Abstract
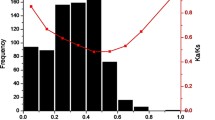
Most disease resistance genes in plants encode NBS-LRR proteins. However, in woody species, little is known about the evolutionary history of these genes. Here, we identified 459 and 330 respective NBS-LRRs in grapevine and poplar genomes. We subsequently investigated protein motif composition, phylogenetic relationships and physical locations. We found significant excesses of recent duplications in perennial species, compared with those of annuals, represented by rice and Arabidopsis. Consequently, we observed higher nucleotide identity among paralogs and a higher percentage of NBS-encoding genes positioned in numerous clusters in the grapevine and poplar. These results suggested that recent tandem duplication played a major role in NBS-encoding gene expansion in perennial species. These duplication events, together with a higher probability of recombination revealed in this study, could compensate for the longer generation time in woody perennial species e.g. duplication and recombination could serve to generate novel resistance specificities. In addition, we observed extensive species-specific expansion in TIR-NBS-encoding genes. Non-TIR-NBS-encoding genes were poly- or paraphyletic, i.e. genes from three or more plant species were nested in different clades, suggesting different evolutionary patterns between these two gene types.





Similar content being viewed by others
References
Ameline-Torregrosa C, Wang BB, O’Bleness MS, Deshpande S, Zhu H, Roe B, Young ND, Cannon SB (2008) Identification and characterization of NBS-LRR genes in the model plant medicago truncatula. Plant Physiol 146:5–21
Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Bai J, Pennill LA, Ning J, Lee SW, Ramalingam J, Webb CA, Zhao B, Sun Q, Nelson JC, Leach JE, Hulbert SH (2002) Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res 12:1871–1884
Bailey TL, Elkan C (2005) The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol 3:21–29
Bent AF, Mackey D (2007) Elicitors, effectors, and R-genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45:399–436
Cannon SB, Zhu H, Baumgarten AM, Spangler R, May G, Cook DR, Young ND (2002) Diversity, distribution, and ancient taxonomic relationships within the TIR and non-TIR NBS-LRR resistance gene subfamilies. J Mol Evol 54:548–562
Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826–833
Ding J, Araki H, Wang Q, Zhang P, Yang S, Chen JQ, Tian D (2007a) Highly asymmetric rice genomes. BMC Genomics 8:154
Ding J, Zhang W, Jing Z, Chen JQ, Tian D (2007b) Unique pattern of R-gene variation within populations in Arabidopsis. Mol Genet Genomics 277:619–629
Holub EB (2001) The arms race is ancient history in Arabidopsis, the wildflower. Nat Rev Genet 2:516–527
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Jaillon O, Aury JM, Noel B, Policriti A, Clepet C et al (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467
Jiang H, Wang C, Ping L, Tian D, Yang S (2007) Pattern of LRR nucleotide variation in plant resistance genes. Plant Sci 173:253–261
Leister D (2004) Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet 20:116–122
Lupas A, Van Dyke M, Stock J (1991) Predicting coiled coils from protein sequences. Science 252:1162–1164
Lynch M, Crease TJ (1990) The analysis of population survey data on DNA sequence variation. Mol Biol Evol 7:377–394
McHale L, Tan X, Koehl P, Michelmore RW (2006) Plant NBS-LRR proteins: adaptable guards. Genome Biol 7:212
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332
Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15:809–834
Meyers BC, Morgante M, Michelmore RW (2002) TIR-X and TIR-NBS proteins: two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J 32:77–92
Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8:1113–1130
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Nobuta K, Ashfield T, Kim S, Innes RW (2005) Diversification of non-TIR class NB-LRR genes in relation to whole-genome duplication events in Arabidopsis. Mol Plant Microbe Interact 18:103–109
Parniske M, Hammond-Kosack KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BB, Jones JD (1997) Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf–4/9 locus of tomato. Cell 91:821–832
Rice Chromosomes 11 and 12 Sequencing Consortia (2005) The sequence of rice chromosomes 11 and 12, rich in disease resistance genes and recent gene duplications. BMC Biol 3:20
Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Shen J, Araki H, Chen L, Chen JQ, Tian D (2006) Unique evolutionary mechanism in R-genes under the presence/absence polymorphism in Arabidopsis thaliana. Genetics 172:1243–1250
Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16:1220–1234
Xiao S, Ellwood S, Calis O, Patrick E, Li T et al (2001) Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291:118–120
Xiao S, Emerson B, Ratanasut K, Patrick E, O’Neill C et al (2004) Origin and maintenance of a broad-spectrum disease resistance locus in Arabidopsis. Mol Biol Evol 21:1661–1672
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I et al (2006) The genome of black cottonwood, Populus trichocarpa (torr. & gray). Science 313:1596–1604
van Ooijen G, van den Burg HA, Cornelissen BJ, Takken FL (2007) Structure and function of resistance proteins in solanaceous plants. Annu Rev Phytopathol 45:43–72
Yang S, Feng Z, Zhang X, Jiang K, Jin X, Hang Y, Chen JQ, Tian D (2006) Genome-wide investigation on the genetic variations of rice disease resistance genes. Plant Mol Biol 62:181–193
Yang S, Jiang K, Araki H, Ding J, Yang YH, Tian D (2007) A molecular isolation mechanism associated with high intra-specific diversity in rice. Gene 394:87–95
Yu J, Wang J, Lin W, Li S, Li H et al (2005) The Genomes of Oryza sativa: a history of duplications. PLoS Biol 3:e38
Zhang P, Gu Z, Li WH (2003) Different evolutionary patterns between young duplicate genes in the human genome. Genome Biol 4:R56
Zhou T, Wang Y, Chen JQ, Araki H, Jing Z, Jiang K, Shen J, Tian D (2004) Genome-wide identification of NBS genes in rice reveals significant expansion of divergent non-TIR NBS genes. Mol Genet Genomics 271:402–415
Acknowledgments
This work was supported by the National Natural Science Foundation of China (30570987 and 30470122), Pre-program for NBRPC (2005CCA02100) and 111 project to D. T., or J-Q. C.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by A. Tyagi.
Sihai Yang and Xiaohui Zhang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, S., Zhang, X., Yue, JX. et al. Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Genet Genomics 280, 187–198 (2008). https://doi.org/10.1007/s00438-008-0355-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-008-0355-0