Abstract
Genetic characterization, diversity analysis and estimate of the genetic relationship among varieties using functional and random DNA markers linked to agronomic traits can provide relevant guidelines in selecting parents and designing new breeding strategies for marker-assisted wheat cultivar improvement. Here, we characterize 20 Moroccan and 19 exotic bread wheat (Triticum aestivum L.) cultivars using 47 functional and 7 linked random DNA markers associated with 21 loci of the most important traits for wheat breeding. The functional marker analysis revealed that 35, 45, and 10 % of the Moroccan cultivars, respectively have the rust resistance genes (Lr34/Yr18/Pm38), dwarfing genes (Rht1b or Rht2b alleles) and the leaf rust resistance gene (Lr68). The marker alleles for genes Lr37/Yr17/Sr38, Sr24 and Yr36 were present only in the exotic cultivars and absent in Moroccan cultivars. 25 % of cultivars had 1BL.1RS translocation. 70 % of the wheat cultivars had Ppo-D1a and Ppo-A1b associated with low polyphenol oxidase activity. 10 % of cultivars showed presence of a random DNA marker allele (175 bp) linked to Hessian fly resistance gene H22. The majority of the Moroccan cultivars were carrying alleles that impart good bread making quality. Neighbor joining (NJ) and principal coordinate analysis based on the marker data revealed a clear differentiation between elite Moroccan and exotic wheat cultivars. The results of this study are useful for selecting suitable parents for making targeted crosses in marker-assisted wheat breeding and enhancing genetic diversity in the wheat cultivars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat (Triticum aestivum L.) is an important staple crop, providing 20 % of all calories consumed by people worldwide. Demand for wheat is predicted to increase in the future as the global population increases. With the world’s population estimated to reach 9.6 billion by 2050, wheat production will have a crucial bearing on food security and the global economy in the coming decades. In Morocco, wheat is the most consumed cereal crop, with a per capita consumption of 258 kg annually (USDA Foreign Agricultural Service 2014). In Morocco, it is cultivated in an area of 3.2 million ha, mostly in rainfed conditions with a production of 6.9 million tonnes in 2013 (FAOSTAT 2014). Its productivity is comparatively low, due to abiotic stresses such as drought, and biotic stresses such as Hessian fly, leaf rust, and yellow rust. Consequently, Morocco is not self sufficient in wheat production in most of the years and imports bread wheat for its domestic consumption. Therefore, the overall objectives of wheat breeding remains the development of wheat genotypes with higher yield, higher adapted to contrasted environment, resistance to the biotic stresses and with acceptable higher grain quality. Even all the effort made for improving wheat, its productivity still depends on traditional breeding and selection using conventional techniques. Currently, the Moroccan breeding program is giving a priority to new technologies such as the use of molecular markers to speed up the development of improved wheat varieties.
The characterization of genetic variability and an estimate of the genetic relationship among varieties are essential to any breeding program; because of artificial crosses among less similar parents allow a larger segregation and the combination of different favorable alleles (Bered et al. 2002). Genetic similarities might be evaluated by means of pedigree analysis (Barrett and Kidwell 1998) or by assessing morphological traits (Schut et al. 1997) as well as biochemical (Metakovsky and Branlard 1998) or, more recently, DNA markers (Barrett and Kidwell 1998; Pagnotta et al. 2005). The use of molecular approaches, particularly molecular markers, has allowed better characterization, maintenance of genetic variation in plant germplasm, identifying genes underlying important traits, and devising optimal breeding strategies for crop improvement (Hayden et al. 2010). Therefore, evaluation of the genetic diversity present in wheat germplasm deployed in the current breeding programs at the molecular level and integration of this information into cultivar development are essential for using genetic resources effectively in breeding programs (Chao et al. 2007).
Identification of molecular markers that cosegregate or closely linked with the agronomic traits is useful for marker-assisted selection (MAS; Mohan et al. 1997). Closely linked random DNA markers (RDMs; Andersen and Lubberstedt 2003) and gene specific or functional markers (Andersen and Lubberstedt 2003) are commonly used for MAS. In contrast to random DNA markers, gene specific or functional markers are ideal for MAS in wheat breeding as they are derived directly from the gene conferring the phenotype. In plant breeding, functional markers can be used for validation of cultivar identity, selection of parental materials to build segregating population, and subsequent selection of lines (Lübberstedt et al. 2005). Several markers were developed and validated for MAS. To date, more than 30 wheat loci associated with end-use quality, agronomic traits, and disease resistance in bread wheat (Liu et al. 2012). 56 functional markers for quality traits such as high- and low-molecular-weight glutenin subunits (HMW-GS and LMW-GS), polyphenol oxidase (PPO) activity, lipoxynase (LOX) activity, yellow pigment content (YPC), kernel hardness (Pin), and starch properties have been developed. 27 functional markers for agronomic traits were developed and reportedly used in wheat breeding programs such as semi-dwarfing genes Rht-B1b (Rht1) and Rht-D1b (Rht2), photoperiod response genes (Ppd), vernalization genes (Vrn) and developmental rate genes. For rust disease resistance, six genes Lr34/Yr18/Pm38, Lr37/Yr17/Sr38, Lr19, Lr47, Lr51 and Yr36 had been cloned in wheat (Feuillet et al. 2003; Huang et al. 2003; Yahiaoui et al. 2004; Fu et al. 2009; Krattinger et al. 2009; Liu et al. 2014) in addition to 1B/1R translocation (Froidmont 1998; Chai et al. 2006; Liu et al. 2008) and functional markers were designed and successfully applied in the breeding. The objective of this work was to genotype 20 Moroccan and 19 exotic bread wheat cultivars using 47 functional markers and 7 random DNA markers closely linked to 21 loci of the most important target traits for breeding and to determine the genetic relationship between them to identify the potential parental lines for the wheat breeding programs.
Materials and methods
Plant materials
A set of 39 wheat lines, which includes 20 improved elite cultivars of Morocco and 19 exotic cultivars (Table 1) were used for the marker analysis. The exotic wheat lines were introduced to Morocco to be used as donors for the specific traits of interest in the wheat breeding program. The Moroccan cultivars were procured from the National Gene Bank of Morocco, whereas, the exotic cultivars were procured from the national or international gene banks of the other countries.
DNA extraction and marker genotyping
Total genomic DNA was extracted by CTAB method of Saghai-Maroof et al. (1984) with some modifications as adapted by Udupa et al. (1999). Fresh young leaves were collected from green house grown plants of individual cultivars. The isolated DNA was estimated both qualitatively and quantitatively using 1.0 % (w/v) agarose gels by comparing bands to known concentrations of lambda DNA.
Total of 47 functional markers and 7 linked random DNA markers (RDMs) to the traits of interest were used for genotyping the bread wheat cultivars. They were Lr34 (Lagudah et al. 2006), Lr68 (Herrera-Foessel et al. 2012), Lr37 (Helguera et al. 2003), Sr24 (Mago et al. 2005), Gpc-B1/Yr36 (Distelfeld et al. 2006), Rht-B1, Rht-D1 (Ellis et al. 2002), Vp1-B3 (Yang et al. 2007), Ppo-A1, Ppo-D1 (He et al. 2007), Ppd-D1 (Beales et al. 2007; Yang et al. 2009), iag95 for 1B/1R (Mago et al. 2002), Pina-D1 (Gautier et al. 1994), Allwaxy (McLauchlan et al. 2001), Glu-A1 (Lafiandra et al. 1997; De Bustos et al. 2000), Glu-B1 (Ahmad 2000; Butow et al. 2004; Lei et al. 2006), Glu-D1 (Ahmad 2000), Glu-A3 (Zhang et al. 2004), Glu-B3 (Wang et al. 2009), Glu-D3 (Zhao et al. 2007) and Xgdm33, closely linked to gene H22 (Zhao et al. 2006). PCR reaction was performed in a reaction volume of 10 μL containing 1× PCR buffer (1.5 mM MgCl2), 200 µM of each dNTPs, 10 pmol of each primer, 0.5U of Taq DNA polymerase (Promega) and approximately 50 ng of genomic DNA. Primers names, sequences and cycling conditions for each molecular marker are detailed in supplementary Table S1. The PCR products were separated 1.2 or 1.5 % (w/v) agarose gels. Except for Allwaxy, Rht-B1, Rht-D1 and Xgdm33 were run in 6 % native polyacrylamide gels, prepared in a vertical electrophoresis unit (CBS Scientific) using 0.5 × TBE buffer. The different gels were stained with ethidium bromide and visualized under UV light.
Analysis of molecular data
PowerMarker software version 3.25 (Liu and Muse 2005) was used to calculate the number of alleles and values of genetic diversity and PIC (Botstein et al. 1980) of each locus. Genetic distances between each pair of cultivars were measured by calculating the shared allele frequencies (Jin and Chakraborty 1993). The Neighbor joining dendrogram was generated using the DARwin software based on the genetic distance calculated using PowerMarker software. The genetic structure was analyzed by performing PCoA (Principal Coordinates analysis) implemented in the program GenAlex 6.5 (Peakall and Smouse 2012).
Results
Genetic diversity analysis
Genetic diversity of 20 elite Moroccan cultivars and 19 potential exotic cultivars to be deployed in the breeding program was evaluated using 47 functional and 7 random DNA markers linked to the target traits of interest. The total number of detected alleles was 48 in Moroccan cultivars and 56 in exotic cultivars. Average number of alleles was slightly higher in exotic cultivars than Moroccan cultivars. Mean number of alleles was 2.5 and 2.9 in Moroccan and exotic cultivars, respectively. Similarly, exotic cultivars had a higher PIC value (0.39) compared to Moroccan cultivars (0.34) (Table 2). The 54 primer pairs for specific alleles linked to 21 loci distributed in 12 chromosomes showed a good polymorphism in Moroccan cultivars with slight difference to the exotic cultivars. The genetic diversity calculated was 0.4. The glutenin genes namely Glu-B1, Glu-A3 and Glu-B3 were the most polymorphic and displayed higher number of alleles (5, 5 and 6) and high genetic diversity (0.735, 0.660 and 0.770), respectively.
Markers based trait analysis
The 20 elite Moroccan cultivars were screened with the functional and the random DNA markers linked with quality, agronomic traits, and disease resistance (Table 3). The frequency of leaf rust resistance functional allele at Lr34 gene was 35 % and at Lr68 gene the linked random DNA marker allele was 10 % (Table 3). The marker alleles for genes Lr37, Sr24 and Yr36 were absent in Moroccan cultivars, whereas they were present only in exotic cultivars. 25 % of cultivars had 1R segment (1BL.1RS translocation) and 10 % of cultivars showed presence of 175 bp size allele of Xgdm33 linked with Hessian fly resistance gene H22. For the other agronomic traits, such as, dwarfing genes Rht1 and Rht2, the frequency was 45 % for each gene. Majority of wheat cultivars had Ppo-D1a and Ppo-A1b alleles associated with low polyphenol oxidase activity (70 %). Only 15 % of the cultivars had photoperiod insensitive allele at Ppd-D1 locus. While, Vp1-B3 STS primer pair amplified 569 bp fragment linked to preharvest sprouting tolerance in all 20 Moroccan cultivars. For the end-use quality traits, the frequency of wx-A1 and wx-B1 associated to improved starch quality was 70 and 75 %, respectively. In addition, wx-D1 (data not shown) existed in all Moroccan and exotic cultivars. Twenty-five percent of cultivars carried Pina-D1a linked to soft grain texture. The Glutenin genes revealed high level of polymorphism related to variable degree of bread making quality.
Genetic relationships and PCoA analysis
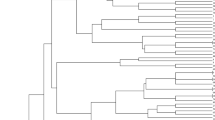
To study the genetic relationships between Moroccan and exotic cultivars for breeding purposes, the allelic data were used to estimate the genetic distance between all cultivars and Neighbor joining dendrogram was generated (Fig. 1). All cultivars were clustered into three major groups (G-I, G-II and G-III). In each group, the Moroccan and exotic cultivars were mainly separated into subgroups. However, Mexican cultivars Pavon-76, Veery and Opata-85 and an Australian cultivar Annuello were grouped with Moroccan cultivars. The most divergent pair was Chinese Spring and Moroccan cultivars Jouda, Mehdia and Saïs, which exhibited highest genetic distance (0.74). The two Moroccan cultivars Aguilal and Saïs were genetically close (0.11). Similarly, genetic distance between exotic cultivars Frontana and Veranopolis were smallest (0.11).
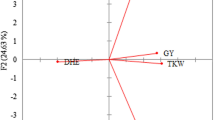
The genetic structure was analyzed using principal coordinates analysis (PCoA). The PCoA of genetic distance between genotypes, based on gene frequencies revealed differentiation between cultivars. The three axes explained 16.38, 13.39 and 9.61 % of the total variance, and separated the cultivars into two clusters, Moroccan cultivars in one cluster and exotic cultivars in another cluster (Fig. 2), except, the exotic Mexican cultivars Pavon-76, Veery and Bobwhite and the American cultivars Yecora Rojo-Gpc-B1/Yr36 were grouped with the Moroccan cluster.
Discussion
The Moroccan wheat cultivars used in this study represent the most advanced breeding lines released for cultivation in Morocco and encompass important gene pools adapted to Morocco and the North Africa region. Therefore, information of genetic diversity, identification of specific alleles, genes or loci and assessment of the genetic relationships among these cultivars can provide relevant guidelines in selecting parents and for designing new breeding strategies for wheat cultivar improvement, especially, against leaf rust, yellow rust and Hessian fly, which are considered as most destructive biotic stresses in Morocco (Elhaddoury et al. 2012). Lombardi et al. (2014) reported that selection of divergent parental genotypes for breeding should be made actively on the basis of systematic assessment of genetic distance between genotypes, rather than passively based on geographical distance.
The total number of alleles detected at 21 loci was 48 alleles in Moroccan cultivars (mean 2.5 alleles) and 56 alleles in the exotic cultivars (mean 2.9 alleles). The PIC value was 0.34 for Moroccan cultivars and 0.39 for exotic cultivars. Similar studies have been conducted by Vanzetti et al. (2013) for 102 Argentinean bread wheat cultivars and reported an average number of alleles and PIC values of 3.26 and 0.458, respectively. In India, Malik et al. (2013) characterized 48 elite Indian wheat genotypes and reported 2.42 alleles per locus and 0.4596 PIC value.
The functional markers and the random DNA markers linked to the target traits such as the rust resistance (Lr34, Lr68, Lr37, Yr36 and Sr24), Hessian fly resistance (H22), 1BL/1RS translocation, growth photoperiod sensitivity (Ppd-D1), plant height (Rht-B1, Rht-D1), grain texture (Pina-D1), starch waxy proteins variants (Wx-A1, Wx-B1), PPO activity (Ppo-A1, Ppo-D1), pre-harvest sprouting tolerance (Vp1-B3), high molecular weight glutenins (Glu-A1, Glu-B1, Glu-D1) and low molecular weight glutenins (Glu-A3, Glu-B3, Glu-D3) shown to be ideal for marker-assisted selection in wheat breeding. The information generated in this study is also useful for selection of parental materials to develop segregating population for marker-assisted selection. The use of gene specific markers permitted to know the genetic structure of Moroccan modern wheat cultivars. The functional alleles of some of these traits were very well related to the respective phenotypes of the cultivars, previously described by the breeders. For instance, the cultivars Arrehane and Aguilal known for their resistance to Hessian fly (Jlibene and Nsarellah 2011), and carrying the H22 gene (Lhaloui et al. 2000), were clearly amplified the allele of the marker Xgdm33 tightly linked to H22 (Zhao et al. 2006). In addition, Arrehane showed the presence of durable resistance gene Lr34/Yr18/Pm38 and dwarfing gene allele Rht-B1b. Other cultivars namely Baraka, Acsad-59, Jouda and Mehdia which were positive for Lr34/Yr18/Pm38 and Rht-B1b dwarfing gene allele are also known for their large adaptation, high yield and tolerance to drought (Jlibene and Nsarellah 2011). However, these cultivars need to be further improved by incorporating the Hessian fly resistance, which is very important problem in arid and semi-arid regions of Morocco and the North Africa. Based on the marker analysis, the cultivars Saada and Massira with resistance to the Hessian fly (H5 gene; Lhaloui et al. 2000) were also found to be carrying Lr34/Yr18/Pm38 slow rusting gene. The linked random DNA analysis also revealed the possibilities of having the second slow rusting gene Lr68 in Saada and Massira, which needs to be further confirmed based on the phenotypic characterization. These two cultivars with two slow rust resistance genes could be a valuable parent in wheat breeding program due the additive resistance effect resulted from combined slow rusting genes (Lillemo et al. 2011). Furthermore, the analysis in this study also revealed that the cultivar Saada also carried photoperiod insensitive allele Ppd-D1a (Yang et al. 2009) and waxy locus allele wx-B1 associated with improved starch quality (McLauchlan et al. 2001). Therefore, Saada is very valuable cultivar for use as donors in molecular breeding program. The cultivars Tilila and Mehdia revealed the presence of iag95 marker specific for 1BL.1RS translocation. Tilila showed also presence of waxy allele wx-A1 and wx-B1, low polyphenol oxidase activity alleles (Ppo-D1a and Ppo-A1b). This cultivar (Tilila) is known in Morocco for its large adaptation, moderate yield and resistance against many diseases (Jlibene 1996).
Estimation of the degree of differentiation between cultivars that are included in a crossing program is useful for selection of parental genotypes. The Mexican cultivars Pavon-76, Bobwhite and Veery were genetically closer to Moroccan cultivars. Based on the knowledge of pedigrees of exotic and Moroccan cultivars and the history of Moroccan breeding, it is known that Mexican cultivars and CIMMYT germplasm were extensively used in Morocco since 1980s (Jlibene and Nsarellah 2011). Most of the Moroccan cultivars had Pavon’s and Veery or their common parents such as Bluebirds and Kalyansona as parents (Skovmand et al. 1997). The NJ dendrogram and PCoA results revealed a clear differentiation between Moroccan and the exotic cultivars deployed in the current breeding program indicating that the exotic cultivars used in this study, were divergent from Moroccan cultivars and can be used to improve disease resistance, quality and also genetic diversity.
References
Ahmad M (2000) Molecular marker-assisted selection of HMW glutenin alleles related to wheat bread quality by PCR-generated DNA markers. Theor Appl Genet 101:892–896. doi:10.1007/s001220051558
Andersen JR, Lubberstedt T (2003) Functional markers in plants. Trends Plant Sci 8:554–560. doi:10.1007/s10529-013-1377-1
Barrett BA, Kidwell KK (1998) AFLP-based genetic diversity assessment among wheat cultivar from the Pacific Northwest. Crop Sci 38:1261–1271. doi:10.2135/cropsci1998.0011183X003800050025x
Beales J, Turner A, Griffiths S, Snape JW, Laurie DA (2007) Pseudo-Response Regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115:721–733. doi:10.1007/s00122-007-0603-4
Bered F, Barbosa-Neto JF, Carvalho FIF (2002) Genetic variability in common wheat germplasm based on coefficients of parentage. Genet Mol Biol 25:211–215. doi:10.1590/S1415-47572002000200015
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Butow BJ, Gale KR, Ikea J, Juhasz A, Bedo Z, Tamas L, Gianibelli MC (2004) Dissemination of the highly expressed Bx7 glutenin subunit (Glu-B1al allele) in wheat as revealed by novel PCR markers and HPLC. Theor Appl Genet 109:1525–1535. doi:10.1007/s00122-004-1776-8
Chai JF, Zhou RH, Jia JZ, Liu X (2006) Development and application of a new codominant PCR marker for detecting 1BL1RS wheat–rye chromosome translocations. Plant Breed 125:302–304. doi:10.1111/j.1439-0523.2006.01186.x
Chao S, Zhang W, Dubcovsky J, Sorrells M (2007) Evaluation of genetic diversity and genome-wide linkage disequilibrium among U.S. wheat (Triticum aestivum L.) germplasm representing different market class. Crop Sci 47:1018–1030. doi:10.2135/cropsci2006.06.0434
De Bustos A, Rubio P, Jouve N (2000) Molecular characterization of the inactive allele of the gene Glu-a1 and the development of a set of AS-PCR markers for HMW glutenins of wheat. Theor Appl Genet 100:1085–1094. doi:10.1007/s001220051390
Distelfeld A, Uauy C, Fahima T, Dubcovsky J (2006) Physical map of the wheat high-grain protein content gene Gpc-B1 and development of a high-throughput molecular marker. New Phytol 169:753–763. doi:10.1111/j.1469-8137.2005.01627.x
Elhaddoury J, Lhaloui S, Udupa SM, Moatassim B, Taiq R, Rabeh M, Kamlaoui M, Hammadi M (2012) Registration of ‘Kharoba’: a bread wheat cultivar developed through doubled haploid breeding. J Plant Regist 6:1–5. doi:10.3198/jpr2011.07.0385crc
Ellis MH, Spielmeyer W, Gale KR, Rebetzke GJ, Richards RA (2002) ‘‘Perfect’’ markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theor Appl Genet 105:1038–1042. doi:10.1007/s00122-002-1048-4
FAOSTAT (2014) http://faostat.fao.org. Accessed July 9 2015
Feuillet C, Travella S, Stein N, Albar L, Nublat A, Keller B (2003) Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc Natl Acad Sci USA 100:15253–15258. doi:10.1073/pnas.2435133100
Froidmont DD (1998) A co-dominant marker for the 1BL 1RS wheat rye translocation via multiplex PCR. J Cereal Sci 27:229–232. doi:10.1006/jcrs.1998.0194
Fu DL, Uauy C, Distelfeld A, Blechl A, Epstein L, Chen XM, Sela H, Fahima T, Dubcovsky J (2009) A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323:1357–1360. doi:10.1126/science.1166289
Gautier M-F, Aleman M-E, Guirao A, Marion D, Joudrier P (1994) Triticum aestivum puroindolines, two basic cysteine-rich seed proteins: CDNA sequence analysis and developmental gene expression. Plant Mol Biol 24:43–47. doi:10.1007/BF00024197
Hayden MJ, Tabone TL, Nguyen TM, Coventry S, Keiper FJ, Fox RL, Chalmers KJ, Mather DE, Eglinton JA (2010) An informative set of SNP markers for molecular characterization of Australian barley germplasm. Crop Past Sci 61:70–83. doi:10.1071/CP09140
He XY, He ZH, Zhang LP, Sun DJ, Morris CF, Fuerst EP, Xia XC (2007) Allelic variation of polyphenol oxidase (PPO) genes located on chromosomes 2A and 2D and development of functional markers for the PPO genes in common wheat. Theor Appl Genet 115:47–58. doi:10.1007/s00122-007-0539-8
Helguera M, Khan IA, Kolmer J, Lijavetzky D, Zhong-Qi L, Dubcovsky J (2003) PCR assays for the Lr37-Yr17-Sr38 cluster of rust resistance genes and their use to develop isogenic hard red spring wheat lines. Crop Sci 43:1839–1847. doi:10.2135/cropsci2003.1839
Herrera-Foessel SA, Singh RP, Huerta-Espino J, Rosewarne GM, Periyannan SK, Viccar L, Calvo-Salazar V, Lan C, Lagudah ES (2012) Lr68: a new gene conferring slow rusting resistance to leaf rust in wheat. Theor Appl Genet 124:1475–1486. doi:10.1007/s00122-012-1802-1
Huang L, Brooks SA, Li WL, Fellers JP, Trick HN, Gill BS (2003) Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics 164:655–664
Jin L, Chakraborty R (1993) Estimation of genetic distance and coefficient of gene diversity from single-probe multilocus DNA fingerprinting Data. Mol Biol Evol 11:120–127
Jlibene M (1996) Mise au point des lignées isogéniques de blé tendre avec différents niveaux de résistance aux rouilles et à la cécidomyie. In: Lyamani A, Farih A, El Yamani M (eds) B Ezzahiri. Symposium régional sur les maladies des céréales et des légumineuses alimentaires, Rabat, pp 257–264
Jlibene M, Nsarellah N (2011) Wheat breeding in Morocco, a historical perspective. In: Angus WJ, Bonjean A, van Ginkel M (eds) The world wheat book, vol 2., A history of wheat breedingLavoisier Publishing, Paris, pp 425–442
Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323:1360–1363. doi:10.1126/science.1166453
Lafiandra D, Tucci GF, Pavoni A, Turchetta T, Margiotta B (1997) PCR analysis of x- and y-type genes present at the complex Glu-A1 locus in durum and bread wheat. Theor Appl Genet 94:235–240. doi:10.1007/s001220050405
Lagudah ES, McFadden H, Singh RP, Huerta-Espino J, Bariana HS, Spielmeyer W (2006) Molecular genetic characterization of the Lr34/Yr18 slow rusting resistance gene region in wheat. Theor Appl Genet 114:21–30. doi:10.1007/s00122-006-0406-z
Lei ZS, Gale KR, He ZH, Gianibelli C, Larroque O, Xia XC, Butow BJ, Ma W (2006) Y-type gene specific markers for enhanced discrimination of high-molecular weight glutenin alleles at the Glu-B1 locus in hexaploid wheat. J Cereal Sci 43:94–101. doi:10.1016/j.jcs.2005.08.003
Lhaloui S, El Bouhssini M, Nsarellah N, Nachit MM, Amri A (2000) Biotic stress limiting durum wheat production in Morocco-Hessian fly and the Russian wheat aphid: Surveys, loss assessment and identification of sources of resistance. In: Royo C, Nachit M, Di Fonzo N, Araus JL (eds) Durum wheat improvement in the Mediterranean region: new challenges. CIHEAM Options Méditerranéennes : Série A. Séminaires Méditerranéens, p 373–379
Lillemo M, Singh RP, William M, Herrera-Foessel SA, Huerta-Espino J, Germán S, Campos P, Chaves M, Madriaga R, Xia X, Liang S, Liu D, Li Z, Lagudah E (2011) Multiple rust resistance and gene additivity in wheat: lessons from multi-location case studies in the cultivars Parula and Saar. In: McIntosh RA (ed) BGRI 2011 Technical Workshop, St. Paul, Minnesota, USA, p 111–120
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129. doi:10.1093/bioinformatics/bti282
Liu C, Yang ZJ, Li GR, Zeng ZX, Zhang Y, Zhou JP, Liu ZH, Ren ZL (2008) Isolation of a new repetitive DNA sequence from Secale africanum enables targeting of Secale chromatin in wheat background. Euphytica 159:249–258. doi:10.1007/s10681-007-9484-5
Liu YN, He ZH, Appels R, Xia XC (2012) Functional markers in wheat: current status and future prospects. Theor Appl Genet 125:1–10. doi:10.1007/s00122-012-1829-3
Liu W, Frick M, Huel R, Nykiforuk CL, Wang X, Gaudet DA, Eudes F, Conner RL, Kuzyk A, Chen Q, Kang Z, Laroche A (2014) The stripe rust resistance gene Yr10 encodes an evolutionary-conserved and unique CC–NBS–LRR sequence in wheat. Mol Plant 7:1740–1755. doi:10.1093/mp/ssu112
Lombardi M, Materne M, Cogan NOI, Rodda M, Daetwyler HD, Slater AT, Forster JW, Kaur S (2014) Assessment of genetic variation within a global collection of lentil (Lens culinaris Medik.) cultivars and landraces using SNP markers. BMC Genet 15:150. doi:10.1186/s12863-014-0150-3
Lübberstedt T, Zein I, Andersen JR, Wenzel G, Krützfeldt B, Eder J, Ouzunova M, Chun S (2005) Development and application of functional markers in maize. Euphytica 146:101–108. doi:10.1007/s10681-005-0892-0
Mago R, Spielmeyer W, Lawrence GJ, Lagudah ES, Ellis JG, Pryor A (2002) Identification and mapping of molecular markers linked to rust resistance genes located on chromosome 1RS of rye using wheat-rye translocation lines. Theor Appl Genet 104:1317–1324. doi:10.1007/s00122-002-0879-3
Mago R, Bariana HS, Dundas IS, Spielmeyer W, Lawrence GJ, Pryor AJ, Ellis JG (2005) Development of PCR markers for the selection of wheat stem rust resistance genes Sr24 and Sr26 in diverse wheat germplasm. Theor Appl Genet 111:496–504. doi:10.1007/s00122-005-2039-z
Malik R, Tiwari R, Arora A, Kumar P, Sheoran S, Sharma P, Singh R, Tiwari V, Sharma I (2013) Genotypic characterization of elite Indian wheat genotypes using molecular markers and their pedigree analysis. Aust J Crop Sci 7:561–567
McLauchlan A, Ogbonnaya FC, Hollingsworth B, Carter M, Gale KR, Henry RJ, Holton TA, Morell MK, Rampling LR, Sharp PJ, Shariflou MR, Jones MGK, Appels R (2001) Development of robust PCR-based DNA markers for each homoeo-allele of granule-bound starch synthase and their application in wheat breeding programs. Aust J Agr Res 52:1409–1416. doi:10.1071/AR01036
Metakovsky EV, Branlard G (1998) Genetic diversity of French common wheat germplasm based on gliadin alleles. Theor Appl Genet 96:209–218. doi:10.1007/s001220050729
Mohan M, Suresh N, Bhagwat A, Krishna TG, Masahiro Y, Bhatia CR, Sasaki T (1997) Genome mapping, molecular markers and marker-assisted selection in crop plants. Mol Breed 3:87–103. doi:10.1023/A:1009651919792
Pagnotta M, Mondini L, Atallah M (2005) Morphological and molecular characterization of Italian emmer wheat accessions. Euphytica 146:29–37. doi:10.1007/s10681-005-8607-0
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539. doi:10.1093/bioinformatics/bts460
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81:8014–8018
Schut JW, Oi X, Stam P (1997) Association between relationship measures based on AFLP markers, pedigree data and morphological traits in barley. Theor Appl Genet 95:1161–1168. doi:10.1007/s001220050677
Skovmand B, Villareal R, van Ginkel M, Rajaram S, Ortiz-Ferrara G (1997) Semi dwarf bread wheat: names, parentages, pedigrees and origins. CIMMYT, Mexico. http://hdl.handle.net/10883/1372. Accessed 17 Aug 2015
Udupa SM, Robertson LD, Weigand F, Baum M, Kahl G (1999) Allelic variation at (TAA)n microsatellite loci in a world collection of chickpea (Cicer arietinum L.) Germplasm. Mol Genet Genom 261:354–363. doi:10.1007/s004380050976
USDA Foreign Agricultural Service (2014) Morocco: grain and feed update (10/22/2014). http://www.fas.usda.gov/data/morocco-grain-and-feed-update. Accessed 8 June 2015
Vanzetti LS, Yerkovich N, Chialvo E, Lombardo L, Vaschetto L, Helguera M (2013) Genetic structure of Argentinean hexaploid wheat germplasm. Genet Mol Biol 36:391–399. doi:10.1590/S1415-47572013000300014
Wang LH, Zhao XL, He ZH, Ma W, Appels R, Peña RJ, Xia XC (2009) Characterization of low-molecular-weight glutenin subunit Glu-B3 genes and development of STS markers in common wheat (Triticum aestivum L.). Theor Appl Genet 118:525–539. doi:10.1007/s00122-008-0918-9
Yahiaoui N, Srichumpa P, Dudler R, Keller B (2004) Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J 37:528–538. doi:10.1046/j.1365-313X.2003.01977.x
Yang Y, Zhao XL, Xia LQ, Chen XM, Xia XC, Yu Z, He Z, Röder MS (2007) Development and validation of viviparous-1 STS marker for pre-harvest sprouting tolerance in Chinese wheats. Theor Appl Genet 115:971–980. doi:10.1007/s00122-007-0624-z
Yang FP, Zhang XK, Xia XC, Laurie DA, Yang WX, He ZH (2009) Distribution of the photoperiod insensitive Ppd-D1a allele in Chinese wheat cultivars. Euphytica 165:445–452. doi:10.1007/s10681-008-9745-y
Zhang W, Gianibelli MC, Rampling LR, Gale KR (2004) Characterisation and marker development for low molecular weight glutenin genes from Glu-A3 alleles of bread wheat (Triticum aestivum L.). Theor Appl Genet 108:1409–1419. doi:10.1007/s00122-003-1558-8
Zhao HX, Liu XM, Chen MS (2006) H22, a major resistance gene to the Hessian fly (Mayetiola destructor), is mapped to the distal region of wheat chromosome 1DS. Theor Appl Genet 113:1491–1496. doi:10.1007/s00122-006-0396-x
Zhao XL, Xia XC, He ZH, Lei ZS, Appels R, Yang Y, Sun QX, Ma W (2007) Novel DNA variations to characterize low molecular weight glutenin Glu-D3 genes and develop STS markers in common wheat. Theor Appl Genet 114:451–460. doi:10.1007/s00122-006-0445-5
Acknowledgments
Authors are grateful to the International Treaty for Plant Genetic Resources for Food and Agriculture/FAO, the European Union, the CRP-Wheat and ICARDA/Morocco Collaborative Grants Program for the financial support. The views expressed herein can in no way be taken to reflect the official opinion of the European Union.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The experiment complies with the current laws of Morocco in which it was performed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Henkrar, F., El-Haddoury, J., Ouabbou, H. et al. Genetic characterization of Moroccan and the exotic bread wheat cultivars using functional and random DNA markers linked to the agronomic traits for genomics-assisted improvement. 3 Biotech 6, 97 (2016). https://doi.org/10.1007/s13205-016-0413-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-016-0413-y