Abstract
Key message
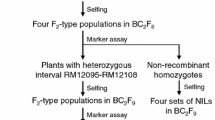
A major QTL for heading date, qHD5, was fine-mapped to a 52.59-kb region on the short arm of rice chromosome 5.
Abstract
Heading date (HD) is one of the most important traits that enables rice to adapt to seasonal differences and specific growth conditions in diverse growing regions. In this study, a major-effect quantitative trait locus (QTL), qHD5, was resolved as a single Medelian factor that causes NIL(BG1) and NIL(XLJ) (two near-isogenic lines (NILs) used in our study) to have at a minimum of 10-day difference in HD under both long-day and short-day conditions in rice. qHD5 was initially mapped to a 309.52-kb genomic region in our previous study. Here, using an advanced BC4F3 population and map-based cloning, we further narrowed the location of qHD5 to a 52.59-kb region between the H71 and RD502 markers. Sequence analysis revealed that Os05g03040, which putatively encodes an AP2 (APETALA2) transcription factor, has six single nucleotide polymorphisms (SNPs) between NIL(BG1) and NIL(XLJ). On this basis, this gene was concluded to be the most probable candidate gene for qHD5. Our results also showed that Hd3a, RFT1, Hd1, Ehd1, and Ghd7 were differentially expressed in the two NILs. Moreover, qHD5 was found to affect yield-related traits such as flag leaf width, flag leaf length, branch number, and 1000-grain weight.







Similar content being viewed by others
References
Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15:2730–2741
Bian XF, Liu X, Zhao ZG, Jiang L, Gao H, Zhang YH, Zheng M, Chen LM, Liu SJ, Zhai HQ, Wan JM (2011) Heading date gene, dth3 controlled late flowering in O. Glaberrima Steud. by down-regulating Ehd1. Plant Cell Rep 30:2243–2254
Borovsky Y, Sharma VK, Verbakel H, Paran I (2015) CaAP2 transcription factor is a candidate gene for a flowering repressor and a candidate for controlling natural variation of flowering time in Capsicum annuum. Theor Appl Genet 128:1073–1082
Chen JY, Guo L, Ma H, Chen YY, Zhang HW, Ying JZ, Zhuang JY (2014) Fine mapping of qHd1, a minor heading date QTL with pleiotropism for yield traits in rice (Oryza sativa L.). Theor Appl Genet 127:2515–2524
Chuck G, Meeley RB, Hake S (1998) The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev 12:1145–1154
Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J (2010) A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 64:936–947
De Belle I, Wu JX, Sperandio S, Mercola D, Adamson ED (2003) In vivo cloning and characterization of a new growth suppressor protein TOE1 as a direct target gene of Egr1. J Biol Chem 278:14306–14312
Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18:926–936
Endo-Higashi N, Izawa T (2011) Flowering time genes heading date 1 and early heading date 1 together control panicle development in rice. Plant Cell Physiol 52:1083–1094
Faris JD, Fellers JP, Brooks SA, Gill BS (2003) A bacterial artificial chromosome contig spanning the major domestication locus Q in wheat and identification of a candidate gene. Genetics 164:311–321
Fujino K, Yamanouchi U, Yano M (2013) Roles of the Hd5 gene controlling heading date for adaptation to the northern limits of rice cultivation. Theor Appl Genet 126:611–618
Gao H, Zheng XM, Fei G, Chen J, Jin M, Ren Y, Wu W, Zhou K, Sheng P, Zhou F, Jiang L, Wang J, Zhang X, Guo X, Wang JL, Cheng Z, Wu C, Wang H, Wan JM (2013) Ehd4 encodes a novel and Oryza-genus-specific regulator of photoperiodic flowering in rice. PLoS Genet 9:e1003281
Gao H, Jin M, Zheng X-M, Chen J, Yuan D, Xin Y, Wang M, Huang D, Zhang Z, Zhou K (2014) Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc Natl Acad Sci USA 111:16337–16342
Glazinska P, Zienkiewicz A, Wojciechowski W, Kopcewicz J (2009) The putative miR172 target gene In APETALA2-like is involved in the photoperiodic flower induction of Ipomoeanil. Plant Physiol 166:1801–1813
Hori K, Ogiso-Tanaka E, Matsubara K, Yamanouchi U, Ebana K, Yano M (2013) Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J 76:36–46
Jung JH, Lee S, Yun J, Lee M, Park CM (2014) The miR172 target TOE3 represses AGAMOUS expression during Arabidopsis floral patterning. Plant Sci 215–216:29–38
Kim SK, Yun CH, Lee JH, Jang YH, Park HY, Kim JK (2008) OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta 228:355–365
Kojima S (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43:1096–1105
Koo BH, Yoo SC, Park JW, Kwon CT, Lee BD, An G, Zhang Z, Li J, Li Z, Paek NC (2013) Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol Plant 6:1877–1888
Lee D-Y, An G (2012) Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J 69:445–461
Lee DY, Lee J, Moon S, Park SY, An G (2007) The rice heterochronic gene supernumerary bract regulates the transition from spikelet meristem to floral meristem. Plant J 49:64–78
Lee YS, Lee DY, Cho LH, An G (2014) Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice 7:1–13
Li D, Yang C, Li X, Gan Q, Zhao X, Zhu L (2009) Functional characterization of rice OsDof12. Planta 229:1159–1169
Lin H, Liang Z-W, Sasaki T, Yano M (2003) Fine mapping and characterization of quantitative trait loci Hd4 and Hd5 controlling heading date in rice. Breed Sci 53:51–59
Lin D, Jiang Q, Zheng K, Chen S, Zhou H, Gong X, Xu J, Teng S, Dong Y (2015) Mutation of the rice ASL2 gene encoding plastid ribosomal protein L21 causes chloroplast developmental defects and seedling death. Plant biology 17:599–607
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408
Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M (2009) Repression of flowering by the miR172 target SMZ. PLoS Biol 7:e1000148
Matsubara K, Yamanouchi U, Nonoue Y, Sugimoto K, Wang ZX, Minobe Y, Yano M (2011) Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J 66:603–612
Matsubara K, Ogiso-Tanaka E, Hori K, Ebana K, Ando T, Yano M (2012) Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol 53:709–716
Moose SP, Sisco PH (1994) Glossy15 controls the epidermal juvenile-to-adult phase transition in maize. Plant Cell 6:1343–1355
Murray M, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326
Ogiso-Tanaka E, Matsubara K, Yamamoto S, Nonoue Y, Wu J, Fujisawa H, Ishikubo H, Tanaka T, Ando T, Matsumoto T, Yano M (2013) Natural variation of the RICE FLOWERING LOCUS T 1 contributes to flowering time divergence in rice. PLoS One 8:e75959
Okamuro JK, Caster B, Villarroel R (1997) The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA 94:7076–7081
Salvi S, Sponza G, Morgante M, Tomes D, Niu X, Fengler KA, Li B (2007) Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Natl Acad Sci USA 104:11376–11381
SAS Institute S Inc (1990) SAS/STAT user’s guide, version 6. SAS institute, Cary
Song J, Wei X, Shao G, Sheng Z, Chen D, Liu C, Jiao G, Xie L, Tang S, Hu P (2014) The rice nuclear gene WLP1 encoding a chloroplast ribosome L13 protein is needed for chloroplast development in rice grown under low temperature conditions. Plant Mol Biol 84:301–314
Sperandio S, Tardito S, Surzycki A, Latterich M, de Belle I (2009) TOE1 interacts with p53 to modulate its transactivation potential. FEBS Lett 583:2165–2170
Takahashi Y, Shomura A, Sasaki T, Yano M (2001) Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the alpha subunit of protein kinase CK2. Proc Natl Acad Sci USA 98:7922–7927
Takeuchi Y, Lin S, Sasaki T, Yano M (2003) Fine linkage mapping enables dissection of closely linked quantitative trait loci for seed dormancy and heading in rice. Theor Appl Genet 107:1174–1180
Tsuji H, Tachibana C, Tamaki S, Taoka K, Kyozuka J, Shimamoto K (2015) Hd3a promotes lateral branching in rice. Plant J 82:256–266
Wei X, Xu J, Guo H, Jiang L, Chen S, Yu C, Zhou Z, Hu P, Zhai H, Wan J (2010) DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol 153:1747–1758
Wu W, Zheng XM, Lu G, Zhong Z, Gao H, Chen L, Wu C, Wang HJ, Wang Q, Zhou K, Wang JL, Wu F, Zhang X, Guo X, Cheng Z, Lei C, Lin Q, Jiang L, Wang H, Ge S, Wan J (2013) Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc Natl Acad Sci USA 110:2775–2780
Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40:761–767
Yamamoto T, Kuboki Y, Lin S, Sasaki T, Yano M (1998) Fine mapping of quantitative trait loci Hd-1, Hd-2 and Hd-3, controlling heading date of rice, as single Mendelian factors. Theor Appl Genet 97:37–44
Yano M, Harushima Y, Nagamura Y, Kurata N, Minobe Y, Sasaki T (1997) Identification of quantitative trait loci controlling heading date in rice using a high-density linkage map. Theor Appl Genet 95:1025–1032
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12:2473–2483
Yano M, Kojima S, Takahashi Y, Lin H, Sasaki T (2001) Genetic control of flowering time in rice, a short-day plant. Plant Physiol 127:1425–1429
Zhan X, Sun B, Lin Z, Gao Z, Yu P, Liu Q, Shen X, Zhang Y, Chen D, Cheng S, Cao L (2015) Genetic mapping of a QTL controlling source-sink size and heading date in rice. Gene 571:263–270
Zhang B, Wang L, Zeng L, Zhang C, Ma H (2015) Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev 29:975–987
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grants 31101209, 31221004, and 31501290) and the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-CNRRI).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest regarding the publication of this paper.
Additional information
Communicated by M. Wissuwa.
B. Sun and X.-D. Zhan contributed equally to this work.
The original version of this article was revised: A value (52.92-kb) under the heading ‘‘Key message’’ was incorrect. This error has been corrected.
An erratum to this article is available at http://dx.doi.org/10.1007/s00122-016-2821-0.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sun, B., Zhan, XD., Lin, ZC. et al. Fine mapping and candidate gene analysis of qHD5, a novel major QTL with pleiotropism for yield-related traits in rice (Oryza sativa L.). Theor Appl Genet 130, 247–258 (2017). https://doi.org/10.1007/s00122-016-2787-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-016-2787-y