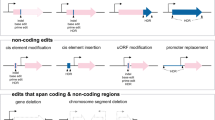
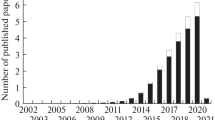
Abstract
Plant tissue culture (PTC) is a set of techniques for culturing cells, tissues, or organs in an aseptic medium with a defined chemical composition, in a controlled environment. Tissue culture, when combined with molecular biology techniques, becomes a powerful tool for the study of metabolic pathways, elucidation of cellular processes, genetic improvement and, through genetic engineering, the generation of cell lines resistant to biotic and abiotic stress, obtaining improved plants of agronomic interest, or studying the complex cellular genome. In this chapter, we analyze in general the use of plant tissue culture, in particular protoplasts and calli, in the implementation of CRISPR/Cas9 technology.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Loyola-Vargas VM, De-la-Peña C, Galaz-Ávalos RM et al (2008) Plant tissue culture. In: Walker JM, Rapley R (eds) Molecular biomethods handbook. Humana Press, Totowa, pp 875–904. https://doi.org/10.1007/978-1-60327-375-6_50
Urnov FD, Rebar EJ, Holmes MC et al (2010) Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11:636–646. https://doi.org/10.1038/nrg2842
Cermak T, Doyle EL, Christian M et al (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39:e82. https://doi.org/10.1093/nar/gkr218
Tsutsui H, Higashiyama T (2017) pKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana. Plant Cell Physiol 58:46–56. https://doi.org/10.1093/pcp/pcw191
Shalem O, Sanjana NE, Hartenian E et al (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. https://doi.org/10.1126/science.1247005
Komaroff AL (2017) Gene editing using CRISPR: why the excitement? JAMA 318:699–700. https://doi.org/10.1001/jama.2017.10159
Doudna JA, Charpentier E (2014) The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096-1–1258096-9. https://doi.org/10.1126/science.1258096
Yamamoto T (2015) Targeted genome editing using site-specific nucleases. ZFNs, TALENs, and the CRISPR/Cas9 system. Springer, Tokyo
Bannikov AV, Lavrov AV (2017) CRISPR/CAS9, the king of genome editing tools. Mol Biol 51:514–525. https://doi.org/10.1134/S0026893317040033
Yin K, Gao C, Qiu JL (2017) Progress and prospects in plant genome editing. Nat Plants 3:17107. https://doi.org/10.1038/nplants.2017.107
Mojica FJM, Díez-Villaseñor C, García-Martínez J et al (2005) Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60:174–182. https://doi.org/10.1007/s00239-004-0046-3
Gasiunas G, Barrangou R, Horvath P et al (2012) Cas-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A 109:E2579–E2586. https://doi.org/10.1073/pnas.1208507109
Nishimasu H, Ran FA, Hsu PD et al (2014) Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156:935–949. https://doi.org/10.1016/j.cell.2014.02.001
Jinek M, Chylinski K, Fonfara I et al (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. https://doi.org/10.1126/science.1225829
Feng Z, Y Mao NX et al (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci U S A 111:4632–4637. https://doi.org/10.1073/pnas.1400822111
Shankaran SS, Dahlem TJ, Bisgrove BW et al (2017) CRISPR/Cas9-directed gene editing for the generation of loss-of-function mutants in high-throughput zebrafish F0 screens. Curr Protoc Mol Biol 119:31.9.1–31.9.22. https://doi.org/10.1002/cpmb.42
Liang Z, Zong Y, Gao C (2016) An efficient targeted mutagenesis system using CRISPR/Cas in monocotyledons. Curr Protoc Plant Biol 1:329–344. https://doi.org/10.1002/cppb.20021
Gao J, Wang G, Ma S et al (2015) CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol Biol 87:99–110. https://doi.org/10.1007/s11103-014-0263-0
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278. https://doi.org/10.1016/j.cell.2014.05.010
Makarova KS, Haft DH, Barrangou R et al (2011) Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477. https://doi.org/10.1038/nrmicro2577
Liu X, S Wu JX et al (2017) Application of CRISPR/Cas9 in plant biology. Acta Pharm Sin B 7:292–302. https://doi.org/10.1016/j.apsb.2017.01.002
Feng Z, Zhang B, Ding W et al (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23:1229–1232. https://doi.org/10.1038/cr.2013.114
Mao Y, Zhang H, Xu N et al (2013) Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant 6:2008–2011. https://doi.org/10.1093/mp/sst121
Osakabe Y, Osakabe K (2015) Genome editing in higher plants. In: Yamamoto T (ed) Targeted genome editing using site-specific nucleases: ZFNs, TALENs, and the CRISPR/Cas9 system. Springer, Tokyo, pp 197–205. https://doi.org/10.1007/978-4-431-55227-7_13
Miao J, Guo D, Zhang J et al (2013) Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res 23:1233–1236. https://doi.org/10.1038/cr.2013.123
Xing HL, Dong L, Wang ZP et al (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14:327. https://doi.org/10.1186/s12870-014-0327-y
Korotkova AM, Gerasimova SV, Shumny VK et al (2017) Crop genes modified using the CRISPR/Cas system. Russ J Gen Appl Res 7:822–832. https://doi.org/10.1134/S2079059717050124
Li X, Xie Y, Zhu Q et al (2017) Targeted genome editing in genes and cis-regulatory regions improves qualitative and quantitative traits in crops. Mol Plant 10:1368–1370. https://doi.org/10.1016/j.molp.2017.10.009
Arora L, Narula A (2017) Gene editing and crop improvement using CRISPR-Cas9 system. Front Plant Sci 8:1932. https://doi.org/10.3389/fpls.2017.01932
Baltes NJ, Gil-Humanes J, Voytas DF (2017) Genome engineering and agriculture: opportunities and challenges. In: PWaB D (ed) Progress in molecular biology and translational science gene editing in plants. Academic Press, Cambridge, pp 1–26. https://doi.org/10.1016/bs.pmbts.2017.03.011
Chilcoat D, Liu ZB, Sander J (2017) Use of CRISPR/Cas9 for crop improvement in maize and soybean. In: PWaB D (ed) Progress in molecular biology and translational science gene editing in plants. Academic Press, Cambridge, pp 27–46. https://doi.org/10.1016/bs.pmbts.2017.04.005
Rouet P, Smih F, Jasin M (1994) Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol 14:8096–8106. https://doi.org/10.1128/MCB.14.12.8096
Puchta H, Dujon B, Hohn B (1996) Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci U S A 93:5055–5060
Puchta H, Fauser F (2014) Synthetic nucleases for genome engineering in plants: prospects for a bright future. Plant J 78:727–741. https://doi.org/10.1111/tpj.12338
Puchta H (2017) Applying CRISPR/Cas for genome engineering in plants: the best is yet to come. Curr Opin Plant Biol 36:1–8. https://doi.org/10.1016/j.pbi.2016.11.011
Jiang W, Zhou H, Bi H et al (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41:e18810. https://doi.org/10.1093/nar/gkt780
Li JF, Norville JE, Aach J et al (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31:688–691. https://doi.org/10.1038/nbt.2654
Nekrasov V, Staskawicz B, Weigel D et al (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31:691–693. https://doi.org/10.1038/nbt.2655
Shan Q, Wang Y, Li J et al (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31:686–688. https://doi.org/10.1038/nbt.2650
Upadhyay SK, Kumar J, Alok A et al (2013) RNA-guided genome editing for target gene mutations in wheat. G3 3:2233–2238. https://doi.org/10.1534/g3.113.008847
Xie K, Yang Y (2013) RNA-guided genome editing in plants using a CRISPR-Cas system. Mol Plant 6:1975–1983. https://doi.org/10.1093/mp/sst119
Belhaj K, Chaparro-Garcia A, Kamoun S et al (2013) Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9:1–10. https://doi.org/10.1186/1746-4811-9-39
Gao X, Chen J, Dai X et al (2016) An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol 171:1794–1800. https://doi.org/10.1104/pp.16.00663
Mao Y, Zhang Z, Feng Z et al (2016) Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol J 14:519–532. https://doi.org/10.1111/pbi.12468
Peterson BA, Haak DC, Nishimura MT et al (2016) Genome-wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in Arabidopsis. PLoS One 11:e0162169. https://doi.org/10.1371/journal.pone.0162169
Ji X, Zhang H, Zhang Y et al (2015) Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants. Nat Plants 1:15144. https://doi.org/10.1038/nplants.2015.144
Baltes NJ, Hummel AW, Konecna E et al (2015) Conferring resistance to geminiviruses with the CRISPR-Cas prokaryotic immune system. Nat Plants 1:15145. https://doi.org/10.1038/nplants.2015.145
Zhang H, Zhang J, Wei P et al (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12:797–807. https://doi.org/10.1111/pbi.12200
Xu R, Qin R, Li H et al (2017) Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol J 15:713–717. https://doi.org/10.1111/pbi.12669
Li M, Li X, Zhou Z et al (2016) Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci 7:377. https://doi.org/10.3389/fpls.2016.00377
Ma J, Chen J, Wang M et al (2018) Disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice. J Exp Bot. https://doi.org/10.1093/jxb/erx458
Wang Y, Cheng X, Shan Q et al (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32:947–951. https://doi.org/10.1038/nbt.2969
Zhang Y, Liang Z, Zong Y et al (2016) Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun 7:12617. https://doi.org/10.1038/ncomms12617
Char SN, Neelakandan AK, Nahampun H et al (2017) An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol J 15:257–268. https://doi.org/10.1111/pbi.12611
Liang Z, Zhang K, Chen K et al (2014) Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genomics 41:63–68. https://doi.org/10.1016/j.jgg.2013.12.001
Shi J, Gao H, Wang H et al (2017) ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J 15:207–216. https://doi.org/10.1111/pbi.12603
Fan D, Liu T, Li C et al (2015) Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Sci Rep 5:12217. https://doi.org/10.1038/srep12217
Jia H, Wang N (2014) Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS One 9:e93806. https://doi.org/10.1371/journal.pone.0093806
Chandrasekaran J, Brumin M, Wolf D et al (2016) Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol 17:1140–1153. https://doi.org/10.1111/mpp.12375
Peng A, Chen S, Lei T et al (2017) Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol J 15:1509–1519. https://doi.org/10.1111/pbi.12733
Jacobs TB, LaFayette PR, Schmitz RJ et al (2015) Targeted genome modifications insoybean with CRISPR/Cas9. BMC Biotechnol 15:1–10. https://doi.org/10.1186/s12896-015-0131-2
Li Z, Liu ZB, Xing A et al (2015) Cas9-guide RNA directed genome editing in soybean. Plant Physiol 169:960–970. https://doi.org/10.1104/pp.15.00783
Pan C, Ye L, Qin L et al (2016) CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci Rep 6:24765. https://doi.org/10.1038/srep24765
Ali Z, Abulfaraj A, Idris A et al (2015) CRISPR/Cas9-mediated viral interference in plants. Genome Biol 16:238. https://doi.org/10.1186/s13059-015-0799-6
Soyk S, Muller NA, Park SJ et al (2017) Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat Genet 49:162–168. https://doi.org/10.1038/ng.3733
Ito Y, Nishizawa-Yokoi A, Endo M et al (2015) CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem Biophys Res Commun 467:76–82. https://doi.org/10.1016/j.bbrc.2015.09.117
Ueta R, Abe C, Watanabe T et al (2017) Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci Rep 7:507. https://doi.org/10.1038/s41598-017-00501-4
Nekrasov V, Wang C, Win J et al (2017) Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep 7:482. https://doi.org/10.1038/s41598-017-00578-x
Butler NM, Atkins PA, Voytas DF et al (2015) Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS One 10:e0144591. https://doi.org/10.1371/journal.pone.0144591
Nishitani C, Hirai N, Komori S et al (2016) Efficient genome editing in apple using a CRISPR/Cas9 system. Sci Rep 6:31481. https://doi.org/10.1038/srep31481
Lawrenson T, Shorinola O, Stacey N et al (2015) Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol 16:258. https://doi.org/10.1186/s13059-015-0826-7
Yang H, Wu JJ, Tang T et al (2017) CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci Rep 7:7489. https://doi.org/10.1038/s41598-017-07871-9
Sugano SS, Shirakawa M, Takagi J et al (2014) CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol 55:475–481. https://doi.org/10.1093/pcp/pcu014
Morineau C, Bellec Y, Tellier F et al (2017) Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol J 15:729–739. https://doi.org/10.1111/pbi.12671
Jiang WZ, Henry IM, Lynagh PG et al (2017) Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol J 15:648–657. https://doi.org/10.1111/pbi.12663
Nejat N, Rookes J, Mantri NL et al (2017) Plant-pathogen interactions: toward development of next-generation disease-resistant plants. Crit Rev Biotechnol 37:229–237. https://doi.org/10.3109/07388551.2015.1134437
Shen L, Wang C, Fu Y et al (2018) QTL editing confers opposing yield performance in different rice varieties. J Int Plant Biol 60(2):89–93. https://doi.org/10.1111/jipb.12501
Yu Q, Wang B, Li N et al (2017) CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci Rep 7:11874. https://doi.org/10.1038/s41598-017-12262-1
Jia H, Orbovic V, Jones JB et al (2016) Modification of the PthA4 effector binding elements in type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccDpthA4:dCsLOB1.3 infection. Plant Biotechnol J 14:1291–1301. https://doi.org/10.1111/pbi.12495
Zhou JP, Deng K, Cheng Y et al (2017) CRISPR-Cas9 based genome editing reveals new insights into microRNA function and regulation in rice. Front Plant Sci 8:1598. https://doi.org/10.3389/fpls.2017.01598
Puchta H (2016) Using CRISPR/Cas in three dimensions: towards synthetic plant genomes, transcriptomes and epigenomes. Plant J 87:5–15. https://doi.org/10.1111/tpj.13100
Thakore PI, D'Ippolito AM, Song L et al (2015) Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Meth 12:1143–1149. https://doi.org/10.1038/nmeth.3630
Kaya H, Mikami M, Endo A et al (2016) Highly specific targeted mutagenesis in plants using Staphylococcus aureus Cas9. Sci Rep 6:26871. https://doi.org/10.1038/srep26871
Steinert J, Schiml S, Fauser F et al (2015) Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J 84:1295–1305. https://doi.org/10.1111/tpj.13078
Zetsche B, Gootenberg J, Abudayyeh O et al (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759–771. https://doi.org/10.1016/j.cell.2015.09.038
Tang X, Lowder LG, Zhang T et al (2017) A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants 3:17018. https://doi.org/10.1038/nplants.2017.18
Ancona V, Barra Caracciolo A, Grenni P et al (2017) Plant-assisted bioremediation of a historically PCB and heavy metal-contaminated area in southern Italy. New Biotechnol 38:65–73. https://doi.org/10.1016/j.nbt.2016.09.006
Sobariu DL, Fertu DIT, Diaconu M et al (2017) Rhizobacteria and plant symbiosis in heavy metal uptake and its implications for soil bioremediation. New Biotechnol 39:125–134. https://doi.org/10.1016/j.nbt.2016.09.002
Vaghari H, Jafarizadeh-Malmiri H, Anarjan N et al (2017) Hairy root culture: a biotechnological approach to produce valuable metabolites. In: Meena VS, Mishra PK, Bisht JK et al (eds) Agriculturally important microbes for sustainable agriculture, Plant-soil-microbe nexus, vol I. Springer, Singapore, pp 131–160. https://doi.org/10.1007/978-981-10-5589-8_7
Vázquez-Flota FA, Monforte-González M, de Lourdes Miranda-Ham M (2016) Application of somatic embryogenesis to secondary metabolite-producing plants. In: Loyola-Vargas VM, Ochoa-Alejo N (eds) Somatic embryogenesis: fundamental aspects and applications. Springer, Cham, pp 455–469. https://doi.org/10.1007/978-3-319-33705-0_25
Loyola-Vargas VM, Ochoa-Alejo N (2012) An introduction to plant cell culture: the future ahead. In: Loyola-Vargas VM, Ochoa-Alejo N (eds) Plant cell culture protocols, methods in molecular biology, vol 877. Humana Press, Heidelberg, pp 1–8. https://doi.org/10.1007/978-1-61779-818-4_1
Loyola-Vargas VM, Ochoa-Alejo N (2016) Somatic embryogenesis. Fundamental aspects and applications. Springer, Switzerland
Shen J, Fu J, Ma J et al (2014) Isolation, culture, and transient transformation of plant protoplasts. Curr Protoc Cell Biol 63:2.8.1–2.8.17. https://doi.org/10.1002/0471143030.cb0208s63
Davey MR, Anthony P, Power JB et al (2005) Plant protoplasts: status and biotechnological perspectives. Biotechnol Adv 23:131–171. https://doi.org/10.1016/j.biotechadv.2004.09.008
Cocking EC (2000) Plant protoplasts. In Vitro Cell Dev Biol Plant 36:77–82. https://doi.org/10.1007/s11627-000-0018-2
Davey MR, Cocking EC, Freeman J et al (1980) Transformation of petunia protoplasts by isolated Agrobacterium plasmids. Plant Sci Lett 18:307–313. https://doi.org/10.1016/0304-4211(80)90121-2
Ahmad MM, Ali A, Siddiqui S et al (2017) Methods in transgenic technology. In: Abdin MZ, Kiran U, Kamaluddin et al (eds) Plant biotechnology: principles and applications. Springer, Singapore, pp 93–115. https://doi.org/10.1007/978-981-10-2961-5_4
Phillips GC (2004) In vitro morphogenesis in plants - recent advances. In Vitro Cell Dev Biol Plant 40:342–345. https://doi.org/10.1079/IVP2004555
Quiroz-Figueroa FR, Rojas-Herrera R, Galaz-Ávalos RM et al (2006) Embryo production through somatic embryogenesis can be used to study cell differentiation in plants. Plant Cell Tissue Org 86:285–301. https://doi.org/10.1007/s11240-006-9139-6
Gaj MD (2004) Factors influencing somatic embryogenesis induction and plant regeneration with particular reference to Arabidopsis thaliana (L.) Heynh. Plant Growth Regul 43:27–47. https://doi.org/10.1023/B:GROW.0000038275.29262.fb
Cabrera-Ponce JL, López L, León-Ramírez CG et al (2015) Stress induced acquisition of somatic embryogenesis in common bean Phaseolus vulgaris L. Protoplasma 252:559–570. https://doi.org/10.1007/s00709-014-0702-4
Ochatt S, Revilla M (2016) From stress to embryos: some of the problems for induction and maturation of somatic embryos. In: Germanà MA, Lambardi M (eds) In vitro embryogenesis in higher plants. Springer, New York, pp 523–536. https://doi.org/10.1007/978-1-4939-3061-6_31
Salo HM, Sarjala T, Jokela A et al (2016) Moderate stress responses and specific changes in polyamine metabolism characterize scots pine somatic embryogenesis. Tree Physiol 36:292–402. https://doi.org/10.1093/treephys/tpv136
Krishnan SRS, Siril EA (2017) Auxin and nutritional stress coupled somatic embryogenesis in Oldenlandia umbellata L. Physiol Mol Biol Plants 23:471–475. https://doi.org/10.1007/s12298-017-0425-z
Loyola-Vargas VM, Ochoa-Alejo N (2016) Somatic embryogenesis. An overview. In: Loyola-Vargas VM, Ochoa-Alejo N (eds) Somatic embryogenesis. Fundamental aspects and applications. Springer, Switzerland, pp 1–10. https://doi.org/10.1007/978-3-319-33705-0_1
Canché-Moor RLR, Kú-González A, Burgeff C et al (2006) Genetic transformation of Coffea canephora by vacuum infiltration. Plant Cell Tissue Org 84:373–377. https://doi.org/10.1007/s11240-005-9036-4
Arroyo-Herrera A, Ku-Gonzalez A, Canche-Moo R et al (2008) Expression of WUSCHEL in Coffea canephora causes ectopic morphogenesis and increases somatic embryogenesis. Plant Cell Tissue Org 94:171–180. https://doi.org/10.1007/s11240-008-9401-1
Bouchabké-Coussa O, Obellianne M, Linderme D et al (2013) Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep 32:675–686. https://doi.org/10.1007/s00299-013-1402-9
Ochoa-Alejo N (2016) The uses of somatic embryogenesis for genetic transformation. In: Loyola-Vargas VM, Ochoa-Alejo N (eds) Somatic embryogenesis: fundamental aspects and applications. Springer, Cham, pp 415–434. https://doi.org/10.1007/978-3-319-33705-0_23
Ikeuchi M, Ogawa Y, Iwase A et al (2016) Plant regeneration: cellular origins and molecular mechanisms. Development 143:1442–1451. https://doi.org/10.1242/dev.134668
Woo JW, Kim J, Kwon SI et al (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33:1162–1164. https://doi.org/10.1038/nbt.3389
Shan Q, Wang Y, Li J et al (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nat Prot 9:2395–2410. https://doi.org/10.1038/nprot.2014.157
Zhou H, Liu B, Weeks DP et al (2014) Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res 42:10903–10914. https://doi.org/10.1093/nar/gku806
Cermák T, Curtin SJ, Gil-Humanes J et al (2017) A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29:1196–1217. https://doi.org/10.1105/tpc.16.00922
Gil-Humanes J, Wang Y, Liang Z et al (2017) High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J 89:1251–1262. https://doi.org/10.1111/tpj.13446
Kim D, Alptekin B, Budak H (2018) CRISPR/Cas9 genome editing in wheat. Funct Integr Genomics 18:31–41. https://doi.org/10.1007/s10142-017-0572-x
Liang Z, Chen K, Li T et al (2017) Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8:14261. https://doi.org/10.1038/ncomms14261
Wang Y, Zong Y, Gao C (2017) Targeted mutagenesis in hexaploid bread wheat using the TALEN and CRISPR/Cas systems. In: Bhalla PL, Singh MB (eds) Wheat biotechnology: methods and protocols. Springer, New York, pp 169–185. https://doi.org/10.1007/978-1-4939-7337-8_11
Zhu J, Song N, Sun S et al (2016) Efficiency and inheritance of targeted mutagenesis in maize using CRISPR-Cas9. J Genet Genomics 43:25–36. https://doi.org/10.1016/j.jgg.2015.10.006
Malnoy M, Viola R, Jung MH et al (2016) DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front Plant Sci 7:1904. https://doi.org/10.3389/fpls.2016.01904
Subburaj S, Chung SJ, Lee C et al (2016) Site-directed mutagenesis in Petunia x hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep 35:1535–1544. https://doi.org/10.1007/s00299-016-1937-7
Xu C, Liberatore KL, MacAlister CA et al (2015) A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat Genet 47:784–792. https://doi.org/10.1038/ng.3309
Andersson M, Turesson H, Nicolia A et al (2017) Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep 36:117–128. https://doi.org/10.1007/s00299-016-2062-3
Xu R, Li H, Qin R et al (2014) Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Rice 7:5. https://doi.org/10.1186/s12284-014-0005-6
Baysal C, Bortesi L, Zhu C et al (2016) CRISPR/Cas9 activity in the rice OsBEIIb gene does not induce off-target effects in the closely related paralog OsBEIIa. Mol Breed 36:108. https://doi.org/10.1007/s11032-016-0533-4
Li J, Du Y Sun J et al (2017) Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol Plant 10:526–529. https://doi.org/10.1016/j.molp.2016.12.001
Minkenberg B, Xie K, Yang Y (2017) Discovery of rice essential genes by characterizing a CRISPR-edited mutation of closely related rice MAP kinase genes. Plant J 89:636–648. https://doi.org/10.1111/tpj.13399
Holme IB, Wendt T, Gil-Humanes J et al (2017) Evaluation of the mature grain phytase candidate HvPAPhy_a gene in barley (Hordeum vulgare L.) using CRISPR/Cas9 and TALENs. Plant Mol Biol 95:111–121. https://doi.org/10.1007/s11103-017-0640-6
Nonaka S, Arai C, Takayama M et al (2017) Efficient increase of g-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci Rep 7:7057. https://doi.org/10.1038/s41598-017-06400-y
Svitashev S, Schwartz C, Lenderts B et al (2016) Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat Commun 7:13274. https://doi.org/10.1038/ncomms13274
Odipio J, Alicai T, Ingelbrecht I et al (2017) Efficient CRISPR/Cas9 genome editing of phytoene desaturase in cassava. Front Plant Sci 8:1780. https://doi.org/10.3389/fpls.2017.01780
Brooks C, Nekrasov V, Lippman ZB et al (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol 166:1292–1297. https://doi.org/10.1104/pp.114.247577
Xu C, Park SJ, Van Eck J et al (2016) Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev 30:2048–2061. https://doi.org/10.1101/gad.288415.116
Wang L, Chen L, Li R et al (2017) Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J Agric Food Chem 65:8674–8682. https://doi.org/10.1021/acs.jafc.7b02745
Wang S, Zhang S, Wang W et al (2015) Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep 34:1473–1476. https://doi.org/10.1007/s00299-015-1816-7
Zhang B, Yang X, Yang C et al (2016) Exploiting the CRISPR/Cas9 system for targeted genome mutagenesis in petunia. Sci Rep 6:20315. https://doi.org/10.1038/srep20315
Braatz J, Harloff HJ, Mascher M et al (2017) CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus L.). Plant Physiol 174(2):935–942. https://doi.org/10.1104/pp.17.00426
Jia H, Zhang Y, Orbovi-ç V et al (2017) Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol J 15:817–823. https://doi.org/10.1111/pbi.12677
Ron M, Kajala K, Pauluzzi G et al (2014) Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol 166:455–469. https://doi.org/10.1104/pp.114.239392
Michno JM, Wang X, Liu J et al (2015) CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web-tool and a modified Cas9 enzyme. GM Crops Food 6:243–252. https://doi.org/10.1080/21645698.2015.1106063
Kirchner TW, Niehaus M, Debener T et al (2017) Efficient generation of mutations mediated by CRISPR/Cas9 in the hairy root transformation system of Brassica carinata. PLoS One 12:e0185429. https://doi.org/10.1371/journal.pone.0185429
Watanabe K, Kobayashi A, Endo M et al (2017) CRISPR/Cas9-mediated mutagenesis of the dihydroflavonol-4-reductase-B (DFR-B) locus in the Japanese morning glory Ipomoea (Pharbitis) nil. Sci Rep 7:10028. https://doi.org/10.1038/s41598-017-10715-1
Loyola-Vargas VM, Miranda-Ham ML (1995) Root culture as a source of secondary metabolites of economic importance. Rec Advan Phytochem 29:217–248. https://doi.org/10.1007/978-1-4899-1778-2_10
Flores HE, Vivanco JM, Loyola-Vargas VM (1999) “Radicle” biochemistry: the biology of root-specific metabolism. Trends Plant Sci 4:220–226. https://doi.org/10.1016/S1360-1385(99)01411-9
David C, Chilton MD, Tempé J (1984) Conservation of T-DNA in plants renegerated from hairy root cultures. Bio Technol 2:73–76. https://doi.org/10.1038/nbt0184-73
Hamill JD, Rhodes MJC (1988) A spontaneous, light independent and prolific plant regeneration response from hairy roots of Nicotiana hesperis transformed by Agrobacterium rhizogenes. J Plant Physiol 133:506–509. https://doi.org/10.1016/S0176-1617(88)80046-4
Jouanin L, Guerche P, Pamboukdjian N et al (1987) Structure of T-DNA in plants regenerated from roots transformed by Agrobacterium rhizogenes strain A4. Mol Gen Genet 206:387–392. https://doi.org/10.1007/BF00428876
Springer NM, Schmitz RJ (2017) Exploiting induced and natural epigenetic variation for crop improvement. Nat Rev Genet 18:563–575. https://doi.org/10.1038/nrg.2017.45
Abudayyeh OO, Gootenberg JS, Essletzbichler P et al (2017) RNA targeting with CRISPR-Cas13. Nature 550:280–284. https://doi.org/10.1038/nature24049
Cox DBT, Gootenberg JS, Abudayyeh OO et al (2017) RNA editing with CRISPR-Cas13. Science 358:1019–1027. https://doi.org/10.1126/science.aaq0180
Acknowledgments
The work from VMLV laboratory was supported by a grant received from the National Council for Science and Technology (CONACyT, Frontiers of Sciences, 1515).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Loyola-Vargas, V.M., Avilez-Montalvo, R.N. (2018). Plant Tissue Culture: A Battle Horse in the Genome Editing Using CRISPR/Cas9. In: Loyola-Vargas, V., Ochoa-Alejo, N. (eds) Plant Cell Culture Protocols. Methods in Molecular Biology, vol 1815. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-8594-4_7
Download citation
DOI: https://doi.org/10.1007/978-1-4939-8594-4_7
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-8593-7
Online ISBN: 978-1-4939-8594-4
eBook Packages: Springer Protocols