Abstract
Background
Durum wheat is considered not suitable for making many food products that bread wheat can. This limitation is largely due to: (i) lack of grain-hardness controlling genes (Puroindoline a and b) and consequently extremely-hard kernel; (ii) lack of high- and low-molecular-weight glutenin subunit loci (Glu-D1 and Glu-D3) that contribute to gluten strength. To improve food processing quality of durum wheat, we stacked transgenic Pina and HMW-glutenin subunit 1Ax1 in durum wheat and developed lines with medium-hard kernel texture.
Results
Here, we demonstrated that co-expression of Pina + 1Ax1 in durum wheat did not affect the milling performance that was enhanced by Pina expression. While stacking of Pina + 1Ax1 led to increased flour yield, finer flour particles and decreased starch damage compared to the control lines. Interestingly, Pina and 1Ax1 co-expression showed synergistic effects on the pasting attribute peak viscosity. Moreover, Pina and 1Ax1 co-expression suggests that PINA impacts gluten aggregation via interaction with gluten protein matrix.
Conclusions
The results herein may fill the gap of grain hardness between extremely-hard durum wheat and the soft kernel durum wheat, the latter of which has been developed recently. Our results may also serve as a proof of concept that stacking Puroindolines and other genes contributing to wheat end-use quality from the A and/or D genomes could improve the above-mentioned bottleneck traits of durum wheat and help to expand its culinary uses.
Similar content being viewed by others
Background
Durum wheat (Triticum turgidum ssp. durum; 2n = 28, AABB) is an allotetraploid species that contributes to ~ 7% global wheat production, while bread wheat (Triticum. aestivum L.; 2n = 42, AABBDD) belongs to allohexaploid species that dominates wheat production worldwide [1]. Durum wheat has been widely grown in low-rainfall and semiarid regions, including Australia, Canada, India, Mexico, the Middle East, the Mediterranean countries and Northern Africa [2,3,4,5]. It is considered as an agronomically vigorous crop with good biotic and abiotic stress tolerance, but unsuitable for making many food products that bread wheat can [6, 7].
The limited culinary application is largely due to two reasons: (i) durum wheat requires specialized mill to process its extremely-hard grains (hardness index, HI, of more than 80); (ii) the milled flour of durum wheat, semolina, has larger flour particle size, higher levels of damaged starch and water absorption compared to bread wheat [8, 9]. Genetically, these inferior qualities of durum wheat can be explained by lack of the D genome. The short arm of chromosome 5D (5DS) contains the grain hardness (Ha) locus largely controlling kernel texture in wheat. Two grain-hardness causal genes on the Ha locus, Puroindoline a and b (Pina and Pinb, respectively) confer soft kernel phenotype when their wild-type alleles are expressed [10,11,12]. The cause-and-effect relationship between Puroindolines and grain hardness has been well studied by genetics and transgenic approaches in bread and durum wheat, as well as in maize and rice, demonstrating mutations in the Pina-D1 or Pinb-D1 genes are related to hard kernel phenotype [13,14,15,16,17,18,19,20,21,22]. Additionally, durum wheat is considered to have weaker gluten strength compared with bread wheat as the high-molecular-weight glutenin subunits (HMW-GS) and low-molecular-weight glutenin subunits (LMW-GS) loci Glu-D1 and Glu-D3 are absent due to lack of the D genome [7]. Particularly, the HMW-GS loci Glu-A1 and Glu-D1 have been shown to positively contribute to enhancing the micro-structures and aggregation of gluten matrix, thus conferring superior rheological properties of wheat dough [23,24,25].
To improve the food processing quality of durum wheat, soft-kernel durum (soft-Svevo) has been generated by introgression of the Ha locus from the soft bread wheat via homoeologous recombination [19, 26, 27]. Development of a series of soft durum wheat lines further demonstrates that the bread-making qualities of the soft-durum lines vary depending on their parental backgrounds and alleles contributing to gluten strength [9]. The soft-kernel durum wheat has similar flour and milling characteristics compared to soft bread wheat (HI < 40). However, no counterpart of hard- or medium-kernel bread wheat (40 < HI < 80) has been achieved in durum wheat via traditional breeding approaches. Previously, we reported the development of Pina-overexpressing transgenic durum wheat with medium-hard kernel texture (HI of ~ 80) [22, 28]. Nevertheless, it is yet to be tested whether stacking of genes or alleles favorable for gluten strength from the A or D genome could improve food processing qualities of durum wheat. As a proof-of-concept, we crossed the Pina-overexpressing line with a 1Ax1-expressing transgenic durum line that shared the same genetic background and showed that stacking transgenic Pina and 1Ax1 in durum wheat has combined effects on dough mixing parameters and could be useful for breeding durum wheat with dual purpose (for pasta and bread) [21]. Still, it is lacking that: (i) whether stacking of HMW-GS would affect milling attributes in the medium-hard durum lines? (ii) what are the effects of stacking HMW-GS and PINA on gluten protein matrix and other aspects of flour food processing qualities except for the dough mixing property that has been studied before? [21] In addition, PINs are starch granule-bound proteins that could influence starch-protein or starch-lipid interactions and possibly impact on starch-related properties, such as pasting property. Evidence also showed that PINs could be located in storage protein matrix during kernel development [29] and could be involved in modifying gluten protein aggregation via hydrophobic interactions [30]. To address the above questions, in the present study, we particularly focused on the effects of 1Ax1 + Pina co-expression on milling characteristics, aggregation pattern of gluten proteins and pasting property.
Results
Grain hardness and milling performance
As shown in Table 1, transgenic lines co-expressing 1Ax1 and Pina (HP-19 and HP-245) and those only expressing Pina (P-121 and P-149) showed similar grain hardness (HI of ~ 60), with the kernel textures of both being softer than those of the control lines (the null-segregant line N-1; the non-transgenic donor Luna; HI > 95) and the 1Ax1 expressing lines (H-182 and H-293 with HI values > 100). The results of kernel texture are consistent with those reported previously [21]. Protein analysis confirmed that HMW-GS 1Ax1 was expressed in lines HP-245, HP-19, H-293 and H-182, while Pina overexpression was detected in HP-245, HP-19, P-149 and P-121 (Fig. 1). In accordance with the results of kernel texture, 1Ax1-Pina- co-expressing lines and lines only expressed Pina exhibited similar distribution patterns of millstream and significantly increased total flour yield compared to the other lines, improved from ~ 44% (H-182, H-293, N-1 and Luna) to over 60% (Table 1). Compared to Pina-absent lines (H-182, H-293, N-1 and Luna), the total flour yield of Pina-expressing lines (HP-19, HP-245, P-121 and P-149) was improved due to the increased amounts of break flour and reduction flour, which were raised from ~ 9% and ~ 34% to ~ 17% and ~ 41%, respectively. These results demonstrated that transgenic 1Ax1 itself did not affect milling property and stacking of 1Ax1 and Pina transgenes did not influence the improvement of milling performance caused by PINA. It is worth notice that the medium-hard lines of durum wheat had a different millstream distribution pattern compared to that of soft-durum wheat [27]. The soft-durum wheat had grain hardness index of less than 40, with drastically increased break flour yield but slightly decreased reduction flour yield compared to hard durum Svevo. Here, the Pina transgenic lines had a medium-hard phenotype with distinct millstream results, which may consequently have different effects on end-use qualities compared with the soft-texture durum lines.
SDS-PAGE and Western blotting analyses of PINA in alcohol-soluble and insoluble proteins extracted from the seeds of transgenic and control lines. a, c SDS-PAGE and Western blotting of PINA detected in soluble gluten protein fractions. Transgenic 1Ax1 is indicated by arrowhead. b, d SDS-PAGE and Western blotting analysis of PINA detected in insoluble gluten protein fractions. ** shows the statistical comparison of PINA abundance between the transgenic lines and positive control line CS. (**P < 0.01 measured by Student’s t-test)
Flour quality
For flour characteristics, it appears that PINA expression was associated with decreased flour protein content, while 1Ax1 expression was not (Table 1). Flour protein content is a trait that is highly impacted by environment and genotype x environment effects [8, 31, 32]. Moreover, studies on near isogenic lines of durum and bread wheat with or without PINs suggested that PINs may be associated with lowered protein content [33, 34]. The association of PINA with flour protein content needs further investigation. Additionally, the lines only expressed Pina showed significantly lower gluten content compared to the remaining lines, while 1Ax1 + Pina co-expressing lines had the highest gluten content, possibly due to both the flour protein content and 1Ax1 expression. For flour color, only Pina expression was correlated to increased whiteness and decreased yellowness, whereas the 1ines only expressing 1Ax1 did not differ from the control lines in color characteristics (Table 1).
We further analyzed several flour traits for the transgenic and control lines, including flour particle size distribution, water binding capacity (WBC) and starch damage. Starch damage and water binding capacity showed similar trend: the decreased starch damage and WBC were associated with transgenic PINA, with 1Ax1 expression seemingly not related to both traits (Table 1). This result is in line with the positive correlation between grain hardness, starch damage and WBC, as hard-textured grains have stronger adhesion between starch granules and protein matrix and hence produce more damaged starch during milling [35,36,37,38,39].
Besides, the lines only expressed Pina and those co-expressed Pina + 1Ax1 showed similar particle size distribution of straight-grade flour, while the lines only expressed 1Ax1 had similar flour particle size distribution compared with the control lines (Table 1; Fig. 2). These results indicated that flour particle size is determined by grain hardness but not HMW-GS 1Ax1.
Effects of 1Ax1 and PINA on flour particles size distribution. a The size distribution curve of flour particles. b Weighted average diameter D32 of flour particles. Data are given as mean ± SEM, calculated from three replicates. The columns labeled by different letters indicate significant difference (P < 0.05)
Pasting property determined by rapid visco-analyzer (RVA)
Interestingly, PINA expression was associated with increased peak and breakdown viscosity, whereas no obvious differences in other RVA attributes were detected between the lines with expression of Pina, 1Ax1 and Pina + 1Ax1 (Table 1; Fig. 3). More importantly, synergistic effects of PINA and 1Ax1 on peak and breakdown viscosity were observed. For example, peak viscosity values of the lines expressed only Pina, or 1Ax1, and co-expressed Pina + 1Ax1 were ~ 484, ~ 355 and ~ 430, respectively. The 1Ax1 expression itself did not have effects on RVA attributes. The trough values were similar between the transgenic and control lines (Table 1; Fig. 3). Previous studies also indicated the association between PINA expression and altered pasting parameters in transgenic rice and near isogenic lines of bread and soft-durum wheat, such as peak viscosity and pasting temperature [34, 40]. Particularly, only peak and breakdown viscosity were increased with PINA expression in the present study. Several factors could impact pasting property, including amylose content, amylose/amylopectin ratio, interactions between starch, starch-associated proteins and lipids [41,42,43,44]. Several explanations of the PINA- peak viscosity association may be possible: (i) Puroindolines are starch granule-binding proteins and could serve as surfactant to improve the swelling power of starch granules [45]; (ii) Puroindolines may impact on starch interaction with polar lipids since PINs are associated with increased starch bound polar lipids [46]; (iii) A side-effect of decreased grain hardness that softer kernel could be associated with altered physiochemical properties of starch and/or damaged starch granules, thus changing the swelling power; (iv) PINA or PINA-lipid complex may also impact on gluten protein matrix that interacts with starch granules. The fourth explanation appears to match with the synergistic effects of PINA+1Ax1: PINA might play roles in both starch granule surface and protein matrix. Moreover, we separated alcohol soluble and insoluble fractions, the latter of which starch granules may be enriched in. PINA were well detected in both fractions (Fig. 1), indicating transgenically overexpressed PINA might have dual locations as discussed above. Indeed, recent evidence of the subcellular localization of Puroindolines and characterization of protein matrix with or without PINs supported the notion that PINs are located in gluten protein matrix and could interact with some gluten proteins via hydrophobic interactions besides its location of starch granule surface [29, 30].
Pasting properties of wheat flours from the transgenic and control lines. a RVA pasting profile of the flour samples from transgenic lines and control lines. b, c and d Comparison of three RVA parameters between the transgenic and control lines. Data are given as mean ± SEM, calculated from three replicates. The columns labeled by different letters indicate significant difference (P < 0.05)
Gluten protein aggregation
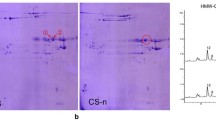
In the present study, SE-HPLC was used to fractionate cold-SDS extracted gluten proteins based on molecular masses without breaking down the inter-chain disulphide bonds [47, 48]. Non-covalent hydrophobic interactions may be dissociated during cold-SDS extraction. The proportion and size distribution of glutenin polymers are important determinants for wheat end-use quality, and accumulated studies have demonstrated that the size distribution of gluten proteins measured by SE-HPLC can serves as a highly reproducible approach to indicate flour functional properties [49, 50]. In Fig. 4, four fractions were detected: large-sized polymers enriched in HMW-GS (F1), medium-sized polymers (F2), oligomers gluten proteins enriched in ω-gliadin (F3), monomeric gluten proteins (mainly α-and γ-type gliadins; F4). Expression of only PINA was associated with increased small monomeric proteins (%F4), while expression of only 1Ax1 was associated with increased polymeric proteins (%F1 + %F2). Moreover, the effects of PINA+1Ax1 were observed as both polymeric and small monomeric proteins were increased (%F1 + %F2, %F4). The relative ratios of several SE-HPLC peaks, namely %F1/%F2 and (%F3 + %F4)/%F1, show strong correlations with dough strength. Generally, %F1/%F2 and (%F3 + %F4)/%F1 are negatively correlated with each other, and %F1/%F2 is positively correlated with the functional parameter G’ (an indication of dough elasticity). Combination of %F1/%F2 and (%F3 + %F4)/%F1 can be used to discriminate the breadmaking quality and dough visco-elasticity between wheat varieties [51,52,53]. In the present study, PINA overexpression resulted in lowered large size polymer (%F1) and increased (%F3 + %F4)/%F1 compared to the control lines. These negative effects on gluten aggregation and dough visco-elasticity were compensated in the Pina + 1Ax1 co-expressing lines (Fig. 4c and d). The ratios and absolute quantities of the four SE-HPLC peaks were clearly reshaped with the expression of PINA and/or 1Ax1 (Fig. 4a). Our results support the hypothesis that PINA interacts with gluten proteins and impacts on gluten aggregation. Previously, Puroindolines were suggestive of changing dough rheological properties and bread parameters based on in vitro reconstitution experiments [54]. Further, it was hypothesized that PINs might interact with gluten proteins by providing some sort of “hydrophobic cores” to modify the structures of gluten aggregates via hydrophobic bonds [30]. Differently, the effects of endogenous PINs in previous study might be less prominent compared to the results shown here, as the transgenically overexpressed PINA are more abundant (Fig. 1). Together with previous studies, the synergistic effect of PINA and 1Ax1 on viscosity reported herein suggests the conclusion that PINs affect several food-processing qualities possibly by interacting with gluten proteins, while the current results do not exclude the possibility that PINs impact on pasting property via their starch/starch-lipid binding ability. Still, it is yet to be established the molecular mechanism of PINs’ interaction with gluten polymers and PINs’ contribution to different food-processing qualities.
Effect of PINA and 1AX1 on gluten protein aggregation. a SE-HPLC analyses of the flour samples from transgenic and control lines. b, c and d Comparisons of three parameters (%F1, %F1/%F2 and (%F3 + %F4)/%F1) between the transgenic and control lines. Data are given as mean ± SEM, calculated from three replicates. The columns labeled by different letters indicate significant difference (P < 0.05)
Discussion
In the present study, the effects of PINA and 1Ax1 on milling property, pasting parameters and gluten aggregation have been investigated using the transgenic lines of durum wheat. We show that PINA and 1Ax1 have synergistic effects on the pasting parameters and PINA overexpression can affect gluten protein aggregation (Table 1; Fig. 1). It is worth notice that the Pina-overexpression studied herein was driven by the ubiquitin promoter, which provides constitutive expression. Indeed, we detected PINA in both alcohol soluble and insoluble gluten protein fractions (Fig. 1). While the endogenous Puroindolines have been suggestive of being located and possibly exerting functions at both starch granule surface and in gluten protein matrix [29, 30], it was unclear whether the effects of PINA on pasting property and gluten aggregation was due to its ectopic expression or the levels of overexpression. Future studies on PINs’ functionality using transgenic lines with endosperm-specific expression as well as with different PINs expression levels are necessary to further dissect the biological roles on starch-bound PINs and gluten-interacting PINs. In addition, we observed variations in PINA expression levels between transgenic lines (Fig. 1). Similarly, variations of some pasting parameters were found between the null-segregant line N-1 and Luna. Variations in pasting parameters were also observed between the lines expressed 1Ax1 or Pina (H-182/H-293, and P-121/P-149, respectively). On one hand, the differences between N-1 and Luna could reflect the variations produced during tissue culture and transgenic process of N-1. The genetic variations could be further accumulated and fixed in the following crossing and multi-generation selfing process when individual transgenic lines expressing Pina and/or 1Ax1 were selected [21]. On the other hand, the variations between H-182 vs H-293 or P-121 vs P-149 could be individual differences between transgenic lines. As these transgenic lines were identified among the F2 segregants from the cross between a Pina-expressing line and a 1Ax1-expressing line followed by selfing, variations from the two original transgenic lines (expressing Pina and 1Ax1, respectively) could be recombined, fixed and accumulated during multi-generation self-pollination process and therefore obvious differences in some traits were found between individual transgenic lines. Backcross of these transgenic lines to the donor variety Luna for two to three generations and comparison between transgenic and their corresponding null segregant lines would provide more rigorous and robust results to accurately quantify the effects of transgene. It is well known that the contribution of Glu-1 loci to end-use qualities is Glu-D1 > Glu-B1 > Glu-A1 [55,56,57,58]. Although the present and our previous studies have demonstrated the synergistic or additive effects of PINA and 1Ax1 on several aspects of food processing quality, such as dough mixing parameters and pasting parameters [21], it would be valuable to stack Glu-D1 HMW-GS with PINs through transgenic approaches in durum wheat. Thus, the present study not only provides useful information for quality improvement in durum wheat, but also suggests significant questions that need further investigations.
Conclusions
Recently, a major advance to expand the culinary use of durum wheat is the development of soft-durum wheat [19, 26, 27, 34, 59, 60]. Further, a series of soft-durum germplasm with varied end-use quality traits have been developed by introgression of the Ha locus into different durum wheat varieties. The durum wheat lines with soft-kernel texture represent a paradigm shift in the industrial use of durum wheat grains [8, 9]. Still, these strategies could have a limitation that the favorable genes/alleles for end-use quality traits from the A and D genomes are unable to be stacked to further enhance the food processing qualities of durum wheat. As a proof-of-concept, we stacked Pina and 1Ax1 together in durum wheat using transgenic approach and our preliminary results showed improvement in dough mixing quality traits [21]. Here, we demonstrate that stacking of Pina and 1Ax1 did not affect the milling performance that was enhanced by PINA expression. Stacking of Pina and 1Ax1 in durum wheat lead to medium-hard kernel texture, increased flour yield and decreased starch damage. Importantly, Pina and 1Ax1 expression show synergistic effects on pasting property traits and gluten aggregation, suggesting a role of PINA in interaction with gluten protein matrix. The results from the present and several previous studies indicate that the effects of PINs on gluten protein aggregation and different end-use quality attributes need further investigations. Further studies should also examine the effects of PINs in different durum wheat backgrounds.
Methods
Plant materials
1Ax1 or Pina transgenic lines of durum wheat was produced in donor variety Luna and described previously [21, 22, 61]. Only HMW-GS pairs 1Bx7 + 1By8 are expressed in Luna [21, 55]. Transgenic 1Ax1 was driven by its endogenous endosperm-specific promoter, while Pina was driven by the maize ubiquitin promoter. After crossing and selfing of the two transgenic lines, the lines of progeny homozygous for Pina (named P-121 and P-149), 1Ax1 (named H-182 and H-293) or Pina + 1Ax1 (named HP-19 and HP-245) as well as a null segregant line (N-1) were identified as F2 individuals and self-pollinated as described elsewhere [21]. The selection of lines with stable expression of 1Ax1 and/or Pina were performed in three continuous generations (F3, F4 and F5) and the F6 generation plants were used for measurement of grain hardness and dough mixing property [21]. These lines and their non-transgenic control Luna were grown in the field in 2014 (F7 generation) for end-use quality analyses using a randomized complete block design with two replicates in Wuhan (Hubei, China).
Grain hardness, flour characteristics and flour milling test measurement
Grain hardness were measured with Perten Single Kernel Characterization System (SKCS) 4100 (Perten, Springfield, USA) using the grain samples harvested from each plot according to the AACC approved method 55–31 [62].
Wheat grain samples (100 g per sample) were milled with a Chopin CD1 mill based on the standard method (NF EN ISO 27971:2008; Additional file 1). Flour characteristics were measured by near-infrared reflectance spectroscopy (NIRS) method.
Determination of damaged starch and water binding capacity (WBC)
Damaged starch content was determined with the amperometric method by using SDmatic by Chopin Technologies (Paris, France; Additional file 1).
The water-binding capacity was measured according to the AACC approved method 56–30 [62] (Additional file 1).
Color analysis
Flour color parameters L*, a* and b*, which correspond to flour lightness, redness and yellowness, respectively, were evaluated with the CR-410 Chromameter (Minolta, Osaka, Japan) according to Gazza et al. [59].
Particle size distribution in suspensions of flour
Particle size distribution of wheat flour was evaluated with laser-light scattering particle size analyzer (Mastersizer 2000, Malvern, UK) under a polydisperse analyzing mode and a 300-mm lens. Size distribution was determined in four replications for the flour samples sampled from each field plot (Additional file 1).
Pasting property determined by rapid Visco-Analyser (RVA)
The flour pasting profile was measured with a Rapid Visco Analyser (RVA-4, Newport Scientific, Australia) (Additional file 1).
Extraction of soluble and insoluble gluten proteins
Wheat flour was dissolved by 50% (v/v) propan-1-ol in a ratio of 10 μL/mg for 30 min. The soluble gluten proteins were obtained by the suspension centrifuged for 5 min at 14,000 g. The insoluble polymeric glutenin proteins were extracted from the residue by using a loading buffer in a ratio of 10 μL/mg. The extracts of soluble and insoluble gluten proteins were analyzed by SDS-PAGE with 10% separating gels and Western blotting with a PINA-specific antibody [22, 47, 63].
Size exclusion high-performance liquid chromatography (SE-HPLC) analysis
Wheat flour was dissolved by 1 mL of 0.05 M sodium phosphate buffer (pH 6.9) with 0.5% (w/v) SDS. The flour protein was extracted by sonication for 15 s and centrifuged for 10 min at 13,000 g. The total protein extracts was filtered through a 0.22-μm PVDF membrane and fractionated by using a Waters high-performance liquid chromatography (HPLC) system. According to Tosi et al. [47], four fractions were used to analyze the chromatographic profiles, F1 (large-sized polymers), F2 (medium-sized polymers), F3 (oligomers glutenin) and F4 (monomeric glutenin and other small non-gluten proteins).
Statistics analysis
All the above-mentioned flour tests, measurements of end-use quality attributes and SE-HPLC analysis were measured in three replicates for each plot. Data obtained from the two replicated plots were used for statistical analysis. Data were calculated using analysis of variance (ANOVA) and significant difference between lines were determined using the least significant difference pairwise comparison and displayed by letters (p < 0.05). Data were expressed as mean ± SEM. The statistical significance for western blotting from lines expressing 1Ax1 and/or Pina was determined using Student’s t test (p < 0.05).
Abbreviations
- AACC:
-
American Association of Cereal Chemists
- ANOVA:
-
analysis of variance
- DAD:
-
photodiode array detection
- Ha:
-
Hardness
- HI:
-
hardness index
- HMW-GS:
-
high-molecular-weight glutenin subunit
- HPLC:
-
high-performance liquid chromatography
- LMW-GS:
-
low-molecular-weight glutenin subunit
- NIRS:
-
near-infrared reflectance spectroscopy
- Pina:
-
Puroindoline a
- Pinb:
-
Puroindoline b
- RVA:
-
rapid visco-analyzer
- SDS-PAGE:
-
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SE-HPLC:
-
size exclusion high-performance liquid chromatography
- SEM:
-
standard error of the mean
- SKCS:
-
single kernel characterization system
- WBC:
-
water binding capacity
References
North Dakota Wheat Commission. World wheat production. 2015 North Dakota wheat Commission. https://www.ndwheat.com/uploads/6/world-web-charts.pdf. Accessed 15 Aug 2016.
Dias AS, Lidon FC. Evaluation of grain filling rate and duration in bread and durum wheat, under heat stress after anthesis. J Agron Crop Sci. 2009;195:137–47.
Karimizadeh R, Mohammadi M. Association of canopy temperature depression with yield of durum wheat genotypes under supplementary irrigated and rainfed conditions. Aust J Crop Sci. 2011;5:138–46.
Marti J, Slafer GA. Bread and durum wheat yields under a wide range of environmental conditions. Field Crop Res. 2014;156:258–71.
Kaur A, Singh N, Kaur S, Katyal M, Virdi AS, Kaur D, et al. Relationships of various flour properties with noodle making characteristics among durum wheat varieties. Food Chem. 2015;188:517–26.
Boyacioglu MH, Appolonia BLD. Characterisation and utilisation of durum wheat for breadmaking. II. Study of flour blends and various additives. Cereal Chem. 1994;71:28–34.
Ammar K, Kronstad WE, Morris CF. Breadmaking quality of selected durum wheat genotypes and its relationship with high molecular weight glutenin subunits allelic variation and gluten protein polymeric composition. Cereal Chem. 2000;77:230–6.
Boehm JD, MIt I, Kiszonas AM, Morris CF. End-use quality of CIMMYT-derived soft-kernel durum wheat germplasm: I. grain, milling, and soft wheat quality. Crop Sci. 2017;57:1–10.
Boehm JD, MIt I, Kiszonas AM, Morris CF. End-use quality of CIMMYT-derived soft-kernel durum wheat germplasm: II. dough strength and pan bread quality. Crop Sci. 2017;57:1485–94.
Morris CF. Puroindolines: the molecular genetic basis of wheat grain hardness. Plant Mol Biol. 2002;48:633–47.
Bhave M, Morris CF. Molecular genetics of puroindolines and related genes: regulation of expression, membrane binding properties and applications. Plant Mol Biol. 2008;66:221–31.
Bhave M, Morris CF. Molecular genetics of puroindolines and related genes: allelic diversity in wheat and other grasses. Plant Mol Biol. 2008;66:205–19.
Giroux MJ, Morris CF. A glycine to serine change in puroindoline b is associated with wheat grain hardness and low levels of starch-surface friabilin. Theor Appl Genet. 1997;95:857–64.
Giroux MJ, Morris CF. Wheat grain hardness results from highly conserved mutations in the friabilin components puroindoline a and b. Proc Natl Acad Sci U S A. 1998;95:6262–6.
Krishnamurthy K, Giroux MJ. Expression of wheat puroindoline genes in transgenic rice enhances grain softness. Nat Biotechnol. 2001;19:162–6.
Beecher B, Bettge A, Smidansky E, Giroux MJ. Expression of wild-type PinB sequence in transgenic wheat complements a hard phenotype. Theor App Genet. 2002;105:870–7.
Hogg AC, Stripo T, Beecher B, Martin JM, Giroux MJ. Wheat puroindolines interact to form friabilin and control wheat grain hardness. Theor Appl Genet. 2004;108:1089–97.
Martin JM, Meyer FD, Smidansky ED, Wanjugi H, Blechl AE, Giroux MJ. Complementation of the pina (null) allele with the wild type Pina sequence restores a soft phenotype in transgenic wheat. Theor Appl Genet. 2006;113:1563–70.
Morris CF, Simeone MC, King GE, Lafiandra D. Transfer of soft kernel texture from Triticum aestivum to durum wheat, Triticum turgidium ssp. durum. Crop Sci. 2011;51:114–22.
Zhang JR, Martin JM, Beecher B, Morris CF, Hannah LC, Giroux MJ. Seed specific expression of the wheat puroindoline genes improves maize wet milling yields. Plant Biotechnol J. 2009;7:733–43.
Li Y, Wang Q, Li XY, Xiao X, Sun FS, Wang C, et al. Coexpression of the high molecular weight glutenin subunit 1Ax1 and puroindoline improves dough mixing properties in durum wheat (Triticum turgidum L. ssp. durum). PLoS One. 2012;7:e50057.
Li Y, Mao X, Wang Q, Zhang JR, Li XY, Ma FY, et al. Overexpression of puroindoline a gene in transgenic durum wheat (Triticum turgidum ssp. durum) leads to a medium-hard kernel texture. Mol Breed. 2014;33:545–54.
Mao X, Li Y, Zhao S, Zhang J, Lei Q, Meng D, et al. The interactive effects of transgenically overexpressed 1Ax1 with various HMW-GS combinations on dough quality by introgression of exogenous subunits into an elite Chinese wheat variety. PLoS One. 2013;8:e78451.
Li X, Liu T, Song L, Zhang H, Li L, Gao X. Influence of high-molecular-weight glutenin subunit composition at Glu-A1 and Glu-D1 loci on secondary and micro structures of gluten in wheat (Triticum aestivum L.). Food Chem. 2016;213:728–34.
Gao X, Liu T, Ding M, Wang J, Li C, Wang Z, et al. Effects of HMW-GS Ax1 or Dx2 absence on the glutenin polymerization and gluten micro structure of wheat (Triticum aestivum L.). Food Chem. 2018;240:626–30.
Heinze K, Kiszonas AM, Murray JC, Morris CF, Lullien-Pellerin V. Puroindoline genes introduced into durum wheat reduce milling energy and change milling behavior similar to soft common wheats. J Cereal Sci. 2016;71:183–9.
Murray JC, Kiszonas AM, Wilson J, Morris CF. The effect of soft kernel texture on the milling properties of soft durum wheat. Cereal Chem. 2016;93:513–7.
Luo LT, Zhang JR, Yang GX, Li Y, Li KX, He GY. Expression of puroindoline a enhances leaf rust resistance in transgenic tetraploid wheat. Mol Biol Rep. 2008;35:195–200.
Lesage VS, Bouchet B, Rhazi L, Elmorjani K, Branlard G, Marion D. New insight into puroindoline function inferred from their subcellular localization in developing hard and soft near-isogenic endosperm and their relationship with polymer size of storage proteins. J Cereal Sci. 2011;53:231–8.
Quayson ET, Marti A, Morris CF, Marengo M, Bonomi F, Seetharamen K, et al. Structural consequences of the interaction of puroindolines with gluten proteins. Food Chem. 2018;253:255–61.
Narasimhamoorthy B, Gill BS, Fritz AK, Nelson JC, Brownguedira GL. Advanced backcross QTL analysis of a hard winter wheat x synthetic wheat population. Theor Appl Genet. 2006;112:787.
Boehm JD, Ibba MI, Kiszonas AM, See DR, Skinner DZ, Morris CF. Genetic analysis of kernel texture (grain hardness) in a hard red spring wheat (Triticum aestivum L.) bi-parental population. J Cereal Sci. 2017;79:57–65.
Hogg AC, Beecher B, Martin JM, Meyer F, Talbert L, Lanning S, et al. Hard wheat milling and bread baking traits affected by the seed-specific overexpression of puroindolines. Crop Sci. 2005;45:871–8.
Quayson ET, Atwell W, Morris CF, Marti A. Empirical rheology and pasting properties of soft-textured durum wheat (Triticum turgidum ssp. durum) and hard-textured common wheat (T. aestivum). J Cereal Sci. 2016;69:252–8.
Greenwell P, Schofield JD. A starch granule protein associated with endosperm softness in wheat. Cereal Chem. 1986;63:379–80.
Schofield JD, Greenwell P. Wheat starch granule proteins and their technological significance. In: Morton JD, editor. Cereals in a European context, Ellis Horwood, New York; 1987. p. 407–20.
Peng M, Gao M, Abdel-Aal ESM, Hucl P, Chibbar RN. Separation and characterization of A- and B-type starch granules in wheat endosperm. Cereal Chem. 1999;76:375–9.
León AE, Barrera GN, Pérez GT, Ribotta PD, Rosell CM. Effect of damaged starch levels on flour-thermal behaviour and bread staling. Eur Food Res Technol. 2006;224:187–92.
Yu JL, Wang SJ, Wang JR, Li CL, Xin QW, Huang W, et al. Effect of laboratory milling on properties of starches isolated from different flour millstreams of hard and soft wheat. Food Chem. 2015;172:504–14.
Wada N, Kajiyama S, Lin L, Akiyama Y, Otani M, Suzuki G, et al. The effects of puroindoline b on the ultrastructure of endosperm cells and physicochemical properties of transgenic rice plant. J Cereal Sci. 2010;51:182–218.
Blazek J, Copeland L. Pasting and swelling properties of wheat flour and starch in relation to amylose content. Carbohyd Polym. 2008;71:380–7.
Wang S, Luo H, Zhang J, Zhang Y, He Z, Wang S. Alkali-induced changes in functional properties and in vitro digestibility of wheat starch: the role of surface proteins and lipids. J Agric Food Chem. 2014;62:3636–43.
Li E, Dhital S, Hasjim J. Effects of grain milling on starch structures and flour/starch properties. Starch. 2014;66:15–27.
Li W, Wu G, Luo Q, Jiang H, Zheng J, Ouyang S, et al. Effects of removal of surface proteins on physicochemical and structural properties of A- and B-starch isolated from normal and waxy wheat. J Food Sci Technol. 2016;53:2673–85.
Dubreil L, Vie V, Beaufils S, Martin D, Renault S. Aggregation of puroindoline in phospholipid monolayers spread at the air-liquid interface. Biophys J. 2003;85:2650–60.
Feiz L, Wanjugi HW, Melnyk CW, Altosaar I, Martin JM, Giroux MJ. Puroindolines co-localize to the starch granule surface and increase seed bound polar lipid content. J Cereal Sci. 2009;50:91–8.
Tosi P, Masci S, Giovangrossi A, D’Ovidio R, Bekes F, Larroque O, et al. Modification of the low molecular weight (LMW) glutenin composition of transgenic durum wheat: effects on glutenin polymer size and gluten functionality. Mol Breed. 2005;16:113–26.
Ma F, Li M, Yu L, Li Y, Liu Y, Li T, et al. Transformation of common wheat (Triticum aestivum L.) with avenin-like b gene improves flour mixing properties. Mol Breed. 2013;32:853–65.
Bangur R, Batey IL, McKenzie E, MacRitchie F. Dependence of Extensograph parameters on wheat protein composition measured by SE-HPLC. J Cereal Sci. 1997;25:237–41.
Shewry PR, Underwood C, Wan Y, Lovegrove A, Bhandari D, Toole G, et al. Storage product synthesis and accumulation in developing grains of wheat. J Cereal Sci. 2009;50:106–12.
Morel MH, Bonicel J, Micard V, Guilbert S. Protein insolubilization and thiol oxidation in sulfite-treated wheat gluten films during aging at various temperatures and relative humidities. J Agric Food Chem. 2000;48:186–92.
Millar SJ. The development of near infrared (NIR) spectroscopy calibrations for the prediction of wheat and flour quality. Project Report No. 310. UK: HGCA; 2003.
Li M, Wang Y, Ma F, Zeng J, Chang J, Chen M, et al. Effect of extra cysteine residue of new mutant 1Ax1 subunit on the functional properties of common wheat. Sci Rep. 2017;7:7510.
Dubreil L, Meliande S, Chiron H, Compoint JP, Quillien L, Branlard G, et al. Effect of puroindoline on the breadmaking properties of wheat flour. Cereal Chem. 1998;75:222–9.
Lawrence GJ, MacRitchie F, Wrigley CW. Dough and baking quality of wheat lines deficient in glutenin subunits controlled by the Glu-A1, Glu-B1 and Glu-D1 loci. J Cereal Sci. 1988;7:109–12.
Uthayakumaran S, Beasley HL, Stoddard FL, Keentok M, Thien-Phan N, Tanner RI, et al. Synergistic and additive effects of three high molecular weight glutenin subunit loci. I. Effects on wheat dough rheology. Cereal Chem. 2002;79:294–300.
Beasley HL, Uthayakumaran S, Stoddard FL, Partridge SJ, Daqiq L, Chong P, et al. Synergistic and additive effects of three high molecular weight glutenin subunit loci. II. Effects on wheat dough functionality and end-use quality. Cereal Chem. 2002;79:301–7.
Yang Y, Li S, Zhang K, Dong Z, Li Y, An X, et al. Efficient isolation of ion beam-induced mutants for homoeologous loci in common wheat and comparison of the contributions of Glu-1 loci to gluten functionality. Theor Appl Genet. 2014;127:359–72.
Gazza L, Sgrulletta D, Cammerata A, Gazzelloni G, Perenzin M, Pogna NE. Pastamaking and bread making quality of soft-textured durum wheat lines. J Cereal Sci. 2011;54:481–7.
Morris CF, Casper J, Kiszonas AM, Fuerst EP, Murray J, Simeone MC, et al. Soft kernel durum wheat: a new bakery ingredient? Cereal Foods World. 2015;60:76–83.
He GY, Rooke L, Steele S, Bekes F, Gras P, Tatham AS, et al. Transformation of pasta wheat (Triticum turgidum L. var. durum) with high-molecular-weight glutenin subunit genes and modification of dough functionality. Mol Breed. 1999;5:377–86.
AACC. American Association of Cereal Chemists. Approved Methods of the AACC. 10th ed. St Paul MN: The Association; 2000.
Miao Y, Chen L, Wang C, Wang Y, Zheng Q, Gao C, et al. Expression, purification and antimicrobial activity of puroindoline a protein and its mutants. Amino Acids. 2012;43:1689–96.
Acknowledgements
We thank the Analytical and Testing Center of Huazhong University of Science and Technology (HUST) for the service of SE-HPLC assay. We also thank Henan University of Technology for the service of damaged starch content, particle size distribution and rapid visco-analysis assay.
Funding
The work was supported by National Genetically Modified New Varieties of Major Projects of China (2016ZX08010004–004), the National Natural Science Foundation of China (No. 31771418, 31570261) and Key Project of Hubei Province (2017AHB041). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The data and plant material that support the findings of this study are available from the corresponding author on request. All data generated or analyzed during this study are included in this published article.
Author information
Authors and Affiliations
Contributions
YL, GY, YW and GH designed the experiments. QW, YL, FS, XL, JC and PW performed the experiments. QW and YL analyzed the data. The article was written by QW, YL and GH, and edited by GY and YW. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Supplementary materials and methods. (DOCX 24 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wang, Q., Li, Y., Sun, F. et al. Co-expression of high-molecular-weight glutenin subunit 1Ax1 and Puroindoline a (Pina) genes in transgenic durum wheat (Triticum turgidum ssp. durum) improves milling and pasting quality. BMC Plant Biol 19, 126 (2019). https://doi.org/10.1186/s12870-019-1734-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-019-1734-x