Abstract.
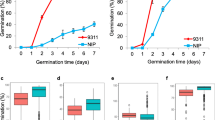
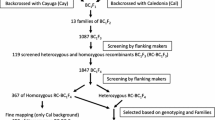
Two quantitative trait loci (QTLs) for seed dormancy (tentatively designated Sdr1) and heading date (Hd8) have been mapped to approximately the same region on chromosome 3 by interval mapping of backcross inbred lines derived from crosses between the rice cultivars Nipponbare (japonica) and Kasalath (indica). To clarify whether Sdr1 and Hd8 could be dissected genetically, we carried out fine-scale mapping with an advanced backcross progeny. We selected a BC4F1 plant, in which a small chromosomal region including Sdr1 and Hd8, on the short arm of chromosome 3, remained heterozygous, whereas all the other chromosomal regions were homozygous for Nipponbare. Days-to-heading and seed germination rate in the BC4F2 plants showed continuous variation. Ten BC4F2 plants with recombination in the vicinity of Sdr1 and Hd8 were selected on the basis of the genotypes of the restriction fragment length polymorphism (RFLP) markers flanking both QTLs. Genotypes of those plants for Sdr1 and Hd8 were determined by advanced progeny testing of BC4F4 families. Sdr1 was mapped between the RFLP markers R10942 and C2045, and co-segregated with C1488. Hd8 was also mapped between C12534S and R10942. Six recombination events were detected between Sdr1 and Hd8. These results clearly demonstrate that Sdr1 and Hd8 were tightly linked. Nearly isogenic lines for Sdr1 and Hd8 were selected by marker-assisted selection.





Similar content being viewed by others
References
Anderson JA, Sorrells ME, Tanksley SD (1993) RFLP analysis of genomic regions associated with resistance to pre-harvest sprouting in wheat. Crop Sci 33:453–459
Cai HW, Morishima H (2000) Genomic regions affecting seed shattering and seed dormancy in rice. Theor Appl Genet 100:840–846
Harushima Y, Yano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin SY, Antonio BA, Parco A, Kajiya H, Huang N, Yamamoto K, Nagamura Y, Kurata N, Khush GS, Sasaki T (1998) A high-density rice genetic linkage map with 2,275 markers using a single F2 population. Genetics 148:479–494
Kojima S, Takahashi Y, Kobayashi Y Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day condition. Plant Cell Physiol 43:1096–1105
Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4:403–410
Kurata N, Nagamura Y, Yamamoto K, Harushima Y, Sue N, Wu J, Antonio BA, Shomura A, Shimizu T, Lin SY, Inoue T, Fukuda A, Shimano T, Kuboki Y, Toyama T, Miyamoto Y, Kirihara T, Hayasaka K, Miyao A, Monna L, Zhong HS, Tamura Y, Wang ZX, Momma T, Umehara Y, Yano M, Sasaki T, Minobe Y (1994) A 300 kilobase interval genetic map of rice including 883 expressed sequences. Nat Genet 8:365–372
Lander ES, Botstein D (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–199
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Li B, Foley ME (1997) Genetic and molecular control of seed dormancy. Trends Plant Sci 2:384–389
Li ZK, Pinson SRM, Park WD (1995) Identification of quantitative trait loci (QTLs) for heading date and plant height in cultivated rice (Oryza sativa L.). Theor Appl Genet 91:374–381
Lin HX, Ashikari M, Yamanouchi U, Sasaki T, Yano M (2002) Identification and characterization of a quantitative trait locus, Hd9, controlling heading date in rice. Breed Sci 52:35–41
Lin HX, Liang ZW, Sasaki T, Yano M (2003) Fine mapping and characterization of quantitative trait loci Hd4 and Hd5 controlling heading date in rice. Breed Sci 53:51–59
Lin SY, Sasaki T, Yano M (1998) Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines. Theor Appl Genet 96:997–1003
Monna L, Lin HX, Kojima S, Sasaki T, Yano M (2002) Genetic dissection of a genomic region for a quantitative trait locus, Hd3, into two loci, Hd3a and Hd3b, controlling heading date in rice. Theor Appl Genet 104:772–778
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Nagato Y, Yoshimura A (1998) Report of the committee on gene symbolization, nomenclature and linkage groups. Rice Genet Newsl 15:13–74
Roberts EH (1962) Dormancy in rice seed. III. The influence of temperature, moisture and gaseous environment. J Exp Bot 13:75–94
Seshu DV, Sorrells ME (1986) Genetic studies on seed dormancy in rice. In: Rice genetics. International Rice Research Institute, Philippines, pp 369–382
Takahashi N (1997) Inheritance of seed germination and dormancy. In: Science of the rice plant. 3. Genetics. Food and Agriculture Policy Research Center, Tokyo, pp 348–359
Takahashi Y, Shomura A, Sasaki T, Yano M (2001) Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α-subunit of protein kinase CK2. Proc Natl Acad Sci USA 98:7922–7927
Wan J, Nakazaki T, Kawaura K, Ikehashi H (1997) Identification of marker loci for seed dormancy in rice (Oryza sativa L.). Crop Sci 37:1759–1763
Wu J, Maehara T, Shimokawa T, Yamamoto S, Harada C, Takazaki Y, Ono N, Mukai Y, Koike K, Yazaki J, Fujii F, Shomura A, Ando T, Kono I, Waki K, Yamamoto K, Yano M, Matsumoto T and Sasaki T (2002) A comprehensive rice transcript map containing 6591 expressed sequence tag sites. Plant Cell 14:525–535
Xiao J, Li J, Yuan L, Tanksley SD (1995) Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics 140:745–754
Xiao J, Li J, Yuan L, Tanksley SD (1996) Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor Appl Genet 92:230–244
Yamamoto T, Kuboki Y, Lin SY, Sasaki T, Yano M (1998) Fine mapping of quantitative trait loci Hd-1, Hd-2 and Hd-3, controlling heading date of rice, as single Mendelian factors. Theor Appl Genet 97:37–44
Yamamoto T, Lin HX, Sasaki T, Yano M (2000) Identification of heading date quantitative trait locus Hd6, and characterization of its epistatic interaction with Hd2 in rice using advanced backcross progeny. Genetics 154:885–891
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12:2473–2483
Yano M, Kojima S, Takahashi Y, Lin HX, Sasaki T (2001) Genetic control of flowering time in rice, a short-day plant. Plant Physiol 127:1425–1429
Yasue T, Asai Y (1968) Studies on the dormancy and viviparous germination in rice seed. Res Bull Fac Agric Gifu University 26:1–12
Zhou Y, Li W, Wu W, Chen Q, Mao D, Worland AJ (2001) Genetic dissection of heading time and its components in rice. Theor Appl Genet 102:1236–1242
Acknowledgements.
We thank Drs. K. Eguchi and Y. Ohtsuki, managing directors of the Rice Genome Research Program at the Institute of the Society for Techno-innovation of Agriculture, Forestry and Fisheries, for their advice and encouragement. This work was supported by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) and partly by the Ministry of Agriculture, Forestry and Fisheries, Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. Mackill
Rights and permissions
About this article
Cite this article
Takeuchi, Y., Lin, S.Y., Sasaki, T. et al. Fine linkage mapping enables dissection of closely linked quantitative trait loci for seed dormancy and heading in rice. Theor Appl Genet 107, 1174–1180 (2003). https://doi.org/10.1007/s00122-003-1364-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-003-1364-3