoS One. 2015;10(12):e0144944.
Tilahun B, Worku B, Tachbele E, Terefe S, Kloos H, Legesse W. High load of multi-drug resistant nosocomial neonatal pathogens carried by cockroaches in a neonatal intensive care unit at Tikur Anbessa specialized hospital, Addis Ababa, Ethiopia. Antimicrob Resist Infect Control. 2012;1:12.
Zenebe T, Kannan S, Yilma D, Beyene G. Invasive bacterial pathogens and their antibiotic susceptibility patterns in Jimma University specialized hospital, Jimma, Southwest Ethiopia. Ethiopian journal of health sciences. 2011;21(1):1–8.
Tibebu M, Embiyale W. Community acquired multi drug resistant Staphylococcus Aureus in a rural setting of north western Ethiopia: a tough challenge. Ethiop Med J. 2014;52(3):147–50.
Truneh M. Phage types and drug susceptibility patterns of Staphylococcus Aureus from two hospitals in northwest Ethiopia. Ethiop Med J. 1991;29(1):1–6.
Teshager L, Asrat D, Gebre-Selassie S, Tamiru S. Catheterized and non-catheterized urinary tract infections among patients attended at Jimma University teaching hospital, southwest, Ethiopia. Ethiop Med J. 2008;46(1):55–62.
Lemma MT, Zenebe Y, Tulu B, Mekonnen D, Mekonnen Z. Methicillin resistant Staphylococcus Aureus among HIV infected pediatric patients in Northwest Ethiopia: carriage rates and antibiotic co-resistance profiles. PLoS One. 2015;10(9):e0137254.
Kejela T, Bacha K. Prevalence and antibiotic susceptibility pattern of methicillin-resistant Staphylococcus Aureus (MRSA) among primary school children and prisoners in Jimma town, Southwest Ethiopia. Ann Clin Microbiol Antimicrob. 2013;12:11.
Gebreyesus A, Gebre-Selassie S, Mihert A. Nasal and hand carriage rate of methicillin resistant Staphylococcus Aureus (MRSA) among health care workers in Mekelle hospital, North Ethiopia. Ethiop Med J. 2013;51(1):41–7.
Dagnew M, Tiruneh M, Moges F, Tekeste Z. Survey of nasal carriage of Staphylococcus Aureus and intestinal parasites among food handlers working at Gondar University, Northwest Ethiopia. BMC Public Health. 2012;12(1):837.
Shibabaw A, Abebe T, Mihret A. Nasal carriage rate of methicillin resistant Staphylococcus Aureus among Dessie referral hospital health care workers; Dessie, Northeast Ethiopia. Antimicrob Resist Infect Control. 2013;2(1):25.
Shibabaw A, Abebe T, Mihret A. Antimicrobial susceptibility pattern of nasal Staphylococcus Aureus among Dessie referral hospital health care workers, Dessie, Northeast Ethiopia. Int J Infect Dis. 2014;25:22–5.
Yismaw G, Abay S, Asrat D, Yifru S, Kassu A. Bacteriological profile and resistant pattern of clinical isolates from pediatric patients, Gondar University teaching hospital, Gondar, Northwest Ethiopia. Ethiop Med J. 2010;48(4):293–300.
Yismaw G, Tiruneh M, Kassu A, Negeri C, Mulu A. A retrospective analysis of prevalence and antimicrobial susceptibility patterns of Staphylococcus Aureus in Gondar teaching hospital, 2001-2005. Ethiop Med J. 2008;46(2):143–8.
Asrat D, Amanuel YW. Prevalence and antibiotic susceptibility pattern of bacterial isolates from blood culture in Tikur Anbassa hospital, Addis Ababa, Ethiopia. Ethiop Med J. 2001;39(2):97–104.
Endalafer N. Bacterial nosocomial infections and their antimicrobial susceptibility patterns in surgical wards and surgical intensive care unit of Tikur Anbessa university hospital. Addis Ababa: Addis Ababa University; 2008.
Neway S, Desta K, Dessie W, Yeshitila B, Lema T. Bacterial profile and antimicrobial susceptibility pattern of external ocular infections with associated risk factors in Alert cente. Ethiopia: Addis Ababa; 2016.
Tadesse S. Antimicrobial resistance profile of Staphylococcus aureus isolated from clinical specimens and nasal swabs of patients at Tikur Anbessa specialized hospital. Addis Ababa: Addis Ababa University; 2014.
DACA. Antimicrobials use, resistance and containment baseline survey: syntheses of findings. Addis Ababa: DACA; 2009.
Hailu D, Mekonnen D, Derbie A, Mulu W, Abera B. Pathogenic bacteria profile and antimicrobial susceptibility patterns of ear infection at Bahir Dar regional Health Research Laboratory center Ethiopia. SpringerPlus. 2016;5:466.
Argaw-Denboba A, Abejew AA, Mekonnen AG. Antibiotic-resistant bacteria are major threats of otitis Media in Wollo Area, Northeastern Ethiopia: A Ten-Year Retrospective Analysis. Int J Microbiol. 2016;2016:8724671.
Muluye D, Wondimeneh Y, Ferede G, Moges F, Nega T. Bacterial isolates and drug susceptibility patterns of ear discharge from patients with ear infection at Gondar University Hospital, Northwest Ethiopia. BMC ear, nose, and throat disorders. 2013;13(1):10.
Wasihun AG, Zemene Y. Bacterial profile and antimicrobial susceptibility patterns of otitis media in Ayder Teaching and Referral Hospital, Mekelle University, Northern Ethiopia. SpringerPlus. 2015;4:701.
Abera B, Kibret M. Bacteriology and antimicrobial susceptibility of otitis Media at Dessie Regional Health Research Laboratory, Ethiopia. Ethiop J Health Dev. 2011;25(2):161–7.
Ferede D, Geyid A, Lulseged S, Melaku A. Drug susceptibility pattern of bacterial isolates from children with chronic suppurative otitis media. Ethiop J Health Dev. 2001;15(2):89–96.
Worku M, Bekele M. Bacterial isolate and antibacterial resistance pattern of ear infection among patients attending at Hawassa university referral hospital, Hawassa, Ethiopia. Indian J Otol. 2014;20:155–9.
Abera B, Biadeglegne F. Antimicrobial resistance patterns of Staphylococcus Aureus and Proteus spp. isolated from otitis media at Bahir Dar regional laboratory, north West Ethiopia. Ethiop Med J. 2009;47(4):271–6.
Shiferaw B, Gelaw B, Assefa A, Assefa Y, Addis Z. Bacterial isolates and their antimicrobial susceptibility pattern among patients with external ocular infections at Borumeda hospital, Northeast Ethiopia. BMC ophthalmology. 2015;15:103.
Muluye D, Wondimeneh Y, Moges F, Nega T, Ferede G. Types and drug susceptibility patterns of bacterial isolates from eye discharge samples at Gondar University Hospital, Northwest Ethiopia. BMC research notes. 2014;7:292.
Tesfaye T, Beyene T, Gelaw T, Bekele S, Saravanan M. Bacterial profile and antimicrobial susceptibility pattern of external ocular infections in Jimma University specialized hospital, Southwest Ethiopia. American Journal of Infectious Diseases and Microbiology. 2013;1(1):13–20.
Negussie A, Mulugeta G, Bedru A, Ali I, Shimeles D, Lema T, Aseffa A. Bacteriological profile and antimicrobial susceptibility pattern of blood culture isolates among septicemia suspected children in selected hospitals Addis Ababa, Ethiopia. Int J Biol Med Res. 2015;6(1):4709–17.
Alebachew G, Teka B, Endris M, Shiferaw Y, Tessema B. Etiologic Agents of Bacterial Sepsis and Their Antibiotic Susceptibility Patterns among Patients Living with Human Immunodeficiency Virus at Gondar University Teaching Hospital, Northwest Ethiopia. Biomed Res Int. 2016;2016:5371875.
Endris M, Takele Y, Woldeyohannes D, Tiruneh M, Mohammed R, Moges F, Lynen L, Jacobs J, van Griensven J, Diro E. Bacterial sepsis in patients with visceral leishmaniasis in Northwest Ethiopia. Biomed Res Int. 2014;2014:361058.
Shitaye D, Asrat D, Woldeamanuel Y, Worku B. Risk factors and etiology of neonatal sepsis in Tikur Anbessa university hospital, Ethiopia. Ethiop Med J. 2010;48(1):11–21.
Wasihun AG, Wlekidan LN, Gebremariam SA, Dejene TA, Welderufael AL, Haile TD, Muthupandian S. Bacteriological profile and antimicrobial susceptibility patterns of blood culture isolates among febrile patients in Mekelle hospital, Northern Ethiopia. SpringerPlus. 2015;4:314.
Ali J, Kebede Y. Frequency of isolation and antimicrobial susceptibility pattern of bacterial isolates from blood culture, Gondar University teaching hospital, Northwest Ethiopia. Ethiop Med J. 2008;46(2):155–61.
Dagnew M, Yismaw G, Gizachew M, Gadisa A, Abebe T, Tadesse T, Alemu A, Mathewos B. Bacterial profile and antimicrobial susceptibility pattern in septicemia suspected patients attending Gondar University Hospital, Northwest Ethiopia. BMC research notes. 2013;6:283.
Gebrehiwot A, Lakew W, Moges F, Moges B, Anagaw B, Yismaw G, Nega T, Unakal C, Kassu A. Bacterial profile and drug susceptibility pattern of neonatal sepsis in Gondar University hospital, Gondar northwest Ethiopia. Pharm Lett. 2012;4(6):1811–6.
Alebachew T, Yismaw G, Derabe A, Sisay Z. Staphylococcus Aureus burn wound infection among patients attending yekatit 12 hospital burn unit, addis ababa, ethiopia. Ethiopian journal of health sciences. 2012;22(3):209–13.
Godebo G, Kibru G, Tassew H. Multidrug-resistant bacterial isolates in infected wounds at Jimma University Specialized Hospital, Ethiopia. Ann Clin Microbiol Antimicrob. 2013;12:17.
Azene MK, Beyene BA. Bacteriology and antibiogram of pathogens from wound infections at Dessie laboratory, north-east Ethiopia. Tanzania journal of health research. 2011;13(4):68–74.
Mulu A, Moges F, Tessema B, Kassu A. Pattern and multiple drug resistance of bacterial pathogens isolated from wound infection at University of Gondar Teaching Hospital, Northwest Ethiopia. Ethiop Med J. 2006;44(2):125–31.
Guta M, Aragaw K, Merid Y. Bacteria from infected surgical wounds and their antimicrobial resistance in Hawassa university referral teaching hospital, southern Ethiopia. Afr J Microbiol Res. 2014;8(11):1118–24.
Mama M, Alemseged A, Tsegaye S. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at Jimma University Specialized Hospital, South-West Ethiopia. Annals of Clinical Microbiology and Antimicrobials. 2014;13(1):14.
Kahsay A, Mihret A, Abebe T, Andualem T. Isolation and antimicrobial susceptibility pattern of Staphylococcus aureus in patients with surgical site infection at Debre Markos Referral Hospital, Amhara Region, Ethiopia. Arch Public Health. 2014;72(1):16.
Mengesha RE, Kasa BG, Saravanan M, Berhe DF, Wasihun AG. Aerobic bacteria in post surgical wound infections and pattern of their antimicrobial susceptibility in Ayder teaching and referral hospital, Mekelle, Ethiopia. BMC research notes. 2014;7:575.
Dessalegn L, Shimelis T, Tadesse E, Gebre-selassie S. Aerobic bacterial isolates from post-surgical wound and their antimicrobial susceptibility pattern: a hospital based cross-sectional study. Journal of Medical Research. 2014;3(2):18–23.
Dessie W, Mulugeta G, Fentaw S, Mihret A, Hassen M, Abebe E. Pattern of bacterial pathogens and their susceptibility isolated from surgical site infections at selected referral hospitals, Addis Ababa, Ethiopia. International journal of microbiology. 2016;2016:2418902.
Sewunet T, Demissie Y, Mihret A, Abebe T. Bacterial profile and antimicrobial susceptibility pattern of isolates among burn patients at Yekatit 12 hospital burn center, Addis Ababa, Ethiopia. Ethiopian journal of health sciences. 2013;23(3):209–16.
Gizachew M, Abdella H, Tiruneh M. Antimicrobial Susceptibility Patterns of Staphylococcus aureus atthe University of Gondar Tertiary Hospital, Northwest Ethiopia: A Retrospective Cross Sectional Study. Bacteriology & Parasitology. 2015;6(3):1000228
Kibret M, Abera B. Antimicrobial resistance trend of bacteria from clinical isolates: an 8-year retrospective study at Dessie regional laboratory, Northeast Ethiopia. Ethiopian pharmaceutical journal. 2010;28:39–46.
Tenssay ZW. Multiple antimicrobial resistance in bacterial isolates from clinical and environmental sources of Jimma hospital, south West Ethiopia. Ethiopia Journal of Science. 2002;25(2):295–302.
Abera B, Alem A, Bezabih B. Methicillin-resistant strains of Staphylococcus Aureus and coagulase-negative staphylococus from clinical isolates at Felege Hiwot Refferal hospital, north West Ethiopia. Ethiop Med J. 2008;46(2):149–54.
Lema T, Woldeamanuel Y, Asrat D, Hunegnaw M, Baraki A, Kebede Y, Yamuah L, Aseffa A. The pattern of bacterial isolates and drug sensitivities of infected ulcers in patients with leprosy in ALERT, Kuyera and Gambo hospitals, Ethiopia. Lepr Rev. 2012;83(1):40–51.
Ramos JM, Perez-Tanoira R, Garcia-Garcia C, Prieto-Perez L, Bellon MC, Mateos F, Tisisano G, Yohannes T, Reyes F, Gorgolas M. Leprosy ulcers in a rural hospital of Ethiopia: pattern of aerobic bacterial isolates and drug sensitivities. Ann Clin Microbiol Antimicrob. 2014;13:47.
Wolday D, Erge W. Increased incidence of resistance to antimicrobials by urinary pathogens isolated at Tikur Anbessa hospital. Ethiop Med J. 1997;35(2):127–35.
Biadglegne F, Abera B. Antimicrobial resistance of bacterial isolates from urinary tract infections at Felge Hiwot referral hospital, Ethiopia. Ethiop J Health Dev. 2009;23(3):236–8.
WHO: Antimicrobial resistance: global report ton surveillance. France; 2014.
Li S, Li J, Qiao Y, Ning X, Zeng T, Shen X. Prevalence and invasiveness of community-acquired methicillin-resistant Staphylococcus Aureus: a meta-analysis. Indian J Pathol Microbiol. 2014;57(3):418–22.
Salgado CD, Farr BM, Calfee DP. Community-acquired methicillin-resistant Staphylococcus Aureus: a meta-analysis of prevalence and risk factors. Clin Infect Dis. 2003;36(2):131–9.
Struelens MJ. The epidemiology of antimicrobial resistance in hospital acquired infections: problems and possible solutions. BMJ. 1998;317(7159):652–4.
Falagas ME, Karageorgopoulos DE, Leptidis J, Korbila IP. MRSA in Africa: filling the global map of antimicrobial resistance. PLoS One. 2013;8(7):e68024.
Sevag MG, Green MN. The mechanism of resistance to sulfonamides: I. Factors controlling the formation of Arylamine from Tryptophane by Staphylococcus Aureus. J Bacteriol. 1944;48(6):615–22.
Steers E, Sevag MG. The mechanism of resistance to sulfonamides; a comparative study of the amino acid metabolism of Staphylococcus Aureus in relation to the mechanism of resistance. Arch Biochem. 1949;24(1):129–43.
Sompolinsky D, Zaidenzaig Y, Ziegler-Schlomowitz R, Abramova N. Mechanism of tetracycline resistance in Staphylococcus Aureus. J Gen Microbiol. 1970;62(3):351–62.
Kono M, Sasatsu M, O'Hara K, Shiomi Y, Hayasaka T. Mechanism of resistance to some cephalosporins in Staphylococcus Aureus. Antimicrob Agents Chemother. 1983;23(6):938–40.
Yamagishi S, Nakajima Y, Inoue M, Oka Y. Decrease in accumulation of macrolide antibiotics as a mechanism of resistance in Staphylococcus Aureus. Japanese journal of microbiology. 1971;15(1):39–52.
Li XF, Fan XJ, Guo XJ, Feng P, Lu XJ, Gao YY, Xiong YL, Yu RJ, Ding X. A study on the mechanism of drug resistance in Staphylococcus aureus. Sichuan Da Xue Xue Bao Yi Xue Ban. 2006;37(3):365–8.
Stein M, Komerska J, Prizade M, Sheinberg B, Tasher D, Somekh E. Clindamycin resistance among Staphylococcus Aureus strains in Israel: implications for empirical treatment of skin and soft tissue infections. Int J Infect Dis. 2016;46:18–21.
Zhang S, Sun X, Chang W, Dai Y, Ma X. Systematic review and meta-analysis of the epidemiology of vancomycin-intermediate and heterogeneous vancomycin-intermediate Staphylococcus Aureus isolates. PLoS One. 2015;10(8):e0136082.
Yadesa TM, Gudina EK, Angamo MT. Antimicrobial use-related problems and predictors among hospitalized medical in-patients in Southwest Ethiopia: prospective observational study. PLoS One. 2015;10(12):e0138385.
Gebretekle GB, Serbessa MK. Exploration of over the counter sales of antibiotics in community pharmacies of Addis Ababa, Ethiopia: pharmacy professionals' perspective. Antimicrobial resistance and infection control. 2016;5:2.
Abera B, Kibret M, Mulu W. Knowledge and beliefs on antimicrobial resistance among physicians and nurses in hospitals in Amhara region, Ethiopia. BMC pharmacology & toxicology. 2014;15:26.
Acknowledgements
Not applicable.
Funding
There was no funding support to conduct this meta-analysis.
Availability of data and materials
The data supporting the conclusions of this article are included within the article and its supporting information.
Author information
Authors and Affiliations
Contributions
SD, SF and AA conceptualized the research idea. SD and SF conducted literature search, selection and data extraction. AA performed the statistical analyses. SD prepared the draft manuscript. All authors revised, edited and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1: S1.
RISMA Checklist. (DOC 63 kb)
Additional file 2: S2.
Egger’s test of publication bias. (DOCX 18 kb)
Additional file 3: S3.
Forest plot of the prevalence of S. aureus resistance to penicillin G. (DOCX 22 kb)
Additional file 4: S4.
Forest plot of the prevalence of S. aureus resistance to Ampicillin. (DOCX 20 kb)
Additional file 5: S5.
Forest plot of the prevalence of S. aureus resistance to amoxacilin-clavulanate. (DOCX 19 kb)
Additional file 6: S6.
Forest plot of the prevalence of S. aureus resistance to carbencilin. (DOCX 16 kb)
Additional file 7: S7.
Forest plot of the prevalence of S. aureus resistance to cephalothin. (DOCX 18 kb)
Additional file 8: S8.
Forest plot of the prevalence of S. aureus resistance to cefoxitine. (DOCX 16 kb)
Additional file 9: S9.
Forest plot of the prevalence of S. aureus resistance to norfloxacilin. (DOCX 17 kb)
Additional file 10: S10.
Forest plot of the prevalence of S. aureus resistance to erythromycin. (DOCX 21 kb)
Additional file 11: S11.
Forest plot of the prevalence of S. aureus resistance to chloramphenicol. (DOCX 22 kb)
Additional file 12: S12.
Forest plot of the prevalence of S. aureus resistance to doxycycline. (DOCX 18 kb)
Additional file 13: S13.
Forest plot of the prevalence of S. aureus resistance to amikacin. (DOCX 16 kb)
Additional file 14: S14.
Forest plot of the prevalence of S. aureus resistance to clindamycin. (DOCX 18 kb)
Additional file 15: S15.
Forest plot of the prevalence of S. aureus resistance to kanamycin. (DOCX 15 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Deyno, S., Fekadu, S. & Astatkie, A. Resistance of Staphylococcus aureus to antimicrobial agents in Ethiopia: a meta-analysis. Antimicrob Resist Infect Control 6, 85 (2017). https://doi.org/10.1186/s13756-017-0243-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-017-0243-7