Abstract
Background
Rice is a facultative short-day plant that flowers under long days (LD) after a lengthy vegetative phase. Although several inhibitors that delay flowering have been identified, the process by which rice eventually flowers under non-permissive LD conditions is not well understood.
Results
Overexpression of miR172 reduced flowering time significantly, suggesting its role as an inducer. Levels of miR172 increased as plants aged, further supporting our findings. Transcripts of SNB and OsIDS1, two members of the AP2 family that have the miR172 target site, were reduced in older plants as the level of miR172 rose. Overexpression of those AP2 genes delayed flowering; overexpression of miR172-resistant forms of SNB or OsIDS1 further delayed this process. This demonstrated that the AP2 genes function downstream of miR172. Two florigen genes -- Hd3a and RFT1 -- and their immediate upstream regulator Ehd1 were suppressed in the AP2 overexpression plants. This suggested that the AP2 genes are upstream repressors of Ehd1. In phytochrome mutants, miR172d levels were increased whereas those of SNB and OsIDS1 were decreased. Thus, it appears that phytochromes inhibit miR172d, an AP2 suppresser.
Conclusions
We revealed that miR172d developmentally induced flowering via repressing OsIDS1 and SNB, which suppressed Ehd1. We also showed that phytochromes negatively regulated miR172.
Similar content being viewed by others
Background
Rice flowers earlier under short day (SD) conditions than under long days (LD). The photoperiodic flowering pathway of rice is controlled by OsGIGANTEA (OsGI) (Yano et al. [2000]; Hayama et al. [2002], [2003]; Kojima et al. [2002]; Doi et al. [2004]). This gene regulates flowering time by promoting Heading date 1 (Hd1), an ortholog of Arabidopsis CONSTANS (CO) (Yano et al. [2000]; Hayama et al. [2002], [2003]). Hd1 enhances Early heading date 1 (Ehd1) expression under SD but inhibits its expression under LD (Wei et al. [2010]; Ishikawa et al. [2011]). Ehd1 is an immediate upstream positive regulator of Heading date 3a (Hd3a) and Rice Flowering Locus T 1 (RFT1), which encode florigens that promote flowering (Doi et al. [2004]; Tamaki et al. [2007]; Komiya et al. [2008], [2009]).
Because Ehd1 is modulated by several regulators, it acts as a mediator of various floral signals. For example, Early heading date 2 (Ehd2)/OsINDETERMINATE 1 (OsId1)/Rice INDETERMINATE 1 (RID1) is a constitutive activator of Ehd1 (Matsubara et al. [2008]; Park et al. [2008]; Wu et al. [2008]). Loss-of-function mutants in the gene do not flower under either SD or LD. OsMADS51 also acts as a positive regulator of Ehd1, specifically under SD (Kim et al. [2007]). One major repressor of Ehd1 is Grain yield and heading date 7 (Ghd7), which functions preferentially under LD (Xue et al. [2008]). Most early-flowering rice cultivars show a disruption in Ghd7 expression (Xue et al. [2008]). A chromatin remodeling factor, OsTrithorax 1 (OsTrx1), suppresses Ghd7 by binding to Early heading date 3 (Ehd3) (Matsubara et al. [2011]; Choi et al. [2014]). In addition, OsLFL1 inhibits Ehd1 when over-expressed (Peng et al. [2007], [2008]). However, expression of the former is suppressed by another chromatin remodeling factor, OsVIN3-like 2 (OsVIL2), when binding to the PRC2 complex (Yang et al. [2013]). OsMADS50 induces Ehd1 by blocking OsLFL1 and Ghd7 (Lee et al. [2004]; Ryu et al. [2009]; Choi et al. [2014]). A third gene, OsCOL4, constitutively inhibits flowering time by suppressing Ehd1 (Lee et al. [2010]). Overexpression of the former delays flowering whereas knockout mutations cause early flowering. Finally, DTH8 and Hd16 preferentially suppress flowering time under LD by inhibiting Ehd1 (Xue et al. [2008]; Wei et al. [2010]; Hori et al. [2013]).
Micro RNAs inhibit expression of target genes by cleaving mRNA or translational suppression (Jones-Rhoades et al. [2006]; Voinnet [2009]). miR172 and miR156 are involved in phase transition (Aukerman and Sakai [2003]; Lauter et al. [2005]; Wu and Poethig [2006]; Poethig [2009]). miR156 plays roles in early vegetative stages, while miR172 functions later stages of develop (Aukerman and Sakai [2003]; Lauter et al. [2005]; Wu and Poethig [2006]; Chuck et al. [2007]; Poethig [2009]). In Arabidopsis, miR156 targets 10 members (SPL2, SPL3, SPL4, SPL5, SPL6, SPL9, SPL10, SPL11, SPL13, and SPL15) of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) family. SPL9 prompts miR172 expression and the other SPL genes redundantly function in regulating miR172. The role of miR172 in controlling flowering time has been reported for Arabidopsis, maize, barley, and soybean (Aukerman and Sakai [2003]; Chen [2004]; Lauter et al. [2005]; Jung et al. [2007]; Mathieu et al. [2009]; Nair et al. [2010]; Yoshikawa et al. [2013]). In Arabidopsis and maize, its temporal expression increases gradually as plants age (Aukerman and Sakai [2003]; Lauter et al. [2005]). In rice, miR172 is most highly expressed during later vegetative stages and in developing panicles (Zhu et al. [2009]; Lee and An [2012]).
AP2 family genes are involved in various processes, including floral organ identity, shattering, and flowering time (Aukerman and Sakai [2003]; Chen [2004]; Lauter et al. [2005]; Lee et al. [2007]; Jung et al. [2007]; Chuck et al. [2008]; Mathieu et al. [2009]; Zhu et al. [2009]; Lee and An [2012]; Zhou et al. [2012]; Yoshikawa et al. [2013]). Six Arabidopsis genes in this family -- APETALA 2 (AP2), TARGET OF EAT 1(TOE1), TOE2, TOE3, SCHLAFMUTZE (SMZ), and SCHNARCHZAPFEN (SNZ) -- delay flowering in an age-dependent manner (Park et al. [2002]; Aukerman and Sakai [2003]; Schmid et al. [2003]; Chen [2004]; Jung et al. [2007]; Mathieu et al. [2009]), and are suppressed by miR172 (Schmid et al. [2003]; Kasschau et al. [2003]; Chen [2004]; Schwab et al. [2005]; Ju ng et al. [2007]; Mathieu et al. [2009]). In maize, enhancement of GLOSSY15 (GL15), an AP2 member, delays phase transition from the vegetative to the reproductive stage (Lauter et al. [2005], Zhu and Helliwell [2011]). Its temporal expression gradually decreases as plants mature, and this gene is also down-regulated by miR172 (Lauter et al. [2005]). In rice, five AP2-like genes (SNB, OsIDS1, SHAT1, Os05g03040, and Os06g43220) contain the miR172 target sites (Sunkar et al. [2005]; Zhu et al. [2009]). While SNB and OsIDS1 control floral organ identity and spikelet development, SHAT1 is involved in seed shattering (Sunkar et al. [2005]; Lee et al. [2007]; Zhu et al. [2009]; Lee and An [2012]; Zhou et al. [2012]).
Although an antagonistic role for miR172 and AP2 genes in floral transition has been described in Arabidopsis and maize, their functions in rice have not been reported. Here, we demonstrated in rice that miR172 induces flowering time by suppressing two AP2 family members - SNB and OsIDS1 - that are negative regulators of Ehd1.
Results
Temporal expression patterns of miR172s and AP2 genes are antagonistic
To examine whether miR172 levels change as plants develop, we monitored the temporal expressions of miR172a and miR172d in leaf blades. The other two miR172 members in rice - miR172b and miR172c – were not measured because they are expressed mainly in panicles and roots, respectively, and are not likely involved in controlling flowering time (Jeong et al. [2011]). Levels of pri-miR172a and pri-miR172d increased gradually as plants aged (Figure 1A and B). To verify this, we conducted northern blot analysis of mature miR172. Because the sequences of mature miR172a and miR172d are identical, we were only able to measure the total amount of both micro RNAs. This analysis confirmed that mature miR172ad levels were gradually increased (Figure 1C).
Temporal expression patterns of miR172 and AP2 genes. (A, B) RT-PCR analysis of pri-miR172a (A) and pri-miR172d (B) in WT plants under LD. Y-axis, relative transcript level compared with OsUbi1. Error bars indicate standard deviations; n = 4 or more. Middle regions of fully emerged uppermost leaves were sampled at 30, 40, 45, 50, 55, and 60 DAG. (C) Northern blot analysis of mature miR172 (top). Ethidium bromide-stained ribosomal RNA served as loading control (bottom). Middle regions were used from leaves sampled at 25, 30, 40, 45, 50, and 55 DAG. (D, E) RT-PCR analysis of transcript levels of SNB (E) and OsIDS1 (E) under LD. Middle regions were used from leaves sampled at 27, 30, 32, 35, 38, 44, 51, 55, and 57 DAG. (F) RT-PCR analysis of Ehd1 transcripts under LD, using leaves sampled at 27, 30, 32, 35, 38, 44, 51, 55, 57, 60, 63, 66, and 73 DAG. Expression was monitored at ZT 18 h for miR172 and at ZT 2 h for AP2 genes and Ehd1. Y-axis, relative transcript level compared with OsUbi1. Error bars indicate standard deviations; n = 4 or more.
The miR172 s target several AP2 genes, with temporal expression of the latter type being slowly diminished in Arabidopsis and maize (Aukerman and Sakai [2003]; Lauter et al. [2005]; Jung et al. [2007]; Mathieu et al. [2009]; Zhu and Helliwell [2011]). In all, six AP2 genes have miR172 target sites (Zhu and Helliwell [2011]). Other miRNAs and their targeted genes also show antagonistic expression patterns (Jeong and Green [2013]; Jung et al. [2013]). We examined the temporal expression of two rice AP2 genes in leaf blades: SNB and OsIDS1. As expected, their transcript levels were high at younger developmental stages but rapidly declined to a low level at 35 days after germination (DAG) (Figure 1D and E). Ehd1 mRNA expression began to increase at 57 DAG (Figure 1F). Under SD, transcript levels of the AP2 genes dropped rapidly at 18 DAG, immediately before the level of Ehd1 began to rise (Additional file 1: Figure S1). This suggested that the AP2 genes are likely targets of miR172a and miR172d.
Overexpression of SNB and OsIDScauses late flowering
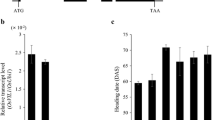
To examine the functional roles of AP2 genes in flowering time, we generated transgenic rice plants that over-express SNB and OsIDS1 (Additional file 1: Figure S2). Such overexpression (OX) of miR172-targeted AP2 causes late-flowering phenotypes in Arabidopsis (Jung et al. [2007]; Mathieu et al. [2009]). Similarly, we noted that SNB OX and OsIDS1 OX plants flowered late when grown in the greenhouse (Figure 2A and B). Among our 11 SNB OX transgenics, flowering was delayed by 2 to 8 weeks for nine of them (Figure 2E). Flowering time for the SNB OX plants was correlated with the degree of SNB expression (Additional file 1: Figure S2A, S2B, and S2C). Likewise, OsIDS1 OX plants flowered 3 to 5 weeks later than usual (Figure 2F). Their heading date was correlated with amounts of OsIDS1 transcript (Additional file 1: Figure S2D, S2E, and S2F). Three SNB OX plants (#3, 4, and 5) and three OsIDS1 OX plants (#2, 3, and 6) were selected for further analysis. In studying photoperiod dependency of those genes, we found that flowering time for SNB OX #3 was delayed by about 12 d under SD and about 7 d under LD (Figure 2A, C, and D). Similarly, OsIDS1 OX #2 flowered 3 weeks later than usual under SD and 2 weeks later under LD (Figure 2B, C, and D). These observations demonstrated that SNB and OsIDS1 function as flowering repressors in rice.
Analysis of SNB and OsIDS1 overexpression plants. (A) Phenotypes of SNB overexpression plant (SNB OX #3) at 97 DAG when grown under SD. (B) Phenotypes of OsIDS1 overexpression plants (OsIDS1 OX #2) at 77 DAG when grown under SD. (C, D) Heading dates for OsIDS1 and SNB OX plants under SD (C) or LD (D). Days-to-heading was scored when first panicle bolted. Error bars indicate standard deviations; n = 10-20 plants. (E, F) Flowering time for SNB (E) and OsIDS1 (Ff) OX plants (T1 generation) in greenhouse. Days-to-heading was scored when first panicle bolted.
SNB and OsIDS1 repress the floral transition by inhibiting Ehd1
To elucidate the roles of SNB and OsIDS1 in controlling flowering time, we measured transcript levels of the previously identified floral regulators in OX plants. Because the delayed-flowering phenotype was more severe in OsIDS1 OX #2, we analyzed those plants. Increased expression of OsIDS1 did not affect transcript levels of the AP2 genes (Figure 3B and C). Transcripts of Hd3a and RFT1, two florigens in rice, were significantly reduced in those plants (Figure 3D and E), as were transcripts of Ehd1 (Figure 3F). However, levels of other flowering activators, i.e., OsGI, Hd1, OsMADS51, and OsCO3, were not altered in the OX plants (Figure 3G, H, I, and J). Overexpression of OsIDS1 also had no influence on flowering time regulators such as OsPhyB, OsMADS50, Ghd7, OsId1, OsCOL4, OsMADS56, Ehd3, and OsTrx1 (Additional file 1: Figure S3). In the SNB OX #3, transcript levels of Ehd1, Hd3a, and RFT1 were also decreased, but those of OsGI, Hd1, OsMADS51, and OsCOL4 were not changed (Additional file 1: Figure S4). These observations indicated that SNB and OsIDS1 repress the floral transition by inhibiting the expression of Ehd1.
Transcript levels of floral regulators in the OsIDS1 OX. Transcript levels of OsIDS1 (A), SNB (B), OsTOE1 (C), Hd3a (D), RFT1 (E), Ehd1 (F), OsGI (G), Hd1 (H), OsMADS51 (I), and OsCO3 (J) in WT (closed boxes) and OsIDS1 OX plants (open boxes) under SD. Middle regions of fully emerged uppermost leaves were sampled at 30 DAG. For OsGI and Hd1, monitoring occurred at ZT 10 h and ZT 14 h, respectively; for others, at ZT 2 h. Y-axis, relative transcript level compared with rice OsUbi1. Error bars indicate standard deviations; n = 4 or more.
Levels of SNB and OsIDS1 did not differ between `Dongjin′ and `Kitaake′ rice. The latter is an early-flowering cultivar and carries mutations in Ghd7 and PRR37 (Kim et al. [2013]). Therefore, this demonstrated that the AP2 genes are not likely influenced by Ghd7 and PRR37. In addition, AP2 expression was not significantly altered in mutants defective in regulatory genes such as Ghd7, OsGI, Ehd1, OsMADS50, OsMADS51, OsId1, OsTrx1, and OsVIL2, all of which control flowering time (Additional file 1: Figure S5). However, expression of SNB and OsIDS1 was affected in the oscol4 and hd1 mutants (Figure 4A and B; Additional file 1: Figure S5A). This suggested that expression of AP2 genes is positively controlled by OsCOL4 and Hd1.
Expression of miR172 s and AP2 genes in oscol4 , osphyB , and osphyA osphyB mutants. (A, B) Transcript levels for SNB (A) and OsIDS1 (B) in WT (black bars) and oscol4 (open bars) plants under LD. (C, D) Transcript levels for miR172d (C) and miR172a (D) in WT (black bars), osphyB (gray bars), and osphyA osphyB (open bars) plants under LD. (E, F) Transcript levels for SNB (E) and OsIDS1 (F) in WT (black bars), osphyB (gray bars), and osphyA osphyB (open bars) plants under LD. Middle regions of fully emerged uppermost leaves were sampled at 35 DAG. Expression was monitored at ZT 11 h for miR172 s and at ZT 2 h for SNB and OsIDS1. Y-axis, relative transcript level compared with OsUbi1. Error bars indicate standard deviations; n = 4 or more.
miR172d induces flowering time by suppressing AP2 genes
The transcripts of SNB and OsIDS1 carry a miR172 target site (Lee and An [2012]). To examine whether miR172 controls flowering time, we generated six transgenic rice plants that over-express miR172d (Figure 5A). In all plants, transcripts were increased (Figure 5B) and the time to flowering was shortened by 9 to 30 d (Figure 5C). Levels of flowering activators Ehd1 and Hd3a were also substantially higher in the miR172d OX lines (Figure 5D and E) while expression of SNB and OsIDS1 was decreased in those lines (Figure 5F and G). These results suggested that miR172d induces flowering by reducing AP2 gene expression.
Phenotypes of miR172d OX plants. (A) Scheme of miR172d OX construct. Spanning region of miR172d was sub-cloned into pGA1611 vector between maize ubiquitin promoter (Pubi) and nopaline synthase terminator (nos T). (B) Northern blot analysis of expression levels of mature miR172 in WT and miR172d OX plants (top). Ethidium bromide-stained ribosomal RNA was used as loading control (bottom). (C) Flowering time for 6 miR172d OX plants compared with WT when grown in greenhouse. Days-to-heading was scored when first panicle bolted. (D-G) Expression levels of Ehd1 (D), Hd3a (E), SNB (F), and OsIDS1 (G) in WT and miR172d OX plants.
To confirm that miR172 controls flowering time via the AP2 genes, we constructed the miR172-resistant form of SNB (rSNB) by changing the miR172 target site CTGCAGCATCATCAGGATTCT to CTGCAGCAATGTCCGGATTCT (Figure 6A). Of the six transgenic rice plants carrying this rSNB construct, five lines (#1, 3, 4, 5, and 6) expressed the transgene at substantially high levels (Figure 6B). The rSNB transcript can be distinguished from the endogenous SNB transcript due to the restriction enzyme site ACC III that is generated in the rSNB OX construct. Here, the RT-PCR products of those rSNB OX plants were digested with ACC III, supporting our findings that most of the SNB transcripts in the OX plants were rSNB (Additional file 1: Figure S7). These rSNB OX plants eventually died without having flowered, even after several months of growth (Figure 6C). Moreover, transgenic plants over-expressing the miR172-resistant form of OsIDS1 (rOsIDS1) did not produce any flowers for more than one year (Additional file 1: Figure S6). These observations further demonstrated that miR172 s induce flowering time by suppressing AP2 transcripts in rice.
Phenotypes of miR172 -resistant SNB OX plants. (A) Scheme of miR172-resistant SNB (rSNB) construct. Four synonymous mutations (red characters) were introduced into miR172 target sequences at C-terminus of SNB full-length cDNA. Primers tSNB F and R were used. Underlined sequence (tccgga) indicates restriction enzyme site by ACC III. Pact, actin promoter; nos T, nopaline synthase terminator. (B) Expression level of SNB in rSNB OX plants. (C) Phenotypes of rSNB OX plants grown under SD. Bolted panicles are shown by red arrows.
The late-flowering phenotype of rSNB OX plants is rescued by overexpression of Ehd1
Because Ehd1 transcripts were reduced in SNB OX plants, we postulated that the AP2 gene functions upstream of Ehd1. To confirm this hypothesis genetically, we generated transgenic plants over-expressing both rSNB and Ehd1 (Figure 7A). It was previously reported that overexpression of Ehd1 causes early flowering (Osugi et al. [2011]), and we also observed early-flowering phenotypes of Ehd1 OX plants (Additional file 1: Figure S8). Thus, if SNB functions downstream of Ehd1, we would expect that transgenic plants expressing both genes do not flower early. Instead, those plants did flower early and their morphology was similar to plants that over-express only Ehd1 (Figure 7B and C). This observation supports that SNB inhibits flowering time by suppressing Ehd1.
Phenotype of transgenic plant overexpressing both r SNB and Ehd1. (A) Diagram of miR172-resistant SNB (rSNB) and Ehd1 double OX construct. pUbi, maize ubiquitin promoter; nos T, nos terminator. (B) Phenotypes of rSNB and Ehd1 OX #1 plants. Photograph was taken at 42 days after transplanting. Red arrows indicate emerged panicles. (C) Heading date of rSNB and Ehd1 OX primary transgenic plants grown in greenhouse.
miR172a and miR172d are negatively regulated by phytochromes in rice
The level of miR172 is increased in phyB mutants of Arabidopsis (Jung et al. [2007]). To study whether miR172 in rice is also regulated by phytochrome, we measured the levels of pri-miR172a and pri-miR172d in mutants (Figure 4). This analysis revealed that pri-miR172a and pri-miR172d transcripts were elevated in the osphyB mutant and further increased in the osphyA osphyB double mutant (Figure 4C and D). This indicated that miR172a and miR172d are negatively regulated by phytochrome. Transcript levels of SNB and OsIDS1 were also significantly reduced in the osphyB single and the osphyA osphyB double mutants (Figure 4E and F). These observations suggested that phytochromes can inhibit flowering time by suppressing miR172a and miR172d, which interfere with SNB and OsIDS1 expression (Figure 8).
Discussion
We have demonstrated that miR172d induces flowering by suppressing SNB and OsIDS1. Overexpression of the AP2 genes delays flowering more significantly under SD, the permissive condition. This phenotype is similar to that of Arabidopsis SMZ- OX plants (Mathieu et al. [2009]). Overexpression of SMZ causes a significant delay in flowers under LD, the permissive condition for that species, but not under SD (Mathieu et al. [2009]). These reports provide evidence that AP2 genes preferentially block the floral transition under inductive conditions.
Although flowering is inhibited under LD, rice plants do flower eventually under normally suppressive conditions, albeit after a certain period of vegetative growth. We showed here that miR172d levels were increased in the leaves as a plant aged and that flowering was induced when that gene was over-expressed. This indicated that the microRNA plays a role in the developmental control of flowering time in rice, as also observed from Arabidopsis (Jung et al. [2007]).
SNB and OsIDS1 are flowering repressors that are highly expressed in young leaves. However, their expression gradually declined to minimal levels at 35 DAG when miR172 transcripts started to increase. Expression of the florigens and Ehd1 began at 60 DAG while that of the AP2 genes remained low. This indicated that the latter are major suppressors of the downstream floral signals.
Ghd7 also functions to repress Ehd1, but its transcription peaks at 2 to 3 weeks before declining to a low level. This occurs much earlier than when the florigen genes are expressed (Matsubara et al. [2011]; Kim et al. [2013]). OsCOL4 is another regulator that suppresses Ehd1 and the florigen genes. In particular, OsCOL4 expression is maintained at a high level in the early vegetative stages but decreases when Ehd1 expression begins (Lee et al. [2010]). This temporal expression pattern is similar to SNB and OsIDS1 except that OsCOL4 is reduced later than the AP2 genes. Therefore, these results suggest that the AP2 genes as well as Ghd7 and OsCOL4 coordinately suppress flowering time.
Except in oscol4 and hd1 plants, AP2 expression is not significantly altered in most flowering mutants, thereby implying that those genes are controlled by OsCOL4 and Hd1, but not by other flowering time regulators. We have previously reported that OsCOL4 is a constitutive repressor of Ehd1 (Lee et al. [2010]). Because this is also true of AP2 genes, it is likely that OsCOL4 suppresses Ehd1 by inducing AP2 expression. Hd1 also suppresses flowering time via repressing Hd3a and RFT1 under LD conditions (Yano et al. [2000]; Hayama et al. [2003]). These indicate that the AP2 genes and OsCOL4 co-operatively suppress flowering time.
We observed that miR172d expression is negatively affected by phytochrome activity. Considering that AP2 genes are controlled by miR172, we might conclude that phytochromes support vegetative growth by maintaining AP2 expression. Both Ghd7 and OsCOL4 are positively modulated by OsPhyB (Lee et al. [2010]; Osugi et al. [2011]). Therefore, it is apparent that phytochromes influence these flowering regulators collectively.
In plants, the miR172/AP2 module is inversely correlated with the miR156/SPL module (Aukerman and Sakai [2003]; [2005]; Wu and Poethig [2006]; Chuck et al. [2007]; Poethig [2009]). In rice, miR156 genes are predominantly expressed in the young shoots, etiolated shoots, and seedling leaves (Xie et al. [2006]) and its target OsSPL genes are expressed high in young panicles (Xie et al. [2006]). Overexpression of miR156 OX causes various phenotypes such as increase of tiller numbers, late flowering, dwarfism, and decrease of spikelet numbers (Xie et al. [2006]). In maize, overexpression of the maize miR156 prevents flowering and increases starch content (Chuck et al. [2007]; [2011]). This late-flowering phenotype is similar to that of SNB OX or OsIDS1 OX plants. Overexpression of the maize miR156 also causes increase of biomass and tiller number in maize and also in Arabidopsis, Brachypodium and switchgrass (Chuck et al. [2011]). These phenotypes are similar to those of OsmiR156 OX (Xie et al. [2006]; Chuck et al. [2011]).
Conclusions
We demonstrated that over-expressions of OsIDS1 and SNB inhibited flowering time in rice by suppressing expression of Ehd1. In addition, expressions of the AP2 genes were repressed by miR172 and the later was increased in the osphyB and osphyA osphyB mutants. Based on these findings, we concluded that miR172 induced flowering by suppressing the AP2 genes and the microRNA gene was inhibited by phytochromes. The facultative LD-flowering phenotype of rice can be explained in part by the miR172-AP2 pathway.
Methods
Plant material and growth conditions
We have previously developed T-DNA-tagging lines in Oryza sativa japonica cv. Dongjin (Jeon et al. [2000]). The flanking sequences were determined via inverse PCR (An et al. [2003]; Ryu et al. [2004]). T-DNA insertional mutants osphyA-2, osphyB-2, osmads50-1, osmads51-1, hd1-1, oscol4-2, osvil2-1, and ostrx1-1 were reported earlier (Lee et al. [2004]; Jeong et al. [2007]; Kim et al. [2007]; Ryu et al. [2009]; Lee et al. [2010]; Yang et al. [2013]; Choi et al. [2014]). RNAi-suppressed plants of Ehd1 RNAi and OsId1 RNAi have been described previously (Kim et al. [2007]; Park et al. [2008]). A near isogenic line (NIL) that carries the ghd7 allele was presented by Kim et al. ([2013]). Seeds were germinated on a half-strength Murashige and Skoog medium containing 3% sucrose. They were incubated for one week at 28°C under constitutive light as previously reported (Yi and An [2013]). Seedlings were transplanted into soil and cultured in growth chambers under either SD (12 h light at 28°C, humidity 70%; 12 h dark at 25°C, humidity 50%) or LD (14.5 h light at 28°C, humidity 70%; 9.5 h dark at 25°C, humidity 50%) as previously reported (Yi and An [2013]).
Construction of osphyA osphyB double mutants
The osphyA osphyB double mutants were obtained by crossing osphyA-2 and osphyB-2 single mutants. This osphyA-2 mutant is a null allele generated by a T-DNA insertion in the fourth exon of OsphyA. The osphyB-2 mutant is also a null allele produced by inserting T-DNA in the third intron of OsphyB (Jeong et al. [2007]). Afterward, F2 segregants were genotyped by PCR (35 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 60 s), using a combination of gene-specific primers and the T-DNA primer 5′-TTGGGGTTTCTACAGGACGTAAC-3′. Those gene-specific primers were 5′-CAGGGAAAAGGGATTAGAGT-3′ and 5′-AGTGGACTCGGGTTAACTTT-3′ for osphyA-2, and 5′-AGTTAAGACGGAGCGACATA-3′ and 5′-GTAAGCGATCAGTTTGTGGT-3′ for osphyB-2. For further analyses, we selected F2 progeny that had homozygous mutant alleles for both genes.
Vector construction and transformation
The full-length cDNA clones of SNB and OsIDS1 were isolated by PCR, using primers SNB-FL-F and SNB-FL-R for SNB, and OsIDS1-FL-F and OsIDS1-FL-R for OsIDS1 (Additional file 2: Table S1). After the amplified fragments were digested with Xba I and Xho I, they were inserted into the pGA3780 binary vector between the rice actin promoter (Pact) and the nopaline synthase terminator (nos T) (Lee et al. [1999]; Kim et al. [2009]). For constructing a miR172-resistant form of SNB (rSNB), we introduced synonymous mutations into the miR172 binding site through site-directed mutagenesis, using primer sets rSNB-N-F/rSNB-N-R and rSNB-C-F/rSNB-C-R (Additional file 2: Table S1). Each amplified fragment was digested with Xba I/ACC III or ACC III/Xho I. Afterward, the digested fragments were inserted into the pGA3780 binary vector between the rice Pact and nos T. Similarly, a miR172-resistant form of OsIDS1 (rOsIDS1) was produced using the primer sets of rOsIDS1-N-F/rOsIDS1-N-R and rOsIDS1-C-F/rOsIDS1-C-R (Additional file 2: Table S1). To obtain double-overexpression plants of rSNB and Ehd1, we amplified their full-length cDNA clones with primer sets rSNB-FL-F/rSNB-FL-R and Ehd1-FL-F/Ehd1-FL-R (Additional file 2: Table S1). The rSNB fragment was digested with Bsi WI and Bsr GI, and the Ehd1 fragment was digested with Mlu I and Hpa I. Afterward, these digested fragments were inserted into the pGA3777 binary vector, a double expression cassette (Kim et al. [2009]). We have previously described plants over-expressing miR172d (Lee and An [2012]). The binary constructs were transformed into Agrobacterium tumefaciens LBA4404 (An et al. [1989]) and transgenic plants were generated via Agrobacterium-mediated co-cultivation (Jeon et al. [1999]).
RNA extraction and quantitative real-time RT-PCR
Total RNA was isolated with RNAiso Plus (Takara, Shiga, Japan) and first-strand cDNA was synthesized using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega, Madison, WI, USA) as previously reported (Ryu et al. [2009]; Lee et al. [2010]; Yang et al. [2013]). Synthesized cDNAs were used as a template for quantitative real-time RT-PCR (qRT-PCR) with SYBR® Premix Ex Taq™ II (Takara, Shiga, Japan) and the Rotor-Gene 6000 (Corbett Research, Sydney, Australia). Osubi1 served to normalize the quantity of cDNA. All experiments were conducted at least three times, with three or more samples at each point. Primer sequences for qRT-PCR are shown in Additional file 2: Table S1. Changes in expression were calculated via the ΔΔCt method. To ensure primer specificity, we performed the experiments when the melting curve showed a single peak. PCR products were sequenced to verify the specificity of the reaction.
Expression analysis of microRNA
For miRNA gel blot analysis, total RNA samples were extracted from plant materials using RNAiso Plus (Takara), with a few modifications as reported previously (Jung et al. [2007]). After isopropanol precipitation, the Eppendorf tube containing the RNA pellets was briefly centrifuged without rinsing with ethanol to improve both the yield and solubility of miRNA in total RNA preparations. RNA gel blot analyses were performed with 5 μg of total RNA. Locked nucleic acid (LNA) 5′-ATgCAgCAtCAtCAaGAtTCT-3′ (upper- and lower-case letters indicate DNA and LNA, respectively) was used as an antisense oligonucleotide probe for miR172 (Varallyay et al. [2008]). The miR172 LNA probe was either labeled with P32 or 3′-end-labeled with DIG-ddUTP (Roche, Mannheim, Germany). Levels of the primary miR172 (pri-miR172) transcripts were measured by qRT-PCR using the primer pairs described in Additional file 2: Table S1.
Authors′ contributions
YSL and DYL designed the project, produced plant materials, monitored expression profiling, observed flowering time, analyzed data and wrote the manuscript. LHC produced Ehd1 OX plants and observed flowering time of those. GA provided an overall direction for this project and helped with the organization and editing of the manuscript. All authors read and approved the final manuscript.
Additional files
References
An G, Ebert PR, Mitra A, Ha SB: Binary vectors. Springer Netherlands, Dordrecht; 1989.
An S, Park S, Jeong DH, Lee DY, Kang HG, Yu JH, Hur J, Kim SR, Kim YH, Lee M, Han S, Kim SJ, Yang J, Kim E, Wi SJ, Chung HS, Hong JP, Choe V, Lee HK, Choi JH, Nam J, Kim SR, Park PB, Park KY, Kim WT, Choe S, Lee CB, An G: Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol 2003, 133: 2040–2047. 10.1104/pp.103.030478
Aukerman MJ, Sakai H: Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 2003, 15: 2730–2741. 10.1105/tpc.016238
Chen X: A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303: 2022–2025. 10.1126/science.1088060
Choi SC, Lee S, Kim SR, Lee YS, Liu C, Cao X, An G: Trithorax group protein OsTrx1 controls flowering time in rice via interaction with Ehd3. Plant Physiol 2014, 164: 1326–1337. 10.1104/pp.113.228049
Chuck G, Cigan AM, Saeteurn K, Hake S: The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet 2007, 39: 544–549. 10.1038/ng2001
Chuck G, Meeley R, Hake S: Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 2008, 135: 3013–3019. 10.1242/dev.024273
Chuck G, Tobias C, Sun L, Kraemer F, Li C, Dibble D, Arora R, Bragg JN, Vogel JP, Singh S, Simmons BA, Pauly M, Hake S: Overexpression of the maize Corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. Proc Natl Acad Sci USA 2011, 108: 17550–17555. 10.1073/pnas.1113971108
Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A: Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-Iike gene expression independently of Hd1. Genes Dev 2004, 18: 926–936. 10.1101/gad.1189604
Hayama R, Izawa T, Shimamoto K: Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method. Plant Cell Physiol 2002, 43: 494–504. 10.1093/pcp/pcf059
Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K: Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 2003, 422: 719–722. 10.1038/nature01549
Hori K, Ogiso-Tanaka E, Matsubara K, Yamanouchi U, Ebana K, Yano M: Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J 2013, 76: 36–46.
Ishikawa R, Aoki M, Kurotani K, Yokoi S, Shinomura T, Takano M, Shimamoto K: Phytochrome B regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol Genet Genomics 2011, 285: 461–470. 10.1007/s00438-011-0621-4
Jeon JS, Chung YY, Lee S, Yi GH, Oh BG, An G: Isolation and characterization of an anther-specific gene, RA8, from rice ( Oryza sativa L.). Plant Mol Biol 1999, 39: 35–44. 10.1023/A:1006157603096
Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Yang K, Nam J, An K, Han MJ, Sung RJ, Choi HS, Yu JH, Choi JH, Cho SY, Cha SS, Kim SI, An G: T-DNA insertional mutagenesis for functional genomics in rice. Plant J 2000, 22: 561–570. 10.1046/j.1365-313x.2000.00767.x
Jeong DH, Green PJ: The role of rice microRNAs in abiotic stress responses. J Plant Biol 2013, 56: 187–197. 10.1007/s12374-013-0213-4
Jeong DH, Lee S, Kim SL, Hwang I, An G: Regulation of brassinosteroid responses by phytochrome B in rice. Plant Cell Environ 2007, 30: 590–599. 10.1111/j.1365-3040.2007.01644.x
Jeong DH, Park S, Zhai J, Gurazada SG, De Paoli E, Meyers BC, Green PJ: Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 2011, 23: 4185–4207. 10.1105/tpc.111.089045
Jones-Rhoades MW, Bartel DP, Bartel B: MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 2006, 57: 19–53. 10.1146/annurev.arplant.57.032905.105218
Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM: The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis . Plant Cell 2007, 19: 2736–2748. 10.1105/tpc.107.054528
Jung HJ, Park SJ, Kang H: Regulation of RNA metabolism in plant development and stress responses. J Plant Biol 2013, 56: 123–129. 10.1007/s12374-013-0906-8
Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC: P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev Cell 2003, 4: 205–217. 10.1016/S1534-5807(03)00025-X
Kim SL, Lee S, Kim HJ, Nam HG, An G: OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol 2007, 145: 1484–1494. 10.1104/pp.107.103291
Kim SR, Lee DY, Yang JI, Moon S, An G: Cloning vectors for rice. J Plant Biol 2009, 52: 73–78. 10.1007/s12374-008-9008-4
Kim SL, Choi M, Jung KH, An G: Analysis of the early-flowering mechanisms and generation of T-DNA tagging lines in Kitaake, a model rice cultivar. J Exp Bot 2013, 64: 4169–4182. 10.1093/jxb/ert226
Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M: Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 2002, 43: 1096–1105. 10.1093/pcp/pcf156
Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K: Hd3a and RFT1 are essential for flowering in rice. Development 2008, 135: 767–774. 10.1242/dev.008631
Komiya R, Yokoi S, Shimamoto K: A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development. 2009, 136: 3443–3450. 10.1242/dev.040170
Lauter N, Kampani A, Carlson S, Goebel M, Moose SP: microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci USA 2005, 102: 9412–9417. 10.1073/pnas.0503927102
Lee DY, An G: Two AP2 family genes, SUPERNUMERARY BRACT (SNB) and OsINDETERMINATE SPIKELET 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J 2012, 69: 445–461. 10.1111/j.1365-313X.2011.04804.x
Lee S, Jeon J, Jung KH, An G: Binary vectors for efficient transformation of rice. J Plant Biol 1999, 42: 310–316. 10.1007/BF03030346
Lee S, Kim J, Han JJ, Han MJ, An G: Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J 2004, 38: 754–764. 10.1111/j.1365-313X.2004.02082.x
Lee DY, Lee J, Moon S, Park SY, An G: The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J 2007, 49: 64–78. 10.1111/j.1365-313X.2006.02941.x
Lee YS, Jeong DH, Lee DY, Yi J, Ryu CH, Kim SL, Jeong HJ, Choi SC, Jin P, Yang J, Cho LH, Choi H, An G: OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J 2010, 63: 18–30.
Mathieu J, Yant LJ, Murdter F, Kuttner F, Schmid M: Repression of flowering by the miR172 target SMZ. PLoS Biol 2009, 7: e1000148. 10.1371/journal.pbio.1000148
Matsubara K, Yamanouchi U, Wang ZX, Minobe Y, Izawa T, Yano M: Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol 2008, 148: 1425–1435. 10.1104/pp.108.125542
Matsubara K, Yamanouchi U, Nonoue Y, Sugimoto K, Wang ZX, Minobe Y, Yano M: Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J 2011, 66: 603–612. 10.1111/j.1365-313X.2011.04517.x
Nair SK, Wang N, Turuspekov Y, Pourkheirandish M, Sinsuwongwat S, Chen G, Sameri M, Tagiri A, Honda I, Watanabe Y, Kanamori H, Wicker T, Stein N, Nagamura Y, Matsumoto T, Komatsuda T: Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proc Natl Acad Sci USA 2010, 107: 490–495. 10.1073/pnas.0909097107
Osugi A, Itoh H, Ikeda-Kawakatsu K, Takano M, Izawa T: Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice. Plant Physiol 2011, 157: 1128–1137. 10.1104/pp.111.181792
Park W, Li J, Song R, Messing J, Chen X: CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana . Curr Biol 2002, 12: 1484–1495. 10.1016/S0960-9822(02)01017-5
Park SJ, Kim SL, Lee S, Je BI, Piao HL, Park SH, Kim CM, Ryu CH, Park SH, Xuan YH, Colasanti J, An G, Han CD: Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J 2008, 56: 1018–1029. 10.1111/j.1365-313X.2008.03667.x
Peng LT, Shi ZY, Li L, Shen GZ, Zhang JL: Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter. Biochem Biophys Res Commun 2007, 360: 251–256. 10.1016/j.bbrc.2007.06.041
Peng LT, Shi ZY, Li L, Shen GZ, Zhang JL: Overexpression of transcription factor OsLFL1 delays flowering time in Oryza sativa . J Plant Physiol 2008, 165: 876–885. 10.1016/j.jplph.2007.07.010
Poethig RS: Small RNAs and developmental timing in plants. Curr Opin Genet Dev 2009, 19: 374–378. 10.1016/j.gde.2009.06.001
Ryu CH, You JH, Kang HG, Hur J, Kim YH, Han MJ, An K, Chung BC, Lee CH, An G: Generation of T-DNA tagging lines with a bidirectional gene trap vector and the establishment of an insertion-site database. Plant Mol Biol 2004, 54: 489–502. 10.1023/B:PLAN.0000038257.93381.05
Ryu CH, Lee S, Kim SL, Lee YS, Choi SC, Jeong HJ, Yi J, Park SJ, Han CD, An G: OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ 2009, 32: 1412–1427. 10.1111/j.1365-3040.2009.02008.x
Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU: Dissection of floral induction pathways using global expression analysis. Development 2003, 130: 6001–6012. 10.1242/dev.00842
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D: Specific effects of microRNAs on the plant transcriptome. Dev Cell 2005, 8: 517–527. 10.1016/j.devcel.2005.01.018
Sunkar R, Girke T, Jain PK, Zhu JK: Cloning and characterization of microRNAs from rice. Plant Cell 2005, 17: 1397–1411. 10.1105/tpc.105.031682
Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K: Hd3a protein is a mobile flowering signal in rice. Science 2007, 316: 1033–1036. 10.1126/science.1141753
Varallyay E, Burgyan J, Havelda Z: MicroRNA detection by northern blotting using locked nucleic acid probes. Nat Protoc 2008, 3: 190–196. 10.1038/nprot.2007.528
Voinnet O: Origin, biogenesis, activity of plant microRNAs. Cell 2009, 136: 669–687. 10.1016/j.cell.2009.01.046
Wei X, Xu J, Guo H, Jiang L, Chen S, Yu C, Zhou Z, Hu P, Zhai H, Wan J: DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol 2010, 153: 1747–1758. 10.1104/pp.110.156943
Wu G, Poethig RS: Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3 . Development 2006, 133: 3539–3547. 10.1242/dev.02521
Wu CY, You CJ, Li CS, Long T, Chen GX, Byrne ME, Zhang QF: RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc Natl Acad Sci USA 2008, 105: 12915–12920. 10.1073/pnas.0806019105
Xie K, Wu C, Xiong L: Genomic organization, differential expression, and interaction SQUAMOSA Promoter-Binding-Like transcription factors and microRNA156 in rice. Plant Physiol 2006, 142: 280–293. 10.1104/pp.106.084475
Xue WY, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q: Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 2008, 40: 761–767. 10.1038/ng.143
Yang J, Lee S, Hang R, Kim SR, Lee YS, Cao X, Amasino R, An G: OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J 2013, 73: 566–578. 10.1111/tpj.12057
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T: Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12: 2473–2483. 10.1105/tpc.12.12.2473
Yi J, An G: Utilization of T-DNA tagging lines in rice. J Plant Biol 2013, 56: 85–90. 10.1007/s12374-013-0905-9
Yoshikawa T, Ozawa S, Sentoku N, Itoh J, Nagato Y, Yokoi S: Change of shoot architecture during juvenile-to-adult phase transition in soybean. Planta 2013, 238: 229–237. 10.1007/s00425-013-1895-z
Zhou Y, Lu D, Li C, Luo J, Zhu BF, Zhu J, Shangguan Y, Wang Z, Sang T, Zhou B, Han B: Genetic control of seed shattering in rice by the APETALA2 transcription factor shattering abortion1. Plant Cell 2012, 24: 1034–1048. 10.1105/tpc.111.094383
Zhu QH, Helliwell CA: Regulation of flowering time and floral patterning by miR172. J Exp Bot 2011, 62: 487–495. 10.1093/jxb/erq295
Zhu QH, Upadhyaya NM, Gubler F, Helliwell CA: Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice ( Oryza sativa ). BMC Plant Biol 2009, 9: 149. 10.1186/1471-2229-9-149
Acknowledgements
We thank Sunghee Hong and Kyungsook An for generating the transgenic lines and managing the transgenic seeds, and Priscilla Licht for critical proofreading of the manuscript. We also thank the members of the Crop Biotech Center at Kyung Hee University for their participation in discussions. This work was supported in part by grants from the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center, No. PJ008128), Rural Development Administration, Republic of Korea; the Basic Research Promotion Fund, Republic of Korea (NRF-2007-0093862); and Kyung Hee University (20130214) to G.A.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Electronic supplementary material
12284_2014_31_MOESM1_ESM.pdf
Additional file 1: Figure S1.: Temporal expressions of AP2 genes and Ehd1 under SD. Figure S2. Construction of SNB and OsIDS1 over-expression plants. Figure S3. Expressions of floral regulators in the OsIDS1 OX. Figure S4. Expressions of floral regulators in the SNB OX. Figure S5. Expression levels of AP2s in the various flowering-time mutants. Figure S6. Phenotypes of miR172-resistant OsIDS1 overexpression. Figure S7. Verification of rSNB construct. Figure S8. Phenotypes of Ehd1 overexpression. (PDF 415 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lee, YS., Lee, DY., Cho, LH. et al. Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice 7, 31 (2014). https://doi.org/10.1186/s12284-014-0031-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-014-0031-4