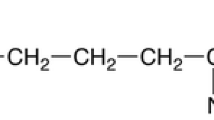Abstract
ε-Poly-l-lysine (ε-PL) has been widely used as food additive. However, the self-inhibition of ε-PL on cell growth limits the accumulation of ε-PL in the wild-type strain. Here, we screened ε-PL-tolerant strain of Streptomyces sp. with higher ε-PL productivity by genome shuffling and studied the mechanism for the improvement. The initial mutant library was constructed by diethyl sulfate mutagenesis. After four rounds of protoplast fusion, a shuffled strain F4-22 with 3.11 g/L ε-PL productivity in shake flask, 1.81-fold in comparison with that of parent strain, was obtained. The higher aspartokinase activity was induced in F4-22 whereas no obvious changes have been found in ε-PL synthetic and degrading enzymes which indicated that the upstream reregulation of the precursor lysine synthesis rather than lysine polymerization or ε-PL degradation in shuffled strain accounted for the higher productivity. The activities of key enzymes in the central metabolic pathway were also enhanced in F4-22 which resulted in increased vigor of the strain and in delayed strain lysis during fermentation. These improved properties of shuffled strain led to the success of combining general two-stage fermentation into one-stage one in 5-L bioreactor with 32.7 % more ε-PL production than that of parent strain. The strategy used in this study provided a novel strain breeding approach of producers which suffered from ε-PL-like self-inhibition of the metabolites.
Similar content being viewed by others
Introduction
ε-Poly-l-lysine (ε-PL) is a microbial metabolite of homo-poly-amino acid where lysine is polymerized between ε-amino and α-carboxyl groups by ε-PL synthetase (PLS) [1]. Its discovery can be traced back to 1977 when Shima and Sakai isolated an alkaloid secretion strain of Streptomyces albulus No. 346 [2]. The advantages of ε-PL over the chemical synthetic homo-poly-lysine (α-PL) have been noted mainly in its broad spectrum of antimicrobial activity, non-toxicity, and biodegradability [3–5]. Nowadays, several countries including Japan, United States, China and Korea have approved the application of ε-PL as food preservative, and the demand for ε-PL has been increasing.
However, the productivity of ε-PL in its microbial producers, such as Streptomyces sp. [6, 7], Kitasatospora sp. [8] and Bacillus subtilis [9], is low. The metabolic flux towards ε-PL is strictly regulated and ε-PL-degrading enzymes (PLDs) are able to degrade ε-PL rapidly to provide self-protection of strain [10, 11]. Efforts have been made to relieve the inhibition of the key enzyme in the ε-PL synthetic pathway by selection of S-(2-aminoethyl)-l-cysteine-resistant mutant, immobilized cell, in situ ε-PL removal [12–14]. Since the self-inhibition on cell growth limits the accumulation of ε-PL in the wild-type strain, it is reasonable to speculate that higher ε-PL production would be expected in ε-PL-tolerant mutant which could survive under high ε-PL concentration. However, no reports have been found to our knowledge to directly relate the ε-PL productivity and ε-PL tolerance and its mechanism remains to be uncovered.
Mutant strains can be obtained with classical mutagenesis which is usually laborious and time-consuming, or with genetic modification which requires the knowledge of genetic information of the concerned strain. In 2002, Zhang et al. [15] proposed an efficient technology of genome shuffling, a recursive protoplast fusion, for rapid evolution of strains toward desirable phenotypes without genome sequence information. Up to now, genome shuffling has been applied to strain breeding in the aspects of stress tolerance [16], phenotype and product yield improvements [17–19]. In this study, we made our effects to select ε-PL-tolerant Streptomyces sp. with increased ε-PL productivity by genome shuffling. Properties in fermentation behavior and ε-PL synthetic pathway between the parent strain and shuffled one were compared to improve the ε-PL fermentation level and to further understand the mechanism of cell growth and ε-PL synthesis regulation.
Materials and methods
Microorganism
Streptomyces sp. M-Z18, isolated from soil as described by Nishikawa and Ogawa [20] and followed by ultraviolet (UV) and nitrosoguanidine mutagenesis as described by Hiraki et al. [12], was used as the parent strain and cryopreserved in 30 % (v/v) glycerol at −80 °C. It was unable to grow on the agar plates containing more than 0.42 g/L ε-PL and its production of ε-PL in shake flask was 1.72 ± 0.03 g/L.
Media and culture conditions
Slant and plate media (BTN) contained (per liter): 10 g glucose, 1 g yeast extract, 2 g peptone, and 20 g agar. The pH value was adjusted to 7.5 with NaOH (1 N) before sterilization.
Seed medium contained (per liter): 50 g glucose, 10 g (NH4)2SO4, 5 g yeast extract, 1.72 g K2HPO4·2H2O, 0.8 g KH2PO4, 0.5 g MgSO4·7H2O, 0.04 g ZnSO4·7H2O, and 0.03 g FeSO4·7H2O. The pH value was adjusted to 6.8 with NaOH (1 N) before sterilization.
Fermentation medium contained (per liter): 60 g glycerol, 10 g (NH4)2SO4, 10 g beef extract, 4 g KH2PO4, 0.8 g MgSO4·7H2O, and 0.05 g FeSO4·7H2O. The pH value was adjusted to 6.8 with NaOH (1 N) before sterilization.
Regeneration medium (RM) consisted (per liter): 103 g sucrose, 15 g glucose, 4 g peptone, 10 g MgCl2·6H2O, 3 g yeast extract, 0.4 g CaCl2·7H2O, 0.25 g KH2PO4, 2 mL trace element solution [21], and 10 mL TES buffer (0.8 g/L Tris, 0.25 g/L EDTA, 0.05 g/L SDS, pH 7.2 adjusted with HCl). The pH value was adjusted to 7.0 with NaOH (1 N) before sterilization.
Both slant and plate cultures were incubated for 8–12 days at the temperature of 30 °C. Two loops full of spores were inoculated in 40 mL seed medium in a 250-mL shake flask. After incubation at 30 °C for 24 h in a rotary shaker at 200 rpm, a 3 mL portion of the seed culture was transferred into 40 mL of fermentation medium in a 250-mL flask. After 72 h of incubation, concentration of ε-PL was measured.
Mutagenesis and selection
Streptomyces sp. M-Z18 was treated with diethyl sulfate (DES) to obtain the starting mutant library as follows: spores (1 × 108–5 × 108) were treated with 2 % (v/v) DES for 30 min at 30 °C. Na2S2O3 was added to stop the reaction. Spores were then diluted and spread onto the BTN agar plates containing different concentrations of ε-PL. The fast-grown colonies were picked out for ε-PL production analysis in shake flask and those with higher ε-PL production were used as the starting strains for genome shuffling (Fig. 1).
Genome shuffling
The preparation and fusion of protoplasts were performed as described by Li et al. [22] with modification. Two loops full of spores of the above starting strains were inoculated 40 mL of seed medium in 250-mL shake flask, respectively. After incubation, mycelia were harvested by centrifugation at 4000×g for 5 min at 4 °C, washed twice with 10 mL of PB buffer (20 mM Tris–HCl buffer, pH 7.5, 103 g/L sucrose, 2 g/L MgCl2) and treated with 5 mg/mL lysozyme for 2 h at 30 °C. After observation of protoplasts formation under a compound light microscope (DM 1000, Leica Microsystems, Wetzlar, Germany), protoplasts from different populations were mixed and then divided equally into two parts. One part was inactivated with UV irradiation for 90 min at a distance of 15 cm from a UV lamp with wavelength of 254 nm and power of 15 W, and the other was heat treated at 70 °C for 50 min. The survival frequency of protoplasts in these two parts was zero. Subsequently, the inactivated protoplasts were mixed in a cell ratio of 1:1, centrifuged at 4000×g for 1 min at 4 °C, and resuspended in 1 mL PB buffer. The protoplast was fused in 5 mL of PB buffer, containing 40 % PEG 6000 and 10 mM CaCl2 for 15 min at 37 °C. After regeneration in liquid RM containing 0.66 g/L ε-PL, the fusant solution was divided equally into two parts, one part was used to obtain colonies on solid RM agar at 28 °C for 7–10 days for further ε-PL productivity assay, the other part was subcultured in liquid RM containing increased 1 g/L ε-PL at 30 °C on a rotary shaker at 100 rpm for 6–8 days. The mycelia collected were used for subsequent rounds of genome shuffling. Four successive rounds of protoplast fusion were performed by the same methods with the increased ε-PL concentration of 2, 4, and 6 g/L in the second, third, and fourth round, respectively (Fig. 1).
ε-PL fermentation of the shuffled strain and parent strain in shake flask
Two loops full of slant cultures of the shuffled strain with the highest ε-PL productivity and the parent strain Streptomyces sp. M-Z18 were transferred into a 250-mL shake flask containing 40 mL seed medium, respectively. After incubation at 30 °C for 24 h on a rotary shaker at 200 rpm, a 3 mL portion of the seed culture was inoculated into 40 mL of fermentation medium in a 250-mL shake flask. During the fermentation, mycelium of each strain was collected for biomass and enzymatic activity analysis.
ε-PL fermentation of the shuffled strain in 5-L fermenter
Two loops full of slant culture of the shuffled strain with the highest ε-PL yield were inoculated into 80 mL of seed medium in a 500-mL shake flask. After incubated at 30 °C and 200 rpm for 24 h, 240 mL of seed culture was transferred into 3.26-L sterilized fermentation medium in a 5-L fermenter (Baoxing Corp., Shanghai, China) with a working volume of 3.5 L at an initial pH of 6.8. 50 % (v/v) NH4OH was supplemented automatically to adjust pH at the desired levels. A sterilized pure glycerol solution was pulsed fed by peristaltic pump to maintain the residual glycerol at about 10 g/L and a sterilized 400 g/L (NH4)2SO4 solution was added to fermentation culture with the same manner of glycerol to maintain the residual NH4 +–N at about 1 g/L in fed-batch fermentation.
Analytical method
The concentration of glycerol was determined by high-performance liquid chromatography (HPLC) as described by Chen et al. [23]. NH4 +–N was analyzed by a colorimetric method. The sample solution was mixed with solution A (0.4 g/L sodium nitroprusside, 35 g/L phenol) and solution B (3.0 g/L NaClO, 18 g/L NaOH) in a ratio of 1:5:5. After reaction for 35 min at 37 °C, absorbance of the resulting solution was measured at 625 nm. The NH4 +–N content was calculated according to the standard curve of ammonium sulfate. The ε-PL concentration was determined according to the procedure described by Kahar et al. [24]. The biomass was determined as dry cell weight (DCW) by filtering culture sample, washing the mycelia twice with distilled water, and drying at 105 °C until constant weight. Enzyme activities of glucose-6-phosphate dehydrogenase (G6PDH), citrate synthetase (CS), aspartokinase (ASK), PLS of the shuffled and parent strains were measured according to Zeng et al. [25] and PLD was measured according to Kito et al. [11].
Results and discussion
Screening of starting strains for genome shuffling
A diverse mutant population with desired phenotype is the starting point for the genome shuffling which imitates the natural evolution process by genetic recombination through recursive protoplast fusion [15]. Since the self-inhibition limits ε-PL accumulation, the target phenotype here was the ε-PL-tolerant mutant with higher ε-PL. DES was used as mutagen and increasing concentration of ε-PL was used as selective pressure (Fig. 1). Since Streptomyces sp. M-Z18 cannot grow on agar plates containing ε-PL more than 0.42 g/L, the initial concentration for mutant selection was determined at 0.48 g/L ε-PL. After three rounds of mutagenesis and selection, the concentration of ε-PL reached 0.6 g/L and four mutants (D3-81, D3-90, D3-239, D3-347) were isolated. The ε-PL productivity of these four mutants in shake flask from triplicate experiments was 2.05 ± 0.04, 2.02 ± 0.02, 2.06 ± 0.02, 1.98 ± 0.04 g/L, respectively. However, when selection of mutants after fourth DES treatment on agar plates containing 0.66 g/L ε-PL was performed, no colony appeared indicating the limitation of mutagenesis on obtaining higher ε-PL-tolerant mutants. Further genome shuffling is necessary and the mutants D3-81, D3-90, D3-239, D3-347 were used as starting strains.
Genome shuffling to improve ε-PL tolerance
Genome shuffling allows the genetic exchange on entire genome level through the successive protoplast fusion of several starting strains. Product-tolerant strains such as ethanol-, lactic acid- and pristinamycin-tolerant stains have been successfully obtained by genome shuffling [16, 26, 27]. In this study, the protoplasts of starting strains were obtained successfully and the protoplast fusion was observed under microscope according to the established genome shuffling methods in our laboratory [22]. However, it takes a long time, 13–15 days, for the fusants to regenerate on the agar plate. Therefore, the regeneration was modified according to the method of Imada et al. [28] prior to the next round of shuffling. As a result, the regeneration time was shortened to 7–9 days. After the first, second, third and fourth round of genome shuffling, 152, 111, 59 and 28 colonies appeared on the agar plates containing 1, 2, 4, and 6 g/L ε-PL, respectively. The ε-PL tolerant ability is much higher than that of the starting mutants. However, no fusant growth appeared when the ε-PL concentration increased to 8 g/L after the fifth round of genome shuffling. As a control, the fusant of the first round of shuffling was subcultured with increased ε-PL 2, 4, and 6 g/L ε-PL to determine whether acclimatization effect could result in adaptive growth of ε-PL tolerance. In contrast to the shuffled strains, no colonies appeared under the same culture condition.
Improvement of ε-PL production in ε-PL-tolerant shuffled strains
During the four rounds of genome shuffling, the colonies formed on solid RM plate were picked out for ε-PL production assay. It can be seen that the ε-PL production increased gradually after each round of genome shuffling (Fig. 2) and the highest production was found using shuffled strain F4-22 obtained in the fourth round of shuffling (Fig. 2). The productivity of F4-22 was 3.11 ± 0.03 g/L in shake flask fermentation, 50.97 and 80.81 % higher than that of the best DES mutant (D-239) and the parent strain M-Z18, respectively. In addition, with the increasing of ε-PL during the successive shuffling, less resistant colonies appeared, however, the ratio of the shuffled strains with higher productivity increased in the population (Table 1). In general, random genome shuffling makes the screening laborious. An effective selection method can help to obtain the desired phenotype conveniently. In this study, the ε-PL was used as selective pressure for obtaining ε-PL-tolerant strain with high productivity efficiently.
The genetic instability leads to the retrogression of the high-producing mutants that originated from various treatments of mutation or recombination. To investigate the genetic stability of high ε-PL-producing shuffled strain, F4-22 was subcultured for successive five generations. The level of ε-PL production ranged from 2.98 ± 0.03 to 3.17 ± 0.02 g/L indicating the genetic stability of F4-22.
Comparison of enzymatic activities and cell vigor between M-Z18 and F4-22
To reveal the mechanisms behind the significant improvement of ε-PL productivity and resistance of F4-22, the enzyme activities of G6PDH, CS, ASK, PLS and PLD, which are supposed to be the key enzymes in glycolytic and ε-PL synthetic pathway [25], were monitored at 16, 24, 28, 32, 48 and 56 h in a 250-mL shake flask, respectively (Fig. 3). In the meantime, the morphologies of these two strains were observed during the fermentation (Fig. 4). It can be seen that the activities of G6PDH, CS were higher in F4-22 than those in M-Z18 during the investigated period. The highest activities of G6PDH, CS were found at 28 h in both strains, whereas in the ε-PL-tolerant F4-22 those activities were 36.4, 42.6 % higher than the parent strain M-Z18, respectively. The higher G6PDH and CS indicated that the pentose phosphate pathway and tricarboxylic acid (TCA) circle were enhanced in F4-22 which might lead to the increased vigor of ε-PL-tolerant strain. This was also verified by morphological changes of M-Z18 and F4-22 during fermentation (Fig. 4). As shown in Fig. 4, the mycelium pellet autolysis of M-Z18 emerged at 48 h and became obviously at 72 h. However, this phenomenon did not occur to F4-22 until 72 h.
ASK is the first key enzyme in lysine, a member of aspartate family amino acid, biosynthesis. As shown in Fig. 3, the activity of ASK in F4-22 significantly increased, 143 and 281 % higher than M-Z18 at 28 and 32 h, respectively, indicating there was a greater pool of precursor lysine for ε-PL biosynthesis. However, no obvious PLS changes have been found during investigated fermentation period (data not shown). Hamano et al. [29] reported that lack of lysine precursor rather than PLS ability limited the level of ε-PL in S. albulus. Our result was consistent with that of Hamano et al. [29]. The ε-PL degradation by PLD might be one reason for ε-PL tolerance. However, the activity of PLD in the two strains expressed no obvious difference (Fig. 3) and, therefore, the accumulation of ε-PL in F4-22 was reasonable. The above results indicated that genome shuffling resulted in high yield by reregulation of ε-PL biosynthetic key enzymes and increasing cell vigor in the ε-PL-tolerant strain. The antimicrobial activity of ε-PL is supposed to disrupt membrane integrity and to arouse oxidative stress by reactive oxygen species due to the expression of various genes [30]. Work is also underway to find out whether the changes in membrane structure or other genes’ expression are contributing to the ε-PL tolerance in our lab.
Fermentation by shuffled F4-22 in 5-L fermenter
To verify the good characters of shuffled strain F4-22, 5-L scale trial was carried out. Since pH is generally regarded as the most important factor on ε-PL fermentation, four different pH values (3.5, 3.8, 4.0, 4.5) were selected to investigate the performance of F4-22 according to our previous study with parent strain M-Z18 [23]. As shown in Fig. 5, the higher pH level, the more DCW and the more rapid glycerol consumption were discovered. The fermentation ended when the glycerol was completely consumed (Fig. 5). However, the ε-PL productivity was not positively related to pH value. At pH 4.5, the DCW was maximum (18.43 g/L), but ε-PL production was minimum (5.34 g/L) compared to other pH values (Fig. 5). These are in accordance with the findings of Kahar et al. [24] that the optimal pH value for cell growth was higher than 4.0 while the ε-PL production was low. Considering the parameters obtained above, pH 4.0 was selected for the highest ε-PL production, relatively high level of biomass formation and rapid glycerol consumption. The glycerol was added to extend the fermentation time and to obtain more ε-PL production. Good effect was found using this strategy. The strain F4-22 was able to grow well (37.74 g/L DCW) and produce 32.7 % more ε-PL (39.96 g/L) than that of M-Z18 (30.11 g/L) which was obtained under optimized two-stage culture [23] at the final fermentation time of 173 h (Fig. 6). Two-stage culture has been widely used in ε-PL fermentation due to the rapid pH decreasing at the initial stage of fermentation when ammonium sulfate was used as nitrogen source [23, 24, 31]. The low pH value inhibits the growth of Streptomyces and consequent ε-PL production. Therefore, generally in the first stage, pH value was maintained at higher than 5.0, and in the second stage, pH value was kept at about 4.0 according to the producer used for ε-PL synthesis. In this study, due to the vigor of cell and more metabolic flux directed to lysine synthesis as discussed above, one-stage culture was sufficient to obtain enough biomass and higher amount of ε-PL which made the fermentation control process simple.
Conclusion
In this study, an ε-PL-tolerant strain with higher productivity was obtained by genome shuffling. The reregulation of central metabolic and ε-PL synthetic pathways by enhancing activities of enzymes in G6PDH, CS and ASK contributed to good cell growth and higher ε-PL productivity under simplified fermentation process. The results suggested that selection of ε-PL-tolerant strain by genome shuffling facilitated the breeding of higher ε-PL-producing strain. The strategy used here might provide an effective approach for the improvements of strains whose metabolites exhibit self-inhibition due to their toxic effects. The mechanism of F4-22 with higher ε-PL production and tolerance than those of M-Z18 is under investigation, especially on aspect of genomic changes.
References
Shima S, Sakai H (1981) Poly-l-lysine produced by Streptomyces. Part II. Taxonomy and fermentation studies. Agric Biol Chem 45(11):2497–2502. doi:10.1080/00021369.1981.10864929
Shima S, Sakai H (1977) Polylysine produced by Streptomyces. Agric Biol Chem 41(9):1807–1809
Shima S, Matsuoka H, Iwamoto T, Sakai H (1984) Antimicrobial action of ε-poly-l-lysine. J Antibiot 37(11):1449–1455
Hiraki J, Ichikawa T, Ninomiya S, Seki H, Uohama K, Seki H, Kimura S, Yanagimoto Y, Barnett JW Jr (2003) Use of ADME studies to confirm the safety of ε-polylysine as a preservative in food. Regul Toxicol Pharmacol 37(2):328–340. doi:10.1016/S0273-2300(03)00029-1
Shih IL, Shen MH, Van YT (2006) Microbial synthesis of poly(ε-lysine) and its various applications. Bioresour Technol 97(9):1148–1159. doi:10.1016/j.biortech.2004.08.012
Hirohara H, Takehara M, Saimura M, Masayuki A, Miyamoto M (2006) Biosynthesis of poly(ε-l-lysine)s in two newly isolated strains of Streptomyces sp. Appl Microbiol Biotechnol 73(2):321–331. doi:10.1007/s00253-006-0479-2
Li S, Tang L, Chen X, Liao L, Li F, Mao Z (2011) Isolation and characterization of a novel ε-poly-l-lysine producing strain: Streptomyces griseofuscus. J Ind Microbiol Biotechnol 38(4):557–563. doi:10.1007/s10295-010-0803-9
Ouyang J, Xu H, Li S, Zhu H, Chen W, Zhou J, Wu Q, Xu L, Ouyang P (2006) Production of ε-poly-l-lysine by newly isolated Kitasatospora sp. PL6-3. Biotechnol J 1(12):1459–1463. doi:10.1002/biot.200600079
EI-Sersy NA, Abdelwahab AE, Abouelkhiir SS, Abou-Zeid DM, Sabry SA (2012) Antibacterial and anticancer activity of ε-poly-l-lysine (ε-PL) produced by a marine Bacillus subtilis sp. J Basic Microbiol 52(5):513–522. doi:10.1002/jobm.201100290
Hamano Y, Yoshida T, Kito M, Nakamori S, Nagasawa T, Takagi H (2006) Biological function of the pld gene product that degrades ε-poly-l-lysine in Streptomyces albulus. App Microbiol Biotechnol 72(1):173–181. doi:10.1007/s00253-006-0396-4
Kito M, Takimoto R, Yoshida T, Nagasawa T (2002) Purification and characterization of an ε-poly-l-lysine-degrading enzyme from an ε-poly-l-lysine-producing strain of Streptomyces albulus. Arch Microbiol 178(5):325–330. doi:10.1007/s00203-002-0459-6
Hiraki J, Hatakeyama M, Morita H, Izumi Y (1998) Improved ε-poly-l-lysine production of an S-(2-aminoethyl)-l-cysteine resistant mutant of Streptomyces albulus. Seibutsu Kogaku Kaishi 76(12):487–493
Zhang Y, Feng X, Xu H, Yao Z, Ouyang P (2010) ε-Poly-l-lysine production by immobilized cells of Kitasatospora sp. MY 5-36 in repeated fed-batch cultures. Bioresour Technol 101(14):5523–5527. doi:10.1016/j.biortech.2010.02.021
Liu S, Wu Q, Zhang J, Mo S (2011) Production of ε-poly-l-lysine by Streptomyces sp. using resin-based, in situ product removal. Biotechnol Lett 33(8):1581–1585. doi:10.1007/s10529-011-0616-6
Zhang YX, Perry K, Vinci VA, Powell K, Stemmer WP, del Cardayré SB (2002) Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415(6872):644–646. doi:10.1038/415644a
Shi DJ, Wang CL, Wang KM (2009) Genome shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 36(1):139–147. doi:10.1007/s10295-008-0481-z
Stephanopoulos G (2002) Metabolic engineering by genome shuffling. Nat Biotechnol 20(7):666–668. doi:10.1038/nbt0702-666
Zhang J, Wang X, Diao J, He H, Zhang Y, Xiang W (2013) Streptomycin resistance-aided genome shuffling to improve doramectin productivity of Streptomyces avermitilis NEAU1069. J Ind Microbiol Biotechnol 40(8):877–889. doi:10.1007/s10295-013-1280-8
Zhang P, Zhang K, Yan Q, Xu Y, Sun Z (2013) Enhanced succinic acid production by Actinobacillus succinogenes after genome shuffling. J Ind Microbiol Biotechnol 40(8):831–840. doi:10.1007/s10295-013-1283-5
Nishikawa M, Ogawa K (2002) Distribution of microbes producing antimicrobial ε-poly-l-lysine polymers in soil microflora determined by a novel method. Appl Environ Microbiol 68(7):3575–3581. doi:10.1128/AEM.68.7.3575-3581.2002
Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate DJ, Smith CP, Ward JM, Schrempf H (1985) Genetic manipulation of Streptomyces-A laboratory manual. The John Innes Foundation, Norwich, UK and Cold Spring Harbour Laboratory. pp 356
Li S, Li F, Chen XS, Wang L, Xu J, Tang L, Mao ZG (2012) Genome shuffling enhanced ε-poly-l-lysine production by improving glucose tolerance of Streptomyces graminearus. Appl Biochem Biotechnol 166(2):414–423. doi:10.1007/s12010-011-9437-2
Chen XS, Li S, Liao LJ, Ren XD, Li F, Tang L, Zhang JH, Mao ZG (2011) Production of ε-poly-l-lysine using a novel two-stage pH control strategy by Streptomyces sp. M-Z18 from glycerol. Bioprocess Biosyst Eng 34(5):561–567. doi:10.1007/s00449-010-0505-8
Kahar P, Iwata T, Hiraki J, Park EY, Okabe M (2001) Enhancement of ε-polylysine production by Streptomyces albulus strain 410 using pH control. J Biosci Bioeng 91(2):190–194. doi:10.1016/S1389-1723(01)80064-5
Zeng X, Chen XS, Ren XD, Liu QR, Wang L, Sun QX, Tang L, Mao ZG (2014) Insights into the role of glucose and glycerol as a mixed carbon source in the improvement of ε-poly-l-lysine productivity. Appl Biochem Biotechnol 173(8):2211–2224. doi:10.1007/s12010-014-1026-8
Wang Y, Li Y, Pei X, Yu L, Feng Y (2007) Genome-shuffling improved acid tolerance and l-lactic acid volumetric productivity in Lactobacillus rhamnosus. J Biotechnol 129(3):510–515. doi:10.1016/j.jbiotec.2007.01.011
Xu B, Jin Z, Wang H, Jin Q, Jin X, Cen P (2008) Evolution of Streptomyces pristinaespiralis for resistance and production of pristinamycin by genome shuffling. Appl Microbiol Biotechnol 80(2):261–267. doi:10.1007/s00253-008-1540-0
Imada C, Ikemoto Y, Kobayashi T, Hamada N, Watanabe E (2002) Isolation and characterization of the interspecific fusants from Streptomycetes obtained using a liquid regeneration method. Fish Sci 68(2):395–402. doi:10.1046/j.1444-2906.2002.00438.x
Hamano Y, Nicchu I, Shimizu T, Onji Y, Hiraki J, Takagi H (2007) ε-P-l-lysine producer, Streptomyces albulus, has feedback-inhibition resistant aspartokinase. Appl Micobiol Biotechnol 76(4):873–882. doi:10.1007/s00253-007-1052-3
Ye R, Xu H, Wan C, Peng S, Wang L, Xu H, Aguilar ZP, Xiong Y, Zeng Z, Wei H (2013) Antibacterial activity and mechanism of action of ε-poly-l-lysine. Biochem Biophys Res Commun 493(1):148–153. doi:10.1016/j.bbrc.2013.08.001
Shih IL, Shen MH (2006) Optimization of cell growth and poly(ε-lysine) production in batch and fed-batch cultures by Streptomyces albulus IFO 14147. Process Biochem 41(7):1644–1649. doi:10.1016/j.procbio.2006.03.013
Acknowledgments
This research was financially supported by the National Natural Science Foundation of China (Grant No. 21376106), the Program of Introducing Talents of Discipline to Universities (111-2-06).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhou, YP., Ren, XD., Wang, L. et al. Enhancement of ε-poly-lysine production in ε-poly-lysine-tolerant Streptomyces sp. by genome shuffling. Bioprocess Biosyst Eng 38, 1705–1713 (2015). https://doi.org/10.1007/s00449-015-1410-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-015-1410-y