Abstract
Background
Sucrose non-fermenting-1-related protein kinase 2 (SnRK2) is a plant-specific serine/threonine kinase family involved in the abscisic acid (ABA) signaling pathway and responds to osmotic stress. A genome-wide analysis of this protein family has been conducted previously in some plant species, but little is known about SnRK2 genes in upland cotton (Gossypium hirsutum L.). The recent release of the G. hirsutum genome sequence provides an opportunity to identify and characterize the SnRK2 kinase family in upland cotton.
Results
We identified 20 putative SnRK2 sequences in the G. hirsutum genome, designated as GhSnRK2.1 to GhSnRK2.20. All of the sequences encoded hydrophilic proteins. Phylogenetic analysis showed that the GhSnRK2 genes were classifiable into three groups. The chromosomal location and phylogenetic analysis of the cotton SnRK2 genes indicated that segmental duplication likely contributed to the diversification and evolution of the genes. The gene structure and motif composition of the cotton SnRK2 genes were analyzed. Nine exons were conserved in length among all members of the GhSnRK2 family. Although the C-terminus was divergent, seven conserved motifs were present. All GhSnRK2s genes showed expression patterns under abiotic stress based on transcriptome data. The expression profiles of five selected genes were verified in various tissues by quantitative real-time RT-PCR (qRT-PCR). Transcript levels of some family members were up-regulated in response to drought, salinity or ABA treatments, consistent with potential roles in response to abiotic stress.
Conclusions
This study is the first comprehensive analysis of SnRK2 genes in upland cotton. Our results provide the fundamental information for the functional dissection of GhSnRK2s and vital availability for the improvement of plant stress tolerance using GhSnRK2s.
Similar content being viewed by others
Background
Plants experience various biotic and abiotic stresses in their natural environment, such as drought and salt stress. Abscisic acid (ABA) plays an important role in plant response to drought and high salinity. ABA was first discovered in the 1960s and isolated from cotton as a promoter of leaf abscission [1, 2]. ABA also regulates diverse plant growth and developmental processes, including seed development, stomatal closure, promotion of seed dormancy and reproduction [3,4,5,6].
Protein phosphorylation/dephosphorylation is an important mechanism for mediating intracellular responses, i.e. responses to hormonal, pathogenic, and environmental stresses [7]. Various studies in plants have indicated that protein kinases are involved in stress signaling via phosphorylation/dephosphorylation, for example, the plant mitogen-activated protein kinase (MAPK) [8], calcium-dependent protein kinase (CDPK) [9], and sucrose non-fermenting1 (SNF1) kinase families. The SNF1 kinase family is known as SNF1 protein kinases in yeasts, AMP-activated protein kinases in mammals, and SNF1-related protein kinases (SnRKs) in plants [10]. All kinases in yeasts and mammals play key roles in carbohydrate metabolism, whereas plant SnRKs may function as interfaces between stress and metabolic signaling pathways [11,12,13].
The SnRK protein kinases are classified into three subfamilies: SnRK1, SnRK2 and SnRK3 [12, 14]. SnRK1 is the plant homolog of the yeast SNF1 gene and plays a role in global regulation of carbon and nitrogen metabolism [15]. The SnRK2 and SnRK3 subfamilies are unique in plants and may be involved in responses to environmental stress. Members of the SnRK2 subfamily contain a highly conserved kinase domain, an ATP-binding domain, and a serine/threonine active site [16]. SnRK3 kinases are known as calcineurin B-like calcium sensor-interacting protein kinases. The kinase domain of this subfamily contains a binding site for calcium-binding proteins and calcium-sensitive calcineurin B-like proteins in the C-terminal region, which combine to activate the protein kinase [17]. The best known member of this group is the Salt Overly Sensitive 2 kinase, which is required for sodium and potassium ion homeostasis and salt tolerance [18, 19]. The SnRK1 and SnRK2 subfamilies of protein kinases are similar in their catalytic domain but have divergent C-terminal domains, whereas the SnRK3 subfamily is structurally distinctive [20]. In the present study we focused on SnRK2 family members in G. hirsutum.
The first SnRK2 cDNA clone (PKABA1) was isolated from an ABA-treated wheat embryo cDNA library, which was induced by dehydration, cold, salinity, and osmotic stresses [21, 22]. In a recent study, overexpression of TaSnRK2.4 resulted in delayed seeding establishment, longer primary roots and enhanced tolerance to abiotic stresses in Arabidopsis [23]. Transgenic experiments indicated that TaSnRK2.7 and TaSnRK2.8 significantly increased tolerance to drought, salt and cold stress in Arabidopsis [24, 25]. In the past decade, SnRK2 kinases have been identified in diverse plants, such as Arabidopsis, rice, maize, soybean and tobacco [26]. There are ten SnRK2 genes in Arabidopsis; among these, nine genes (except SnRK2.9) are activated by hyperosmotic and salinity stress, and five out of nine are also activated by ABA, whereas none of these genes are activated by cold stress [27]. Moreover, SnRK2.6/SRK2E//OST1 is involved in ABA regulation during stomatal closure and ABA-mediated gene expression [28, 29]. Overexpression of SnRK2.8 enhances drought tolerance in Arabidopsis [30]. In rice, all ten SnRK2 members have been isolated and found to be activated by hyperosmotic stress but only three (SAPK8, SAPK9 and SAPK10) are activated by ABA [31]. Overexpression of SAPK4 significantly enhances the salt tolerance of transgenic plants [32]. In maize, 10 SnRK2 genes have been cloned and most of these are activated by one or more abiotic stresses [33]. ZmSPK1, a SnRK2 homolog in maize, is expressed in different plant tissues including roots, leaves and reproductive organs as well [34]. In soybean, four SnRK2 members have been isolated, where all these are induced by hyperosmotic stress [35, 36]. The expression level of only SPK3 and SPK4 increases in response to dehydration and high salinity [35]. A 42 kDa protein kinase (NtOSAK), belonging to the SnRK2 family, is activated by osmotic stress in tobacco [31]. Overexpression of NtSnRK2.1 confers enhanced salt tolerance in tobacco, which suggests that NtSnRK2s are involved in abiotic stress response pathways [37].
Cotton is an economically important and a model polyploid crop [38, 39]. Upland cotton (G. hirsutum), an allotetrapoid species (AtAtDtDt, 2n = 4× = 52, where ʻt’ stands for tetraploid), is the widely cultivated cotton species that accounts 90% of total world cotton production [40,41,42]. Only one SnRK2 gene has been identified from upland cotton [42]. Nevertheless, systematic analysis of SnRK2 family genes has not been performed in upland cotton. Currently, the G. hirsutum genome has been sequenced and provides the opportunity to perform a genome-wide analysis of the SnRK2 protein kinases family.
In the present study, we conducted a genome-wide survey of G. hirsutum molecular databases using SnRK2 protein sequences from Arabidopsis. We detected 20 putative SnRK2 sequences in the G. hirsutum genome, named GhSnRK2.1 to GhSnRK2.20. We analyzed their gene structures, chromosomal locations, expression profiles under exposure to abiotic stresses, and phylogenetic relationships. This is the first study to undertake a genome-wide analysis of GhSnRK2s. These results provide valuable information on SnRK2 genes in G. hirsutum and will facilitate future studies of evolutionary relationships among cotton species.
Results
Identification of SnRK2 genes in the G. hirsutum genome
A local BLASTP search was performed to identify SnRK2 members in the G. hirsutum genome using Arabidopsis SnRK2 protein sequences as the query sequence. The Hidden Markov Model profile of the Pkinase domain (PF00069.22) (http://pfam.sanger.ac.uk/) was used as a query to search the Pfam database using the program HMMER 3.0 with the default E-value. The serine/threonine protein kinase catalytic domains were determined using the online program SMART (http://smart.embl-heidelberg.de/) with the default E-value. We detected 20 predicted candidate SnRK2 family proteins that have complete serine/threonine protein kinase catalytic domains in the G. hirsutum proteome (Table 1). Eighteen SnRK2 genes were mapped onto chromosomes but two SnRK2 genes could not be conclusively localized to a chromosome. The genes were designated GhSnRK2.1 to GhSnRK2.20. The length of the SnRK2 coding sequence ranged from 999 to 1092 bp.
The SnRK2 genes differed substantially by the encoded protein size and its biophysical properties (Table 1). The SnRK2 protein sequences ranged from 332 to 363 amino acids in the case of GhSnRK2.2 and GhSnRK2.13, respectively. The isoelectric point (pI) of all SnRK2 proteins was acidic, which indicates that SnRK2 proteins in cotton are rich in acidic amino acids. The GRAVY value of a protein is calculated as the sum of the hydropathy value of each residue, divided by the total number of residues present in the sequences. Positive and negative GRAVY scores reflect hydrophobicity and hydrophilicity, respectively [43]. The GRAVY scores of all G. hirsutum SnRK2 proteins were negative, which indicated that all were hydrophilic proteins, however, the degree of hydrophilicity exhibited a higher variability.
Cis-element analysis in the promoter regions of GhSnRK2 genes
To identify the putative cis-acting regulatory elements, 2000 bp of sequence upstream from the start codon was isolated. These regulatory elements included ABA-responsive elements (ABREs), low-temperature responsive elements (LTRs), defense and stress responsiveness elements (TC-rich repeats), salicylic acid responsive elements (TCA-elements), heat stress responsive elements (HSEs), MeJA-responsive elements (TGACG-motifs), MYB-binding sites (MBS), which are involved in drought-inducibility (Table 2).
Plant hormone-responsive elements, including ABREs, TCA-elements and TGACG-motifs, were enriched in the upstream promoter regions of GhSnRK2 genes. ABRE was the most abundant cis-acting hormone responsive element in the promoters of GhSnRK2 genes. The genes, except GhSnRK2.7 and GhSnRK2.19, contained ABRE-elements. TCA-elements and TGACG-motifs are involved in SA and MeJA responsiveness, respectively. In total, 16 members contained a TCA-element, whereas 11 members contained a TGACG-motif element. This indicates that GhSnRK2 genes may respond to ABA, SA and JA.
The other important type of cis-acting elements in the upstream regions of GhSnRK2 genes are environmental stress-related elements. In total, four types of elements were found that respond to four respective kinds of external environmental stresses. These were low-temperature responsive (LTR), stress-responsive TC-rich repeats, heat-stress-responsive HSEs and drought-responsive MBSs. Over half of the 20 GhSnRK2 genes contained an HSE, MBS and TC-rich repeat (for defense and stress responses). Only six members (GhSnRK2.2, GhSnRK2.3, GhSnRK2.4, GhSnRK2.12, GhSnRK2.14 and GhSnRK2.19) contained LTR-element. We hypothesized that external environmental stresses could induce the expression of GhSnRK2 genes through their responsive cis-acting elements, further heightening the plants’ resistance to environmental stresses.
Phylogenetic analysis and classification of the GhSnRK2 gene family
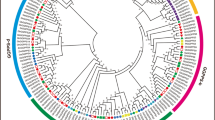
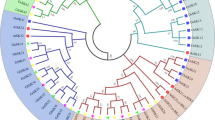
We constructed a phylogenetic tree from a multiple alignment of SnRK2 protein sequences, comprising 20 GhSnRK2s from G. hirsutum, 10 SAPKs from rice, 10 SnRK2s from Arabidopsis, 11 ZmSnRK2s from maize, and 1 NtOSAK from tobacco. The SnRK2 proteins were clustered into three groups (Groups I, II and III; Fig. 1). Consistent with previous classification, all of the Arabidopsis SnRK2s were distributed among the three groups [27]. Our phylogenetic reconstruction showed that the GhSnRK2s were more closely grouped with SnRK2s from the dicots Arabidopsis and tobacco than with proteins from the monocots rice and maize. GhSnRK2.5 and GhSnRK2.15 belonged to Group I. Group II comprised of 8 members (GhSnRK2.2, GhSnRK2.4, GhSnRK2.6, GhSnRK2.8, GhSnRK2.12, GhSnRK2.14, GhSnRK2.17 and GhSnRK2.18). 10 members (GhSnRK2.1, GhSnRK2.3, GhSnRK2.7, GhSnRK2.9, GhSnRK2.10, GhSnRK2.11, GhSnRK2.13, GhSnRK2.16, GhSnRK2.19 and GhSnRK2.20) were included in Group III.
Sequence alignment and C-terminal conserved motifs analysis
We analyzed all the conserved motifs of GhSnRK2s and identified the pattern of amino acid residues conservation in their domains (Fig. 2). The N-terminal catalytic domain was highly conserved, containing an ATP-binding site and a protein kinase-activating site. The ATP-binding region contained I/LGXGXFGVA where a lysine residue serving as a binding site for ATP (red box). The serine/threonine protein kinase active-site signature V/ICHRDLKLENTLL contained an aspartic acid residue (the active site) (purple box). These two parts constituted the serine/threonine kinase domain. The regulatory C-terminal domain contained stretches of acidic amino acids. On the basis of these structural features, the SnRK2 kinases were divided into two classes, namely SnRK2a and SnRK2b, either poly-Asp (D) (SnRK2a) or poly-Glu (E) (SnRK2b).
Multiple sequence alignment of the GhSnRK2 and AtSnRK2 proteins was obtained by DNAMAN software. Protein kinases ATP-binding region signature and serine/threonine protein kinases active-site signature are indicated by a red box and a purple box, respectively. DomainIand DomainIIat C terminus are marked by green box and blue box, respectively
The C-terminal domain consisted of two subdomains. Domain I was characteristic for all SnRK2 family members and is needed for activation by osmotic stress, i.e. the SnRK2-specific box (Gln-303 to Pro-318) (green box). Domain II was specific to the ABA-dependent SnRK2s and is needed for ABA responses, i.e. the ABA-specific box (Leu-333 to Met-362) (blue box) [44]. The ABA-specific box was present only in the members of Group III.
The GhSnRK2s showed 76.75% amino acid sequence identity, with highly conserved N-terminal kinase domains but divergent C-terminal domains. We employed MEME to detect conserved motifs in the GhSnRK2 family. Ten conserved motifs were scattered among each GhSnRK2 family (Fig. 3). All of the GhSnRK2 proteins shared the same seven motifs, designated motifs 1 to 7. Motif 4 contained the ATP-binding site. Motif 1 contained the serine/threonine protein kinase active-site. These two motifs constituted the N-terminal kinase domain. Motif 7 was located in the C-terminal Domain I, the SnRK2-specific box (Gln-303 to Pro-318). GroupIIISnRK2s, which are strongly activated by ABA, contain the ABA-specific box (Leu-333 to Met-362) involved in Motif 8. On the basis of previous reports, other conserved sequences in SNF1 were Arg(R)-Asp(D)-Leu(L) (residues 176 to 178 in the SNF1 protein), Asp(D)-Phe(F)-Gly(G) (residues 195 to 197) and Ala(A)-Pro(P)-Glu(E) (residues 219 to 221) [15], which were located in motif 1 and motif 6, respectively. Information for each motif, including site number and motif logo, is shown in (Additional file 1: Figure S1).
Phylogenetic relationship, exon-intron organizations, and motif analysis of GhSnRK2s. a Phylogenetic analysis of GhSnRK2s using by UPGMA method (b) Gene structure of the GhSnRK2 family. Exons and introns are shown by filled boxes and single lines, respectively. c Analysis with MEME to investigate 10 conserved motifs of GhSnRK2 proteins. Each colored box represents conservation. Details are included with Supplementary
Exon/intron organization of the cotton GhSnRK2 gene family
Analysis of exons and introns arrangement can provide important insights into the evolution of gene families [45]. To investigate the exon/intron structure of the GhSnRK2 genes, the coding and genomic sequences were compared, which showed that the genes contained nine exons interrupted by multiple introns (Fig. 3). In contrast to phylogenetic analysis, most members within the same group showed similar exon–intron structure and gene length. For example, in Group I, GhSnRK2.5 and GhSnRK2.15 showed 99.07% nucleotide sequence identity as indicated by the number of exons and similar intron phase patterns. This conservation of exon and intron numbers in each group strongly support the close evolutionary relationships of GhSnRK2 genes. In addition, GhSnRK2 genes in different groups usually varied in intron phase patterns and gene lengths.
Chromosomal location and evolutionary history of the GhSnRK2s gene family
Among 20 cotton SnRK2 genes, 18 genes were mapped onto 13 chromosomes and were named according to their order on the chromosomes (i.e., GhSnRK2.1, GhSnRK2.2, GhSnRK2.3, GhSnRK2.4, GhSnRK2.5, GhSnRK2.6, GhSnRK2.7, GhSnRK2.8, GhSnRK2.10, GhSnRK2.11, GhSnRK2.12, GhSnRK2.13, GhSnRK2.14, GhSnRK2.15, GhSnRK2.16, GhSnRK2.17, GhSnRK2.18 and GhSnRK2.20) (Fig. 4). Two GhSnRK2 genes that could not be conclusively mapped to any chromosome were named GhSnRK2.9 and GhSnRK2.19 (Fig. 4).
Nine GhSnRK2 genes were located on A-subgenome chromosomes. Among these GhSnRK2.1, GhSnRK2.2, GhSnRK2.3, GhSnRK2.4, GhSnRK2.5 and GhSnRK2.10 were located on chromosome A01, A02, A03, A05, A10 and A12, respectively. GhSnRK2.6, GhSnRK2.7 and GhSnRK2.8 were all present on A11.Nine GhSnRK2 genes were mapped to D-subgenome chromosomes. GhSnRK2.11, GhSnRK2.14, GhSnRK2.15 and GhSnRK2.20 were located on D01, D05, D10 and D12, respectively. D02 hosted two genes (GhSnRK2.12 and GhSnRK2.13). Three genes were allocated to chromosome D11 (GhSnRK2.16, GhSnRK2.17 and GhSnRK2.18). GhSnRK2.9 and GhSnRK2.19 were mapped to scaffold2734_A11 and scaffold4551_D11, respectively. This unbalanced distribution of GhSnRK2 genes on chromosomes suggested that genetic variation existed in the evolutionary process.
In eukaryotes, presumably there might be a high similarity to among duplication mechanisms during the expansion of gene families; these mechanisms include tandem duplication, segmental duplication and whole-genome duplication [46]. To infer possible relationships between GhSnRK2 genes and potential gene duplication within the G. hirsutum genome, we analyzed the occurrence of tandem duplication and segmental duplication during the evolution of this gene family. By calculating the similarity and the sequence coverage of the 20 GhSnRK2 genes, we identified 10 pairs of genes (GhSnRK2.1/GhSnRK2.11, GhSnRK2.2/GhSnRK2.12, GhSnRK2.3/GhSnRK2.13, GhSnRK2.4/GhSnRK2.14, GhSnRK2.5/GhSnRK2.15, GhSnRK2.6/GhSnRK2.17, GhSnRK2.7/GhSnRK2.19, GhSnRK2.8/GhSnRK2.18, GhSnRK2.9/GhSnRK2.16 and GhSnRK2.10/GhSnRK2.20) that undergo segmental duplication (Table 3).
To assess the evolutionary history of the GhSnRK2 gene family, synonymous (d S) and non-synonymous (d N) values were calculated using PAL2NAL. Nucleotide substitutions in protein-coding genes can be classified as synonymous or non-synonymous. Therefore, d S and d N values may allow ascertainment of whether selection has acted on protein-coding genes [47]. We estimated the d N/d S values for the 10 pairs of segmental-duplicated genes (Table 3). The d N/ds ratios for the 10 pairs of genes were lower than 1, which suggested that purified selection acted on these duplicated gene pairs [48].
Expression profiles of the GhSnRK2 genes under abiotic stresses and ABA treatment
SnRK2 genes were shown to respond to abiotic stress [30, 32]. To understand expression patterns of these 20 GhSnRK2 genes in G.hirsutum, we used publicly available transcriptome data to assess the expression under abiotic stresses conditions, i.e. cold, heat, salt and drought (Fig. 5). The analysis revealed that GhSnRK2 genes (GhSnRK2.3/2.7/2.8/2.9/2.10/2.13/2.14/2.16/2.18/2.19/2.20) responded to abiotic stresses, whereas the expression of other genes was not significantly altered under different stresses.
Expression analysis of GhSnRK2 genes in G.hirsutum TM-1 under abiotic stresses. The RNA-Seq expression profiles of G.hirsutum TM-1 [51] were used to identify the relative expression levels of GhSnRK2 genes. Levels of gene expression are depicted in different color on the scale. Red color represents high expression and green color represents low expression.The detail FPKM values are present in Additional file 2: Table S1 and Additional file 3: Table S2
Five GhSnRK2 genes (GhSnRK2.3/2.7/2.8/2.9/2.10) were notably up-regulated under salt and PEG (Polyethylene Glycol) treatment based on the transcriptome data, were selected for further analysis by qRT-PCR. Furthermore we detected the expression patterns of five GhSnRK2 selected genes in vegetative tissues (root, stem and leaf) (Fig. 6).
Expression analysis of five selected GhSnRK2 genes using quantitative real-time RT-PCR (qRT-PCR) under Nacl, PEG, ABA treatment. The relative expression level of five selected GhSnRK2 genes was normalized to the reference gene histone3 in different tissues. The transcripts in non-stressed was set as 1 for each gene in different tissues. The bars show the standard deviation of three technical repeats
In response to NaCl treatment, the transcription of GhSnRK2.3, GhSnRK2.7 and GhSnRK2.8 peaked at 3 h. While based on the transcriptome data the expression of these three genes was maximum at 24 h under salt stress. The expression level of GhSnRK2.10 in stem increased until 6 h, and then decreased at 12 h. This is consistent with transcriptome data that the expression of GhSnRK2.10 peaked at 6 h after salinity treatment.
Under drought stress, the transcription of GhSnRK2.3 in stem was increased until 3 h as well as the expression of GhSnRK2.3 that peaked at 3 h under PEG treatment based on the transcriptome data. Transcript level of GhSnRK2.10 in stems was slightly increased in response to dehydration. Meanwhile the expression level of GhSnRK2.10 was maximum at 1 h in leaves. In contrast, GhSnRK2.10 was notably up-regulated under PEG treatment in transcriptome data.
In response to exogenous ABA application, the transcript levels of GhSnRK2.3 and GhSnRK2.10 in stems were dramatically induced at 1 h. In leaves transcription of GhSnRK2.10 decreased within the first 6 h, however transcript levels peaked at 12 h, and then declined at 24 h. Additionally, GhSnRK2.8 expression was mainly induced in leaves and peaked at 12 h.
Discussion
An understanding of the response of plants to water deficit is important in efforts to model cotton (G.hirsutum) growth [49]. Therefore it is necessary to identify novel genes that confer significantly enhanced high water-use efficiency. The SnRK2 protein family members are plant-specific serine/threonine kinases involved in response to abiotic stresses, especially drought and salinity [50]. Such protein kinases have been identified in many plant species. However, little information is available on this protein family in G.hirsutum. The release of the G. hirsutum genome sequence laid the foundation for comprehensive identification and characterization of the SnRK2 family in G. hirsutum [51]. Identification and analysis of SnRK2 genes in G. hirsutum will prove to be a valuable tool for genetic improvement of allotetraploid Gossypium species.
Identification of GhSnRK2 genes
We identified 20 SnRK2 genes in the G. hirsutum genome, where 10 genes belonged to the A subgenome and the others to the D subgenome. Compared with other comprehensively surveyed plant SnRK2s (10 SnRK2 family genes have been identified in Arabidopsis, 10 in rice, 11 in maize, four in soybean, six in grape, three in tobacco, 10 in sorghum and 12 in apple), the GhSnRK2 family is the largest with 20 phylogenetically expanded genes. The striking expansion and diversification of the GhSnRK2 family suggests that these SnRK2s play crucial roles in the environmental adaptability and physiological maintenance in G. hirsutum, which can survive drought stress and other stressful environments.
Evolution of the SnRK2 family genes
Phylogenetic analysis and comparative genomic analyses are usually conducted to gain insight into evolutionary relationships of genes or species. To further evaluate the evolutionary relationships of GhSnRK2s with related genes from other plant species, we constructed an unrooted phylogenetic tree from an alignment of the full-length protein sequences. The phylogenetic analysis divided the GhSnRK2 family members into three different groups. The orthologous genes from rice, maize, Arabidopsis, tobacco and cotton were clustered in the same group, implying that SnRK2s originated before the divergence of monocots and dicots. Evolutionary studies based on SnRK2 protein sequences from green algae (Chlamydomonas reinhardtii), moss (Physcomitrella patens), fern (Selaginella moellendorffii), and angiosperms (including Arabidopsis and rice) showed that higher plants possess all three SnRK2 groups. Moss only possesses SnRK2s belonging to Group III and fern kinases belong to Group II. Green algal SnRK2s are distinct from those of land plants [52,53,54,55]. These results indicate a possible evolutionary pattern of plant SnRK2s: Group III is an ancient form of land plant SnRK2, whereas Group I arising before angiosperms of most recent origin. The size of Group I SnRK2s in cotton is smaller when compared with that of Arabidopsis, rice and maize, which indicates that the number of novel Group I proteins may have increased in these herbaceous plants during evolution.
From the perspective of the primary structure of the protein, phylogenetic analysis of the amino acid sequence provides information on evolutionary relationships. Previous reports have indicated that most SnRK2s from higher plants include nine exons [33]. We found that our GhSnRK2s also have nine exons while intron phase patterns and gene lengths were contrary to other species. This finding may indicate that in dicot and monocot plant lineages the SnRK2 members have experienced uneven deletion of introns leading to the variation in intron size and number. In addition, on the basis of the chromosomal arrangement of genes and comparative analysis of gene location, length, and structure, we identified 10 paired segmental-duplicated SnRK2 genes in the G. hirsutum genome. Preliminary analyses revealed that gene duplication and subsequent divergence are the main contributors to evolutionary momentum [56]. Usually, the criteria for inferring a gene duplication event are (1) the length of the alignment sequence covers ≥80% of the longest gene, and (2) the similarity of the aligned regions is ≥70% [57]. Two or more genes located on the same chromosome result from tandem duplication, whereas gene duplication between different chromosomes is designated as a segmental duplication event [58]. Two pairs of SnRK2 genes in Arabidopsis (AtSnRK2.1/AtSnRK2.5, and AtSnRK2.2/AtSnRK2.3) were identified as segmental duplication pairs. In rice, SAPK1/SAPK2 and SAPK4/SAPK5 have undergone segmental duplication [31]. The current study suggests that G. hirsutum is similar to Arabidopsis and rice in having experienced segmental duplication events. Gene duplication plays a key role facilitating the evolution of organisms by creating the raw genetic material that is modified by subsequent natural selection pressure [59]. These segmental duplications may have contributed greatly to the expansion and evolution of the SnRK2 gene family in G. hirsutum. It is speculated that the existence of such genes in the G. hirsutum genome might have been conducive to the structural and functional innovations by which cotton has been able to adapt to adverse environments.
Regulatory elements in the promoters of GhSnRK2 genes
Gene expression pattern can provide important clues to gene functions, which are believed to be associated with divergence in the promoter region [60]. Cis-acting regulatory elements contained in gene’ promoter regions play key roles in conferring the developmental and environmental regulation of gene expression. Based on previous reports, Group III genes were strongly activated by ABA, and in our group classification, 10 genes (GhSnRK2.1/2.3/2.7/2.9/2.10/2.11/2.13/2.16/2.19/2.20) belonged to Group III. ABA-responsive elements were found all of the members of Group III, expect GhSnRK2.7 and GhSnRK2.19. ABA-responsive elements were also found in the promoters of GhSnRK2s in other groups. For example, GhSnRK2.5 and GhSnRK2.15, which contain ABRE elements, belong to Group I.
Environmental stress-related elements were abundant in the GhSnRK2s. The regulation of adapting to certain external environmental conditions, such as drought, heat and low temperature, through SnRK2s has been well studied [23, 30, 37]. For GhSnRK2s, most members contained LTR, HSE and MBS elements. Moreover, many cis-elements were located at adjacent sites. There are two MBS elements in the promoter of GhSnRK2.3, which may enhance its drought responsiveness.
C-terminal motifs might contribute to the ABA response
The amino acid sequences of all SnRK2s contain highly conserved N-terminal kinase domain and a regulatory C-terminal domain with an ‘acidic patch’ region. It is further divided into two subgroups based on the composition of the C-terminal domain: SnRK2a (rich in glutamic acid) and SnRK2b (rich in aspartic acid) [20]. In Arabidopsis, the acidic patch is aspartate-rich in SnRK2 members of Groups II and III but glutamate-rich in Group I members [53, 61]. Only aspartate-rich SnRK2s can be activated by ABA. In our protein motif analysis, an acidic patch region was also present in the C-terminal motifs of GhSnRK2s. Motif 10 (specific to Group I) is poly-glutamate and GhSnRK2.5/GhSnRK2.15 may not be activated by ABA. On the basis of previous reports, Group I comprised kinases were not activated by ABA, Group II kinases were not activated or activated very weakly by ABA (depending on plant species), and Group III proteins were strongly activated by ABA [28, 62,63,64]. In Group III, C-terminal Domain II is poly-aspartate involved in motif 8. Moreover, members of Group III in cotton (GhSnRK2.1/2.3/2.7/2.9/2.10/2.11/2.13/2.16/2.19/2.20) contain an ABA-specific box (Leu-333 to Met-362) and may be strongly activated by ABA. Although members of Group II lacked an ABA-specific box, they were rich in aspartate, thus Group II SnRK2s may be weakly activated by ABA.
Some GhSnRK2 genes respond to abiotic stresses and ABA treatment
Numerous studies demonstrated that SnRK2s are involved in multiple abiotic stress responses. In the present study we determined the dynamic transcriptional changes of GhSnRK2 genes under drought and salinity stresses and ABA treatment. Drought resistance of Arabidopsis plants is influenced by AtSnRK2.8, which belongs Group II [30]. As determined by our real-time PCR analysis, GhSnRK2.8, which belongs to Group II, was induced in leaves at 3 h and 6 h after PEG treatment. In Arabidopsis, members of Groups II and III are reported to be involved in ABA-dependent signaling pathways [27, 65]. GhSnRK2.10, a member of Group III, also responded quickly to ABA treatment at 1, 3 and 6 h in stems. In rice, many SAPK genes are differentially regulated by mannitol, NaCl and ABA in different organs [62]. Salinity and drought induced elevated expression levels of SAPK3, SAPK8 and SAPK9 in rice [32]. We noted that GhSnRK2.3 showed 82.31% amino acid identity with SAPK8. Thus, we speculate that GhSnRK2.3 involved in regulation of salt stress tolerance in cotton. In many plant species, such as Arabidopsis, rice and maize, it is evident that SnRK2 proteins function as transcriptional activators in ABA-signaling mechanisms in response to abiotic stresses such as drought and salinity [26]. The expression of GhSnRK2 genes is induced by drought, salinity and ABA treatments, which may be indicative of potential roles in stress responses.
Conclusions
Although the function of certain SnRK2 genes of Arabidopsis and rice has been clearly demonstrated, the functions of SnRK2 genes in G. hirsutum are still elusive. In the present research, we performed a genome-wide analysis of the SnRK2 gene family in G. hirsutum, including examination of gene structure, evolutionary relationships, and transcriptional changes in response to stress treatments. GhSnRK2 genes were identified, which are distributed in different evolutionary lineages among higher plants. The expansion of the SnRK2 family in G. hirsutum might be as result of segmental duplication. Our expression analysis revealed differential responses among the GhSnRK2 genes to abiotic stresses. The present genomic and bioinformatics analyses of GhSnRK2 genes provide a solid foundation for further investigation of GhSnRK2 gene functions. In addition, these results may help in genetic improvement of cotton stress tolerance.
Methods
Identification of SnRK2 genes in Gossypium hirsutum
We downloaded the G. hirsutum genome sequence and the proteome sequences from the CottonGen database (http://www.cottongen.org/specises/Gossypium_hirsutum/nbi-AD1_genome_v1.1, 51]. A local BLASTP algorithm-based search with the blast-2.2.9 program, which was downloaded from the National Center for Biotechnology Information (NCBI) (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/2.2.19/), was performed to identify complete SnRK2 members, using Arabidopsis SnRK2 sequences as query. The E- value threshold for BLASTP was set at 1e−10 to obtain the final dataset of SnRK2 proteins. Redundant sequences were removed. Then, the Pfam (http://pfam.sanger.ac.uk/search) and SMART (http://smart.embl-heidelberg.de/) databases were used to confirm each predicted GhSnRK2 protein sequence [66, 67]. The sequences of 10 rice and 10 Arabidopsis SnRK2 proteins were obtained from the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) and the Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/) databases, respectively. The biophysical properties of the encoded proteins were computed using the ExPASy ProtParam tool (http://us.expasy.org/tools/protparam.html).
Gene structure and C-terminal conserved motifs analysis
Structural information for the GhSnRK2 genes, including chromosomal location and gene length, were obtained from the CottonGen databases. Exons and introns were predicted by comparing the coding sequences with genomic sequences using the gene structure display server (GSDS program 2.0) (http://gsds.cbi.pku.edu.cn/) [68]. Conserved motif prediction was performed using the MEME (http://meme-suite.org/) program. The analysis was performed with the following parameters: number of unique motifs: 10; and maximum and minimum search widths: 50 and 16, respectively.
Mapping SnRK2 genes on cotton chromosomes, and estimating synonymous (d S) and non-synonymous (d N) substitution rates
The chromosomal distributions of GhSnRK2 genes were obtained based on genome annotation. The Mapchart 2.2 software was used to visualize the distribution of the GhSnRK2 genes on the 13 G. hirsutum chromosomes.
The dS and dNsubstitution rates were calculated with the PAL2NAL web server (http://www.bork.embl.de/pal2nal#RunP2N), which uses the CODEMAL program of PAML [69].
Sequence alignment and phylogenetic tree construction
An alignment of multiple SnRK2 protein sequences from Arabidopsis thaliana, Zea mays, Oryza sativa and Nicotiana tabacum was generated using the ClustalW program [27, 31, 33, 62]. A neighbor-joining analysis of the generated alignment was performed using the unweighted pair-group method with arithmetic mean algorithm to construct an unrooted phylogenetic tree. Support for the tree topology was assessed by performing a bootstrap analysis with 1000 replicates. The tree was visualized with MEGA 5.05 software [70].
Retrieval and analysis of promoter sequences
The G. hirsutum genome sequences were used to retrieve the promoter sequences (2 kb upstream of the start codon) of the GhSnRK2 genes. The analysis of the GhSnRK2 promoters was carried out using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [71].
Gene expression analysis
The expression levels of GhSnRK2 genes were measured using the RNA-sequencing data of G.hirsutum TM-1 acquired under four different stresses (cold, heat, salt and drought), which were downloaded from the NCBI Gene Expression repository under accession number PRJNA248163 (Additional files 2 and 3) (http://www.ncbi.nlm.nih.gov/bioproject/PRJNA248163/).
RNA isolation and the qRT-PCR analysis
To examine expression profiles of GhSnRK2 genes in different tissues and in response to abiotic stresses, upland cotton with three or four leaves were submerged in 10% PEG6000, 150 mM NaCl, or 100 μM ABA solutions. Samples were collected from the root, stems and leaves at 0, 1, 3, 6, 12, and 24 h after treatment. Samples collected at 0 h were used as controls. All samples were frozen immediately in liquid nitrogen and kept at −80 °C for total RNA extraction. Total RNA was extracted from the samples using the RNAprep Pure Kit (For Plants) (TIANGEN, Beijing, China). To remove genomic DNA contamination, the RNA samples were treated with DNase I. Gel electrophoresis and a Nanodrop2000 nucleic acid analyzer were employed to detect the quality of the RNA. First-strand cDNA was synthesized based on reverse transcription of 1 μg RNA digested by DNase I using the PrimeScript™ RT Reagent Kit (Takara, Dalian, China). PCR amplifications were performed using SYBR® Premix Ex Taq™ (Takara). For real-time PCR, gene-specific primers were designed using Primer 5.0 (Additional file 4). The cotton histone3 gene (GenBank accession no. AF024716) was used as the internal reference gene [72].
For the qRT-PCR assay, cDNA was diluted to 100 ng/μl with ddH2O. The qRT-PCR reaction was carried out on an ABI 7900 HT Real-time PCR System (Applied Biosystems). The reaction system (in a total volume of 20 μl) contained 10 μl SYBR® Premix Ex Taq™ (2×), 0.4 μl of each primer (10 μM), 0.4 μl ROX Reference Dye (50×), 1 μl template (about 100 ng/μl), and ddH2O to make up the total volume. The protocol was performed as follows: pre-denaturation at 95 °C for 30 s (step 1), denaturation at 95 °C for 5 s (step 2), primer annealing/extension and gathering of the fluorescent signal at 60 °C for 1 min (step 3), followed by 40 cycles of step 2. Three biological replicates, each consisting of three technical replicates, were analyzed. The relative expression levels of the SnRK2 genes were calculated using the 2−△△Ct method [73].
Abbreviations
- GhSnRK2s :
-
Genes in Gossypium hirsutum L.(AADD)
- SnRK2:
-
Sucrose non-fermenting-1-related protein kinase 2
References
Ohkuma K, Lyon JL, Addicott FT, Smith OE. Abscisin II, an abscission-accelerating substance from young cotton fruit. Science. 1963;142(3599):1592–3.
Addicott FT, Lyon JL. physiology of abscisic acid and related substances. Annu Rev Plant Physiol. 1969;20(20):139–64.
Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell. 2002;14(11):2723–43.
Barrero JM, Piqueras P, González-Guzmán M, Serrano R, Rodríguez PL, Ponce MR, Micol JL. A mutational analysis of the ABA1 gene of Arabidopsis thaliana highlights the involvement of ABA in vegetative development. J Exp Bot. 2005;56(418):2071–83.
Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. P Natl Acad Sci USA. 2009;106(20):8380–5.
Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50(7):1345–63.
Cohen P. Review lecture: protein phosphorylation and hormone action. Proc R Soc Lond. 1988;234(234):115–44.
Wrzaczek M, Hirt H. Plant MAP kinase pathways: how many and what for? Biol Cell. 2001;93(1–2):81.
Ludwig AA, Romeis T, Jones JDG. CDPK-mediated signalling pathways: specificity and cross-talk. J Exp Bot. 2004;55(395):181.
Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003;132(2):666–80.
Halford NG, Hardie DG. SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Mol Biol. 1998;37(5):735.
Hardie DG, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem. 1998;67(1):821–55.
Halford NG, Hey SJ. Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem J. 2009;419(2):247–59.
Hrabak EM, Dickmann LJ, Satterlee JS. Characterization of eight new members of the calmodulin-like domain protein kinase gene family from Arabidopsis thaliana. Plant Mol Biol. 1996;31(2):405–12.
Celenza JL, Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986;233(233):1175–80.
Dong XF, Cui N, Wang L, Zhao XC, Qu B, Li TL, Zhang GL. The SnRK protein kinase family and the function of SnRK1 protein kinase. Int J Agric Biol. 2012;14(4):575–9.
Guo Y, Halfter U, Ishitani M, Zhu JK. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell. 2001;13(6):1383–400.
Halfter U, Zhu JK. The arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. P Natl Acad Sci USA. 2000;97(7):3735–40.
Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. P Natl Acad Sci USA. 2000;97(7):3730.
Shukla V, Mattoo AK. Sucrose non-fermenting 1-related protein kinase 2 (SnRK2): a family of protein kinases involved in hyperosmotic stress signaling. Physiol Mol Biol Plants. 2008;14(1):91.
Anderberg RJ, Walkersimmons MK. Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. P Natl Acad Sci USA. 1992;89(21):10183–7.
Holappa LD, Walkersimmons MK. The wheat abscisic acid-responsive protein kinase mrna, pkaba1, is up-regulated by dehydration, cold temperature, and osmotic stress. Plant Physiol. 1995;108(3):1203–10.
Mao X, Zhang H, Tian S, Chang X, Jing R. TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. J Exp Bot. 2010;61(3):683–96.
Zhang H, Mao X, Jing R, Chang X, Xie H. Characterization of a common wheat (Triticum aestivum L.) TaSnRK2.7 gene involved in abiotic stress responses. J Exp Bot. 2011;62(3):975–88.
Zhang H, Mao X, Wang C, Jing R. Overexpression of a common wheat gene tasnrk2.8 enhances tolerance to drought, salt and low temperature in arabidopsis. PLoS One. 2010;5(12):e16041.
Wawer I, Krzywińska E. SnRK2 protein kinases--key regulators of plant response to abiotic stresses. Omics J Integr Biol. 2011;15(12):859–72.
Boudsocq M, Barbierbrygoo H, Laurière C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem. 2004;279(40):41758–66.
Mustilli AC, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14(12):3089–99.
Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem. 2006;281(8):5310–18.
Umezawa T, Yoshida R, Maruyama K, Yamaguchishinozaki K, Shinozaki K. SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. P Natl Acad Sci USA. 2004;101(49):17306.
Muszyńska G, Hardie DG, Dobrowolska G. Biochemical characterization of the tobacco 42-kD protein kinase activated by osmotic stress. Plant Physiol. 2004;136(2):3255–65.
Diédhiou CJ, Popova OV, Dietz KJ, Golldack D. The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice. BMC Plant Biol. 2008;8(1):49.
Huai J, Wang M, He J, Zheng J, Dong Z, Lv H, Zhao J, Wang G. Cloning and characterization of the SnRK2 gene family from Zea mays. Plant Cell Rep. 2008;27(12):1861–8.
Zou H, Zhang X, Zhao JR, Yang Q, Wu Z, Wang F, Huang C. Full Length Research-Cloning and characterization of maize ZmSPK1, a homologue to nonfermenting1-related protein kinase2. Afr J Biotechnol. 2006;5(6):490–96.
Yoon HW, Kim MC, Shin PG, Kim JS, Kim CY, Lee SY, Hwang I, Bahk JD, Hong JC, Han C. Differential expression of two functional serine/threonine protein kinases from soyabean that have an unusual acidic domain at the carboxy terminus. Mol Gen Genomics. 1997;255(4):359–71.
Monks DE, Aghoram K, Courtney PD, Dewald DB, Dewey RE. Hyperosmotic stress induces the rapid phosphorylation of a soybean phosphatidylinositol transfer protein homolog through activation of the protein kinases SPK1 and SPK2. Plant Cell. 2001;13(5):1205–19.
Zhang H, Jia H, Liu G, Yang S, Zhang S, Yang Y, Yang P, Cui H. Cloning and characterization of SnRK2 subfamily II genes from Nicotiana tabacum. Mol Biol Rep. 2014;41(9):5701.
Chen ZJ, Scheffler BE, Dennis E, Triplett BA, Zhang T, Guo W, Chen X, Stelly DM, Rabinowicz PD, Town CD. Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 2008;145(4):1303–10.
Yu-Xian F-G. The Gossypium raimondii Genome, a huge leap forward in cotton genomics. J Integr Plant Biol. 2013;55(7):570–1.
Tiwari SC, Wilkins TA. Cotton (Gossypium hirsutum) seed trichomes expand via diffuse growing mechanism. Can J Bot. 2011;73(5):746–57.
Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ, Ma Z, Shang H, Ma X, Wu J. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol. 2015;33(5):524.
Bello B, Zhang X, Liu C, Yang Z, Yang Z, Wang Q, Ge Z, Li F. Cloning of Gossypium hirsutum Sucrose non-fermenting 1-related protein kinase 2 gene ( GhSnRK2 ) and its overexpression in transgenic arabidopsis escalates drought and low temperature tolerance. PLoS One. 2014;9(11):e112269–9.
Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. Jmolbiol. 1982;157(1):105–32.
Belin C, de Franco PO, Bourbousse C, Chaignepain S, Schmitter JM, Vavasseur A, Giraudat J, Barbierbrygoo H, Thomine S. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 2006;141(4):1316.
Mach J. Alternative splicing produces a JAZ protein that is not broken down in response to jasmonic acid. Plant Cell. 2009;21(1):14.
Wei K, Wang Y, Xie D. Identification and expression profile analysis of the protein kinase gene superfamily in maize development. Mol Breeding. 2014;33(1):155–72.
De LRB, Marcobal A, Muñoz R. Allelic diversity and population structure in Oenococcus oeni as determined from sequence analysis of housekeeping genes. Appl Environ Microbiol. 2004;70(12):7210.
Hurst LD. The K a/ K s ratio: diagnosing the form of sequence evolution. Trends in Genetics Tig. 2002;18(9):486.
Pace PF, Cralle HT, Shm EH, Cothren JT, Senseman SA. Drought-induced changes in shoot and root growth of young cotton plants. J Cotton Sci. 1999;3(4):183–7.
Hey SJ, Byrne E, Halford NG. The interface between metabolic and stress signalling. Ann Bot. 2010;105(2):197.
Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33(5):531.
Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci. 2009;106(41):17588–93.
Mizoguchi M, Umezawa T, Nakashima K, Kidokoro S, Takasaki H, Fujita Y, Yamaguchishinozaki K, Shinozaki K. Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression. Plant Cell Physiol. 2010;51(5):842–7.
Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K. Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 2010;51(51):1821–39.
Hauser F, Waadt R, Schroeder JI. Evolution of abscisic acid synthesis and signaling mechanisms. Curr Biol. 2011;21(9):R346.
Chothia C, Gough J, Vogel C, Teichmann SA. Evolution of the protein repertoire. Science. 2003;300(5626):1701–3.
Yang S, Zhang X, Yue JX, Tian D, Chen JQ. Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Gen Genomics. 2008;280(3):187–98.
He H, Dong Q, Shao Y, Jiang H, Zhu S, Cheng B, Xiang Y. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 2012;31(7):1199–217.
Cronk QC. Plant evolution and development in a post-genomic context. Nat Rev Genet. 2001;2(8):607.
Stephen J, Fei N, Juergen E, Zhang S, Dong W, Xue T, Zheng C, Yuan Z. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics. 2008;9(1):1–21.
Halford NG, Hardie DG. SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Mol Biol. 1998;37(5):735–48.
Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell. 2004;16(5):1163–77.
Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005;44(6):939–49.
Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19(2):485–94.
Mao X, Zhang H, Tian S, Chang X, Jing R. TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. J Exp Bot. 2009;61(3):683–96.
Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43(Database issue):D257.
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2015;44(D1):D279.
Bo H, Jin J, Guo AY, He Z, Luo J, Ge G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296.
Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34(Web Server issue):W609.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9.
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325.
Wang JW, Wang S, Chen XY. Characterization of GaWRKY1, a Cotton Transcription Factor That Regulates the Sesquiterpene Synthase Gene (+)-δ-Cadinene Synthase-A. Plant Physiol. 2004;135(1):507–15.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–8.
Acknowledgments
We thank Peng Huo (Zhengzhou Research Center, Institute of Cotton Research of CAAS, Zhengzhou) for technical assistance. We also appreciate the reviewers and editors for the patience to the work.
Funding
This work was supported by grants from Major Program of Joint Funds (Sinkiang) of the National Natural Science Foundation of China (Grant No. U1303282) and the Young Elite Scientist Sponsorship Program by CAST.
Availability of data and materials
The data sets supporting the results of this article are included within the article and its additional file.
Authors’ contributions
ZL, XZ, CZ, ZY and FL conceived and designed the experiments. ZL, GZ, EC performed the experiments and analyzed the data. ZL drafted the manuscript. ZL, ZY, GZ, XG and JL revised the manuscript. All authors have read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Additional files
Additional file 1: Figure S1.
Details of conserved motifs detected among members of the GhSnRK2 protein family by MEME. (TIFF 1466 kb)
Additional file 2: Table S1.
FPKM values under abiotic stresses. (XLSX 17 kb)
Additional file 3: Table S2.
Gene accession number and samples information of transcriptome data used in our research. (XLSX 10 kb)
Additional file 4: Table S3.
List of the primers used for quantitative real-time PCR in this study. (XLSX 10 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liu, Z., Ge, X., Yang, Z. et al. Genome-wide identification and characterization of SnRK2 gene family in cotton (Gossypium hirsutum L.). BMC Genet 18, 54 (2017). https://doi.org/10.1186/s12863-017-0517-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12863-017-0517-3