Abstract
Key message
A minor QTL for heading date located on the long arm of rice chromosome 1 was delimitated to a 95.0-kb region using near isogenic lines with sequential segregating regions.
Abstract
Heading date and grain yield are two key factors determining the commercial potential of a rice variety. In this study, rice populations with sequential segregating regions were developed and used for mapping a minor QTL for heading date, qHd1. A total of 18 populations in six advanced generations through BC2F6 to BC2F11 were derived from a single BC2F3 plant of the indica rice cross Zhenshan 97 (ZS97)///ZS97//ZS97/Milyang 46. The QTL was delimitated to a 95.0-kb region flanked by RM12102 and RM12108 in the terminal region of the long arm of chromosome 1. Results also showed that qHd1 was not involved in the photoperiodic response, having an additive effect ranging from 2.4 d to 2.9 d observed in near isogenic lines grown in the paddy field and under the controlled conditions of either short day or long day. The QTL had pleiotropic effects on yield traits, with the ZS97 allele delaying heading and increasing the number of spikelets per panicle, the number of grains per panicle and grain yield per plant. The candidate region contains ten annotated genes including two genes with functional information related to the control of heading date. These results lay a foundation for the cloning of qHd1. In addition, this kind of minor QTLs could be of great significance in rice breeding for allowing minor adjustment of heading date and yield traits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In rice, heading date is a crucial determinant for adaption to different cultivation area and cropping seasons, and grain yield is an immediate indicator of the productivity. They are key factors determining the commercial potential of a rice variety. Mapping of quantitative trait loci (QTLs) for heading date, grain yield and yield component traits in rice has resulted in remarkable progresses on the genetic basis underlying the natural variation of these traits. A total of 711 QTLs for heading date and 2060 QTLs for yield traits have been documented in the Gramene database (http://archive.gramene.org/qtl/; Monaco et al. 2014). Nevertheless, only a small proportion of these QTLs have been fine mapped or cloned (Bai et al. 2012; Guo et al. 2013b).
Generally, genetic loci chosen for fine mapping and cloning have been those considered major QTLs due to large effects observed across different genetic backgrounds and environments. Many QTLs considered minor for having smaller individual effect which are often inconsistent across environments, thus remain poorly characterized. However, major QTLs for heading date and grain yield have shown a common association between delayed heading and increased grain yield, such as Ghd7 (Xue et al. 2008; Weng et al. 2014), DTH8/Ghd8/qHY-8/LH8 (Wei et al. 2010; Yan et al. 2011; Cai et al. 2012; Chen et al. 2014), Hd1 (Zhang et al. 2012) and Ghd7.1 (Yan et al. 2013). The utilization of these QTLs would significantly influence the regional and seasonal adaption of a rice variety.
On the other hand, minor QTLs could be of great significance in rice breeding for allowing minor adjustment or fine tuning of the traits. In this regard, minor QTLs that slightly delay or promote heading can allow plants to sometimes flower later, making full use of the temperature and sunlight of longer growing seasons, but other times allow earlier heading times as needed to avoid abiotic stress from high or low temperature during flowering or grain filling. For example, the study of DTH2 has revealed the great value of such a minor QTL in the historical expansion of rice into northern production areas of Asia noted for long-day growing seasons (Wu et al. 2013). In addition, slow increase of grain yield in the past few decades has been witnessed for rice varieties released in China (Yang et al. 2010; Yu et al. 2012), suggesting that the pyramiding of minor QTLs might be a main approach to increase the yield potential before a new breakthrough on the germplasm exploitation and gene deployment turns up.
We herein report the detection and fine mapping of a minor QTL for heading date of rice, qHd1, which was located in the terminal region of the long arm of chromosome 1. This QTL was not detected in primary QTL mapping using recombinant inbred lines of an indica rice cross between maintainer line Zhenshan 97 (ZS97) and restorer line Milyang 46 (MY46) (Zhang et al. 2011), but its effect was observed in two BC2F6 populations of the cross ZS973/MY46 and validated in populations with higher homogenous background.
Materials and methods
Plant materials
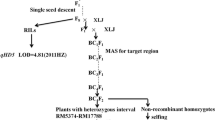
A total of 18 segregating populations were used, including 17 grown in the paddy field (Table 1) and one tested in the phytotron. They were constructed from the rice cross ZS973/MY46 as described below and illustrated in Fig. 1.
In a previous study (Guo et al. 2013a), three sets of populations were derived from a single BC2F3 plant of ZS973/MY46. Among them, two BC2F6 populations were selected as the starting materials for the present study. They were segregated in the intervals RM12026–RM12285 and RM12026–RM12108, respectively (Fig. 2a). The two BC2F6 populations, as well as their BC2F6:7 offspring, were used for QTL analysis on heading date. Henceforth, results of QTL detection were followed in each generation to develop new populations for validating and fine mapping qHd1.
Segregating populations used for mapping qHd1, showing sequential segregating regions in each generation and the delimitation of qHd1 to a 95.0-kb region flanked by RM12012 and RM12108. a Two sets of BC2F6 and BC2F6:7 populations; b four F2-type populations in BC2F8 and four sets of near isogenic lines in BC2F9; c three F2-type populations in BC2F10 and three sets of near isogenic lines in BC2F11
In BC2F7, four plants with sequential heterozygous segments extending from RM12026 to RM12108 were identified, from which four BC2F8 populations were derived and named C8001, C8002, C8003 and C8004, respectively (Fig. 2b). They were assayed with DNA markers in the target interval. In each population, non-recombinant homozygotes were identified and selfed to produce homozygous lines. Four sets of near isogenic lines (NILs) in BC2F9 were established and named C1, C2, C3 and C4, respectively (Fig. 2b).
In the mean time, BC2F8 plants carrying a heterozygous segment extending from RM12095 to RM12108 were selected. The resultant BC2F9 populations were assayed with DNA markers in the target interval. Three plants with sequential heterozygous segments were identified, from which three BC2F10 populations were developed and named CJ101, CJ102 and CJ103, respectively (Fig. 2c). Then, non-recombinant homozygotes were identified and selfed to produce homozygous lines. Three sets of NILs in BC2F11 were developed and named CJ1, CJ2 and CJ3, which were segregated in regions covering RM12095–Wn40348, Wn40348–RM12108 and RM12108, respectively.
Field experiments
The rice populations were tested at experimental fields of the China National Rice Research Institute located either in Hangzhou, Zhejiang, or Lingshui, Hainan (Table 1). In all the trials, the planting density was 16.7 cm between plants and 26.7 cm between rows.
Heading date (HD) was scored for each of the populations. For the two BC2F6, four BC2F8 and three BC2F10 populations, HD was measured on a single-plant basis. For the remaining populations, a randomized complete block design with two replications was applied. In each replication, one line was grown in a single row of 12 plants. HD was scored for each plant and averaged for each replication.
Four yield traits, including number of spikelets per panicle (NSP), number of grains per panicle (NGP), 1,000-grain weight (TGW) and grain yield per plant (GY), were also measured for one of the NIL sets in BC2F9 and all the three NIL sets in BC2F11. For NIL set C3 in BC2F9, five middle plants of each row were harvested in bulk and measured for the four yield traits. For the three NIL sets in BC2F11, two main panicles in each of the five middle plants were bulk-harvested and measured for NSP, NGP and TGW. The remaining panicles of the five middle plants were also harvested and added for the measurement of GY.
Phytotron experiments
To determine the photoperiodic response of qHd1, NIL set C4 consisting of two homozygous genotypes differing in a 120.2 kb region flanked by RM12095 and RM12108 (Fig. 2b) was grown in controlled chambers, under short-day (SD, 10 h light/14 h dark, 12 h 28 °C/12 h 23 °C) and long-day (LD, 14 h light/10 h dark, 12 h 28 °C/12 h 23 °C) conditions, respectively. Eight lines for each genotype with eight plants per line were grown. HD was scored for each plant.
DNA marker analysis
Total DNA was extracted following the method of Zheng et al. (1995). PCR amplification was performed according to Chen et al. (1997). The products of the SSR markers were visualized on 6 % non-denaturing polyacrylamide gels using silver staining, and that of the InDel marker were visualized on 2 % agarose gels using Gelred staining. All the SSR markers were selected from the Gramene database (http://www.gramene.org/). The InDel marker Wn40438 was designed according to the difference of the genomic sequences between ZS97 and MY46 detected by the whole-genome re-sequencing (Forward primer: ACATGTGTAGCATTAACAAC; Reverse primer: ATGATTTTGTTCAACCTTGG).
Parental survey of polymorphism was performed using SSR markers located in the terminal region of chromosome 1. Polymorphic markers were used to determine new segregating regions of the two BC2F6 populations, by testing two DNA bulks of 10 plants from each population. DNA markers shown in Fig. 2a were used for the selection of sequential heterozygotes in BC2F7, and those presented in Fig. 2b, c for constructing the NIL sets in BC2F9 and BC2F11, respectively.
In QTL mapping using the two BC2F6 populations and their BC2F7 progenies, RM12026 and RM12285 were used to test the 161 plants of GL6001, while RM12026 and RM12063 were chosen to assay the 222 plants of GL6002. In QTL mapping using the BC2F8 and BC2F10 populations, one marker was applied for each population. In BC2F8, the markers used were RM12026 for C8001, RM12063 for C8002, and RM12102 for C8003 and C8004. In BC2F10, the markers used were RM12102 for CJ101, and RM12108 for CJ102 and CJ103.
Data analysis
Mapmaker/Exp 3.0 (Lander et al. 1987) was used for the map construction of the two BC2F6 populations, and the genetic distances in centiMorgan (cM) were derived by Kosambi function. QTL analysis was performed with Windows QTL Cartographer 2.5 (Wang et al. 2012), in which the interval mapping was used for the BC2F6 and BC2F6:7 populations, while the single-marker analysis was used for the BC2F8 and BC2F10 populations. Using 1,000 permutation tests, the critical LOD values at P = 0.01 were determined, ranging from 1.9 to 4.1.
Two-way ANOVA was performed for the six NIL sets in BC2F9 or BC2F11 which were planted in the paddy field in Hangzhou (Table 1). Phenotypic differences between the two homozygous genotypic groups in each NIL set were tested using SAS procedure GLM (SAS Institute Inc 1999) as described previously (Dai et al. 2008). When significant differences were detected (P < 0.01), the same model was applied to estimate the genetic effect of the QTL, including additive effect and the proportion of phenotypic variance explained.
For NIL set C4 which was grown in the controlled chamber, one-way ANOVA was conducted to test the phenotypic differences between the two homozygous genotypic groups under SD and LD conditions, respectively.
Results
Detection of qHd1 using BC2F6 and BC2F6:7 populations
The two BC2F6 populations were previously found to be segregated in the intervals RM11448–RM11615 and RM11448–RM11787, and meanwhile to be ZS97 homozygous in the subsequent regions extended to RM11982, respectively (Guo et al. 2013a). An additional region towards the terminal end of the long arm of chromosome 1 was determined in the present study, extending from RM12026 that is 696 kb apart from RM11982, to RM12285 having a distance of 106 kb from the terminal end.
The regions RM12026–RM12285 and RM12026–RM12108 were found to be segregated in GL6001 and GL6002, respectively (Fig. 2a), and the segregation is highly distorted. No heterozygotes were found in GL6001, with the genotype ratio of ZS97:MY46 appearing to be 56:105 at RM12026 and 32:129 at RM12285. Three genotypes were detected in GL6002, with the genotype ratio of ZS97:MY46:heterozygote appeared to be 72:102:48 at RM12026, and 73:102:47 at RM12063.
Heading date was continuously distributed in the BC2F6 and BC2F6:7 populations, but it exhibited a single-QTL segregation when each population was classified based on the genotype of the common segregating marker RM12026 (Fig. 3). In the GL6001 population, the MY46 homozygous plants tended to flower earlier than the ZS97 homozygous plants, and the separation between the two groups was much more obvious in GL7001 when the phenotype was measured in the replicated trial. In GL6002 and GL7002, the MY46 homozygotes also tended to flower earlier than the ZS97 homozygotes, while the heterozygotes were more evenly distributed and scattered in the whole range.
Two segmental linkage maps were constructed for GL6001 and GL6002, spanning 15.4 cM for the interval RM12026–RM12285 and 1.1 cM for the interval RM12026–RM12063, respectively. They were used to detect the QTL responsible for the segregation of heading date in the two sets of BC2F6 and BC2F6:7 populations. A QTL was detected in each population, with the MY46 allele always promoting flowering (Table 2). The phenotypic variance explained was 49.5 and 25.0 % in the BC2F6 populations GL6001 and GL6002, and 68.3 and 48.3 % in the BC2F6:7 populations GL7001 and GL7002, respectively, with the additive effect ranging from 1.2 to 3.0 d. This putative QTL was named qHd1. In addition to the common segregating region RM12026–RM12108, the two flanking cross-over regions RM11982–RM12026 and RM12108–RM12138 may also segregated due to their unknown genotypes, thus qHd1 was located at RM11982–RM12138 (Fig. 2a).
Validation of qHd1
The effect of qHd1 on heading date was validated in BC2F8 and BC2F10 populations which were segregated in the expected 1:2:1 ratio. Results of the χ 2 test and the average heading date of the three genotypic groups in each population are presented in Table S1.
In BC2F8, significant association between marker genotypes and heading date was detected in all the population except C8001 (Table 2), suggesting that qHd1 was located in the common segregating region of C8002, C8003 and C8004. This QTL explained 11.0–24.6 % of the phenotypic variance in the three populations, with the MY46 allele promoting heading by 1.2–1.7 d and the dominance effect ranging from −0.1 d to 0.4 d. These effects were consistent with those detected in the BC2F6 and BC2F6:7 populations. Similar results were obtained using the three BC2F10 populations. The effects of qHd1 were again highly consistent with those detected in previous generations (Table 2). It could be concluded that qHd1 is a minor QTL with the additive genetic action mode.
Fine mapping of qHd1
The six NIL sets tested in the replicated trials in Hangzhou in 2012 or 2013 (Table 1) were used to measure the effect of qHd1 with lower random error and fine map qHD1. Results of two-way ANOVA for the heading date difference between two homozygous genotypic groups in each of the NIL sets are shown in Table 3.
Among the three NIL sets in BC2F9, significant variations on heading date were observed in C2 and C3 but not in C1, indicating that the allelic difference at qHd1 between ZS97 and MY46 was present in C2 and C3 but absent in C1. Therefore, qHd1 could be delimited to the region within the interval RM12072–RM12138, which included the region RM12095–RM12108 segregated in C2 and C3 but homozygous in C1, as well as the two flanking cross-over regions RM12072–RM12095 and RM12108–RM12138 due to their unknown genotypes (Fig. 2b). As estimated from C2 and C3, qHd1 explained 77.4 and 79.1 % of the phenotypic variance, with the MY46 allele reducing heading date by 2.6 d and 2.4 d, respectively (Table 3).
Among the three NIL sets in BC2F11 which were segregated within the candidate interval RM12072–RM12138, significant genotypic effects on heading date were detected in CJ1 and CJ2 but not in CJ3. Obviously, qHd1 was located in a place between RM12102 and RM12108 since no parts of this region were homozygous in CJ1 and CJ2 (Fig. 2c). This interval corresponds to a 95.0-kb region in the Nipponbare genome (www.gramene.org). As estimated from CJ1 and CJ2, qHd1 explained 74.2 and 69.8 % of the phenotypic variance, with the MY46 allele reducing heading date by 2.8 d and 2.5 d, respectively.
Photoperiodic response of qHd1
To investigate whether the qHd1 was involved in the photoperiodic response of rice, NIL set C4 which consisted of two homozygous genotypes differing in the interval RM12095-RM12108 covering qHd1 was examined in the controlled chambers. Under both the SD (10 h light/14 h dark) and LD (14 h light/10 h dark) conditions, highly significant difference (P < 0.0001) was shown between the heading date of the two genotypic groups (Table 4). The MY46 allele promoting flowering in both the SD and LD conditions, with the additive effect estimated as 2.9 d and 2.4 d, respectively. These results suggest that the allelic variation at qHd1 did not affect the photoperiodic sensitivity of rice.
Effect of qHd1 on yield traits
The effect of qHd1 on yield traits was first tested using one of the three NIL sets in BC2F9, C3 which consisted of two homozygous genotypes differing in the interval RM12095-RM12108 covering Hd1 (Fig. 2b), and then validated using the three NIL sets planted in BC2F11 (Table 1). Results of the two-way ANOVA on the four yield traits are presented in Table 3.
In the NIL set C3 which was planted in the paddy field in 2012, significant genotypic effect was observed for all the four yield traits analyzed except TGW, indicating that qHd1 had pleiotropic effects on yield traits. A consistent allelic direction was observed, with the ZS97 allele increasing NGP by 4.0, NSP by 5.2, and GY by 1.2 g, explaining 12.9 % of the phenotypic variance for NGP, 18.0 % for NSP, and 19.7 % for GY.
In the three NIL sets in BC2F11 which were planted in the paddy field in 2013, significant variations on NGP, NSP and GY were found in CJ1 and CJ2 which were segregated for qHd1, and it was not detected in CJ3 which was not segregated for qHd1. As estimated from CJ1 and CJ2, the contribution to the phenotypic variance was 20.6 and 36.4 % for NGP, 22.3 and 41.5 % for NSP, 10.0 and 10.2 % for GY, with the ZS97 allele increasing NGP by 7.7 and 8.2, NSP by 9.2 and 8.8, and GY by 1.1 and 1.0 g, respectively. Thus, the pleiotropism of qHd1 on yield traits was confirmed.
Discussion
In QTL analysis for heading date and yield traits of rice, a great attention has been paid to characterize major QTLs, whereas minor QTLs are poorly investigated. In this study, a minor QTL for heading date in rice, qHd1, was delimited into a 95.0-kb region flanked by RM12102 and RM12108 on the long arm of chromosome 1. It has also been shown that qHd1 did not respond to photoperiod and had pleiotropic effects on yield traits including grain number, spikelet number and grain yield. It is noteworthy that the minor effect of qHd1 on heading date has been stably detected across different generations, years and locations. Its allelic direction remained unchanged in all the trials, and the additive effect ranged from 2.4 d to 2.9 d as estimated from NIL sets which were either in the generation of BC2F9 or BC2F11, tested in 2012 or 2013, and examined in the paddy field or controlled environments.
Candidate genes for qHd1
According to the Rice Annotation Project database (http://rapdb.dna.affrc.go.jp/) (Sakai et al. 2013), ten genes in the target region for qHd1 defined by RM12102 and RM12108 were predicted. Six genes of them contain functional domains, and the others are hypothetical genes/proteins.
Two of the six genes with functional information are related to the control of heading date. Os01g0922800 encodes a protein containing a conserved SRF-like MADS domain and corresponds to the cloned heading date gene OsMADS51. However, OsMADS51 has shown a photoperiodic sensitivity and affects heading date in SD conditions only (Kim et al. 2007). This is obviously different from qHd1 which was found to be photoperiodic insensitive, suggesting that OsMADS51 is non-allelic to qHd1. The other gene, Os01g0922600, is a member of SQUAMOSA Promoter-Binding Protein-Like (SPL) gene family that is known to participate in the regulation of multiple plant developmental processes, such as phase transition and flowering (Huijser and Schmid 2011) and branch formation (Jiao et al. 2010; Miura et al. 2010). Thus, Os01g0922600 could be a potential candidate for qHd1.
The remaining four genes with functional information are Os01g0923200 encoding a AT.I.24-6-like protein, Os01g0923300 encoding a cystathionine beta-synthase, Os01g0923600 encoding an ankyrin domain-containing protein, and Os01g0923700 encoding a histidine kinase-type protein. More work is needed to examine whether these genes could be candidate for qHd1.
Sequence comparison between ZS97 and MY46 showed no polymorphism for the coding sequence in each of the ten annotated genes (data not shown), suggesting that the minor effect at qHd1 might result from the difference of the expression levels between the ZS97 allele and MY46 allele. Sequencing analysis of the promoter regions and expression analysis for each of the ten genes are underway to determine the most potential candidate underlying qHd1.
Application of sequential residual heterozygotes in QTL fine mapping
Determination of critical recombination breakpoints based on the genotypes of phenotypic extremes has been frequently applied to fine map major QTLs such as Hd1, Ghd7 and DTH8/Ghd8 (Yano et al. 2000; Xue et al. 2008; Wei et al. 2010; Yan et al. 2011), and this is greatly relied on the clear classification of the phenotypes. More generally, substitution mapping using multiple NILs with introgressions covering different portions of the target region was used for QTL fine mapping, e.g., gw3.1, qSS7, qLTG-9, qSV-1 and qSV-5c (Li et al. 2004, 2013; Qiu et al. 2012; Xie et al. 2014). Identification of sufficient amount of introgression lines for each segment required a great effort on phenotyping and genotyping.
In our study, a new strategy of QTL fine mapping was employed. A single residual heterozygote identified was selfed to produce a population segregated in the target region. Plants carrying heterozygous segments in a sequential order were identified from the population, which could be named sequential residual heterozygotes (SeqRHs). Populations with sequential segregating regions were produced from the selfed seeds of the SeqRHs. NIL sets of which each consisted of two homozygous genotypes differing in the corresponding heterozygous regions were developed. Significant and insignificant phenotypic variation between the two genotypic groups would be an indication of existence and absence of QTL segregation in the target region, respectively.
Generally, a step-by-step approach could be applied in QTL fine mapping using SeqRHs. First, a segregating population is derived from one residual heterozygote that carried a relatively large heterozygous segment. This population is used for validating the QTL effect in an isogenic background, and in the same time for the identification of SeqRHs with smaller heterozygous segments. Then, new populations are produced to narrow down the QTL region, and to identify new SeqRHs covering the narrowed region if necessary. Eventually, a QTL might be delimited in a region containing one or a few candidate genes.
This approach has an advantage of greatly reducing the cost of QTL fine mapping. While large populations are needed to screen sufficient recombinants in substitution mapping, as few as one residual heterozygote is required for a target region. Another advantage of this approach is the easy development of NILs with sufficient sample size for reliable phenotyping. A considerable quantity of lines could be easily identified for each genotype, since generally a few hundred seeds would be produced from a single rice plant.
Potential of qHd1 in rice breeding
Association of longer heading date with large panicle has been frequently observed in the fine mapping and cloning of major QTL for heading date in rice (Xue et al. 2008; Wei et al. 2010; Yan et al. 2011, 2013; Cai et al. 2012; Zhang et al. 2012). It has also been reported that the duration of panicle differentiation was closely related to the panicle size (Huang et al. 2006). The pre-flowering development in rice includes three successive phases (Chang et al. 1969): the basic vegetative phase (BVP), the photoperiod-sensitive phase (PSP), and the panicle-differentiation phase (PDP). Since qHd1 is not involved in the photoperiodic response, it might affect heading through its influence on BVP or PDP. Moreover, qHd1 exhibits pleiotropism for spikelet number and grain number, providing another evidence for the involvement of this QTL in the panicle differentiation during PDP.
In terms of the breeding application, selection of the genotypes at qHd1 would be helpful for fine tuning heading date, so as to make a full use of the temperature and sunlight while avoiding abiotic stresses. In this field, the two SSR markers flanking qHd1, RM12102 and RM12108, and the InDel marker Wn40348 located between them, could be used to increase the breeding efficiency by marker-assisted selection.
Author contributions
JYZ designed the experiments and selected the rice materials. JYC and LG performed most of the experiments. HM and JZY developed new DNA markers. YYC and HWZ performed some of the phenotyping. JYC and JYZ analyzed the data and wrote the paper.
References
Bai XF, Wu B, Xing Y (2012) Yield-related QTLs and their applications in rice genetic improvement. J Integr Plant Biol 54:300–311
Cai HY, Diao S, He YG, Zhang LP, Liu SJ, Zhu YG, Zhang ZH (2012) Genetic and physical mapping of qHY-8, a pleiotropic QTL for heading date and yield-related traits in rice. Euphytica 184:109–118
Chang TT, Li CC, Vergara BS (1969) Component analysis of duration from seeding to heading in rice by the basic vegetative phase and the photoperiod-sensitive phase. Euphytica 18:79–91
Chen X, Temnykh S, Xu Y, Cho GY, McCouch SR (1997) Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theor Appl Genet 95:553–567
Chen J, Li X, Cheng C, Wang Y, Qin M, Zhu H, Zeng R, Fu X, Liu Z, Zhang G (2014) Characterization of epistatic interaction of QTLs LH8 and EH3 controlling heading date in rice. Sci Rep 4:4263. doi:10.1038/srep04263
Dai WM, Zhang KQ, Wu JR, Wang L, Duan BW, Zheng KL, Cai R, Zhuang JY (2008) Validating a segment on the short arm of chromosome 6 responsible for genetic variation in the hull silicon content and yield traits of rice. Euphytica 160:317–324
Guo L, Wang K, Chen JY, Huang DR, Fan YY, Zhuang JY (2013a) Dissection of two quantitative trait loci for grain weight linked in repulsion on the long arm of chromosome 1 of rice (Oryza sativa L.). Crop J 1:70–76
Guo L, Zhang ZH, Zhuang JY (2013b) Quantitative trait loci for heading date and their relationship with genetic control of yield traits in rice (Oryza sativa). Rice Sci 20(1):1–12
Huang Y, Zhang LD, Zhang JW, Yuan DJ, Xu CG, Li XH, Zhou DX, Wang SP, Zhang QF (2006) Heterosis and polymorphisms of gene expression in an elite rice hybrid as revealed by a microarray analysis of 9198 unique ESTs. Plant Mol Biol 62:579–591
Huijser P, Schmid M (2011) The control of developmental phase transitions in plants. Development 138:4117–4129
Jiao YQ, Wang YH, Xue DW, Wang J, Yan MX, Liu GF, Dong GJ, Zeng DL, Lu ZF, Zhu XD, Qian Q, Li JY (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42:541–544
Kim SL, Lee S, Kim HJ, Nam HG, An G (2007) OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14 and Hd3a. Plant Physiol 145:1484–1494
Lander E, Green P, Abrahamson J, Barlow A, Daly M, Lincoln S, Newburg L (1987) MAPMAKER: an interactive computer program for constructing primary genetic maps of experimental and natural populations. Genomics 1:174–181
Li JM, Thomson M, McCouch S (2004) Fine mapping of a grain-weight quantitative trait locus in the pericentromeric region of rice chromosome 3. Genetics 168:2187–2195
Li LF, Liu X, Xie K, Wang YH, Liu F, Lin QY, Wang WY, Yang CY, Lu BY, Liu SJ, Chen LM, Jiang L, Wan JM (2013) qLTG-9, a stable quantitative trait locus for low-temperature germination in rice (Oryza sativa L.). Theor Appl Genet 126:2313–2322
Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42:545–549
Monaco MK, Stein J, Naithani S, Wei S, Dharmawardhana P, Kumari S, Amarasinghe V, Youens-Clark K, Thomason J, Preece J, Pasternak S, Olson A, Jiao Y, Lu Z, Bolser D, Kerhornou A, Staines D, Walts B, Wu G, D’Eustachio P, Haw R, Croft D, Kersey PJ, Stein L, Jaiswal P, Ware D (2014) Gramene 2013: comparative plant genomics resources. Nucleic Acids Res 42(D1):D1193–D1199
Qiu XJ, Gong R, Tan YB, Yu SB (2012) Mapping and characterization of the major quantitative trait locus qSS7 associated with increased length and decreased width of rice seeds. Theor Appl Genet 125:1717–1726
Sakai H, Lee SS, Tanaka T, Numa H, Kim J, Kawahara Y, Wakimoto H, Yang CC, Iwamoto M, Abe T, Yamada Y, Muto A, Inokuchi H, Ikemura T, Matsumoto T, Sasaki T, Itoh T (2013) Rice Annotation Project Database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol 54(2):e6 (1–11)
SAS Institute Inc (1999) SAS/STAT User’s Guide. SAS Institute, Cary
Wang S, Basten CJ, Zeng ZB (2012) Windows QTL cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC
Wei XJ, Xu JF, Guo HN, Jiang L, Chen SH, Yu CY, Zhou ZL, Hu PS, Zhai HQ, Wan JM (2010) DTH8 Suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol 153:1747–1758
Weng X, Wang L, Wang J, Hu Y, Du H, Xu C, Xing Y, Li X, Xiao J, Zhang Q (2014) Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol 164:735–747
Wu W, Zheng XM, Lu GW, Zhong ZZ, Gao H, Chen LP, Wu CY, Wang HJ, Wang Q, Zhou KN, Wang JL, Wu FQ, Zhang X, Guo XP, Cheng ZJ, Lei CL, Lin QB, Jiang L, Wang HY, Ge S, Wan JM (2013) Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc Natl Acad Sci USA 110:2775–2780
Xie LX, Tan ZW, Zhou Y, Xu RB, Feng LB, Xing YZ, Qi XQ (2014) Identification and fine mapping of quantitative trait loci for seed vigor in germination and seedling establishment in rice. J Integr Plant Biol. doi:10.1111/jipb.12190
Xue WY, Xing YZ, Weng XY, Zhao Y, Tang WJ, Wang L, Zhou HJ, Yu SB, Xu CG, Li XH, Zhang QF (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40:761–767
Yan WH, Wang P, Chen HX, Zhou HJ, Li QP, Wang CR, Ding ZH, Zhang YS, Yu SB, Xing YZ, Zhang QF (2011) A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol Plant 4:319–330
Yan WH, Liu HY, Zhou XC, Li QP, Zhang J, Lu L, Liu TM, Liu HJ, Zhang CJ, Zhang ZY, Shen GJ, Yao W, Chen HX, Yu SB, Xie WB, Xing YZ (2013) Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res 23:969–971
Yang S, Liao Q, Gu T, Hu X, Cheng B (2010) Review and analysis on rice cultivars registered in China. Chin Rice 16(2):1–4
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12:2473–2483
Yu YQ, Huang Y, Zhang W (2012) Changes in rice yields in China since 1980 associated with cultivar improvement, climate and crop management. Field Crop Res 136:65–75
Zhang ZH, Guo L, Zhu YJ, Fan YY, Zhuang JY (2011) Mapping of quantitative trait loci for heading date and plant height in two populations of Indica rice. Sci Agric Sin 44:3069–3077
Zhang ZH, Wang K, Guo L, Zhu YJ, Fan YY, Cheng SH, Zhuang JY (2012) Pleiotropism of the photoperiod-insensitive allele of Hd1 on heading date, plant height and yield traits in rice. PLoS One 7(12):e52538
Zheng KL, Huang N, Bennett J, Khush GS (1995) PCR-based marker-assisted selection in rice breeding. IRRI Discussion Paper Series No.12. Philippines: International Rice Research Institute
Acknowledgments
This work was supported by grants for the National Natural Science Foundation of China (31371605, 31221004) and the Ministry of Science and Technology of China (2012AA101102). We are also grateful to Dr. SR Pinson of the USDA-ARS National Rice Research Center for valuable discussion and comments.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Thomas H. Tai.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Chen, JY., Guo, L., Ma, H. et al. Fine mapping of qHd1, a minor heading date QTL with pleiotropism for yield traits in rice (Oryza sativa L.). Theor Appl Genet 127, 2515–2524 (2014). https://doi.org/10.1007/s00122-014-2395-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-014-2395-7