Abstract
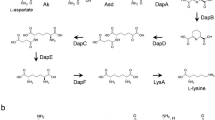
ε-Poly-l-lysine (ε-PL) is one of the few naturally occurring biopolymers and is characterized by a peptide bond between the α-carboxyl and ε-amino groups. Previously, we purified and characterized the ε-PL-degrading enzyme (Pld) from Streptomyces albulus, which is an ε-PL producer, and this enzyme was expected to confer self-resistance to the ε-PL produced by the organism itself. The gene encoding Pld was cloned based on the N-terminal amino acid sequence determined in this study, and a sequencing analysis revealed eight open reading frames (ORFs), i.e., ORF1 to ORF8 in the flanking region surrounding the pld gene (present in ORF5). To investigate the biological function of Pld, we constructed a knockout mutant in which the pld gene is inactivated. Studies on ε-PL susceptibility, ε-PL-degrading activity, and ε-PL productivity demonstrated that the pld gene does play a partial role in self-resistance and that S. albulus was found to produce other ε-PL-degrading enzyme(s) in addition to Pld. To the best of our knowledge, this is the first report on a self-resistance gene for a biopolymer possessing antibacterial activity.






Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Anzai H, Kumada Y, Hara O, Murakami T, Itoh R, Takano E, Imai S, Satoh A, Nagaoka K (1988) Replacement of Streptomyces hygroscopicus genomic segments with in vitro altered DNA sequences. J Antibiot (Tokyo) 41:226–233
Chater KF (1990) The improving prospects for yield increase by genetic engineering in antibiotic-producing Streptomycetes. Biotechnology (N Y) 8:115–121
Hamano Y, Nicchu I, Hoshino Y, Kawai T, Nakamori S, Takagi H (2005) Development of gene delivery systems for the ε-poly-l-lysine producer, Streptomyces albulus. J Biosci Bioeng 99:636–641
Hiraki J, Hatakeyama M, Morita H, Izumi Y (1998) Improved ε-poly-l-lysine production of an S-(2-aminoethyl)-l-cysteine resistant mutant of Streptomyces albulus. Seibutu Kougaku Kaishi 76:487–493
Itzhaki RF (1972) Colorimetric method for estimating polylysine and polyarginine. Anal Biochem 50:569–574
Kahar P, Iwata T, Hiraki J, Park EY, Okabe M (2001) Enhancement of ε-polylysine production by Streptomyces albulus strain 410 using pH control. J Biosci Bioeng 91:190–194
Kawai T, Kubota T, Hiraki J, Izumi Y (2003) Biosynthesis of ε-poly-l-lysine in a cell-free system of Streptomyces albulus. Biochem Biophys Res Commun 311:635–640
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. The John Innes Foundation, Norwich
Kito M, Takimoto R, Yoshida T, Nagasawa T (2002) Purification and characterization of an ε-poly-l-lysine-degrading enzyme from an ε-poly-l-lysine-producing strain of Streptomyces albulus. Arch Microbiol 178:325–330
Martin JF, Liras P (1989) Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu Rev Microbiol 43:173–206
Nilsson I, von Heijne G (1991) A de novo designed signal peptide cleavage cassette function in vivo. J Biol Chem 266:3408–3410
Nishikawa M, Ogawa K (2002) Distribution of microbes producing antimicrobial ε-poly-l-lysine polymers in soil microflora determined by a novel method. Appl Environ Microbiol 68:3575–3581
Oppermann Sanio FB, Steinbuchel A (2002) Occurrence, functions and biosynthesis of polyamides in microorganisms and biotechnological production. Naturwissenschaften 89:11–22
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Shima S, Sakai H (1977) Polylysine produced by Streptomyces. Agric Biol Chem 41:1807–1809
Shima S, Sakai H (1981a) Poly-l-lysine produced by Streptomyces. II. Taxonomy and fermentation studies. Agric Biol Chem 45:2497–2502
Shima S, Sakai H (1981b) Poly-l-lysine produced by Streptomyces. III. Chemical studies. Agric Biol Chem 45:2503–2508
Shima S, Matsuoka H, Sakai H (1982) Inactivation of bacteriopharges by ε-poly-l-lysine produced by Streptomyces. Agric Biol Chem 46:1917–1919
Shima S, Oshima S, Sakai H (1983) Biosynthesis of ε-poly-l-lysine by washed mycelium of Streptomyces albulus No. 346. Nippon Nogei Kagakukaishi 57:221–226
Shima S, Matsuoka H, Iwamoto T, Sakai H (1984) Antimicrobial action of ε-poly-l-lysine. J Antibiot (Tokyo) 37:1449–1455
Acknowledgement
This work was supported by a grant from the Chisso Corporation.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Hamano, Y., Yoshida, T., Kito, M. et al. Biological function of the pld gene product that degrades -poly-l-lysine in Streptomyces albulus . Appl Microbiol Biotechnol 72, 173–181 (2006). https://doi.org/10.1007/s00253-006-0396-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-006-0396-4