Abstract
Background
Due to progress in science and technology, several harmful polycyclic aromatic hydrocarbons are synthesized and released into the environment. In the present investigation, a phenanthrene- and pyrene-degrading white rot fungi Ganoderma lucidum strain CCG1 was isolated from the Janjgir Champa district of Chhattisgarh, India, and then the degradation of phenanthrene and pyrene was estimated by high-performance liquid chromatography.
Results
It was found that G. lucidum able to degrade 99.65% of 20 mg/L of phenanthrene and 99.58% of pyrene in mineral salt broth after 30th day of incubation at 27 °C. G. lucidum produced significant amounts (p < 0.0001) of ligninolytic enzymes (Laccase, lignin peroxidase and manganese peroxidase) in the phenanthrene- and pyrene-containing mineral salt broth. G. lucidum produced maximum 10,788.00 U/L laccase, 3283.00 U/L Lignin peroxidase and 47,444.00 U/L Manganese peroxidase enzymes in the presence of phenanthrene and produced maximum 10,166.00 U/L laccase, 3613.00 U/L lignin peroxidase and 50,977.00 U/L manganese peroxidase enzymes in the presence of pyrene. Therefore, G. lucidum will be a potent phenanthrene and pyrene degrader from the environment.

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Background
Polycyclic aromatic hydrocarbons (PAHs) are a type of xenobiotic compounds that are released into the environment due to incomplete combustion of organic materials such as oil, petroleum gas, wood, municipal and urban waste (Juhasz and Naidu 2000; Ijoma and Tekere 2017; Kadri et al. 2017). PAHs are made up of three or more fused benzene rings, arranged in the linear, cluster or angular arrangements (Di Toro et al. 2000). PAHs are thermodynamically stable due to their strong negative resonance energy. PAHs are highly bioaccumulative compound in soil and sediments because of their high hydrophobicity and low volatility (Singh 2006). The exposure to PAHs causes acute symptoms such as eye irritation, vomiting, and nausea. High concentration of PAHs can also cause kidney and liver damage, skin inflammation, suppress immune reactions, an embryotoxic effect during pregnancy and also show genotoxic, carcinogenic, mutagenic and teratogenic effect (Rostami and Juhasz 2011; Rengarajan et al. 2015). Thus, it is necessary to remediate the PAH compound from the environment. The United States Environmental Protection Agency (USEPA) has been listed 16 PAHs as priority pollutants, including phenanthrene and pyrene (Jin et al. 2007; Puglisi et al. 2007; Wu et al. 2016). Phenanthrene and pyrene are considered as model compounds for the PAH degradation study because they were found most abundantly in the PAH-polluted environment (Bezalel et al. 1996; Makkar and Rockne 2003; Mrozik and Piotrowska-Seget 2010).
Bioremediation is an efficient technique which utilizes the metabolic potential of microorganisms to degrade and transform of PAHs compound to non-toxic compound with less input of chemicals, energy and time (Haritash and Kaushik 2009; Adenipekun et al. 2012). Bioremediation technique removes the toxic compound permanently. It is a less-expensive technique can be used in the site directly (Boopathy 2000). Many bacterial species were able to degrade PAHs (Aitken et al. 1998; Singh 2006) but now researchers concentrate on the degradation of PAHs from the terrestrial and aquatic environment by fungi, because fungi remove PAHs by their cometabolic pathway. Fungi do not use PAHs as a source of their carbon and energy (Wunder et al. 1994; Pothuluri et al. 1995; Casillas et al. 1996). Fungi play robust roles in ecosystem functioning by regulating the flow of nutrients and energy over their mycelia networks; they act like natural ecosystem engineers (Lawton and Jones 1995; Tisma et al. 2010). White rot fungi perform a significant role in the process of mycoremediation (Dominguez et al. 2005; Casas et al. 2009; Wong 2009). White rot fungi produced nonspecific extracellular ligninolytic enzymes: laccase, lignin peroxidase (LiP) and manganese peroxidase (MnP) (Kitamura et al. 2005). These ligninolytic enzymes play an important role in the transformation and mineralization of various organic pollutants (Pointing 2001; Lee et al. 2004; Casas et al. 2009; Wang et al. 2009). The common white rot fungi used for the degradation of aromatic ring or pretreatment of lignin through ligninolytic enzymes are Phanerochaete chrysosporium, Pleurotus ostreatus, Trametes versicolor, Armillaria sp., and Bjerkandera adusta (Rigas et al. 2009; Balesteros et al. 2014; Kumar and Sharma 2017).
The objective of this study is to isolate polycyclic aromatic hydrocarbon-degrading white rot fungi G. lucidum from Chhattisgarh, India, identified by 18S rRNA sequencing and used for the analysis of phenanthrene and pyrene degradation on the basis of their ligninolytic activity. Degradation of phenanthrene and pyrene was estimated by HPLC and the ligninolytic enzyme production was quantified in phenanthrene- and pyrene-containing media.
Methods
Chemicals
Phenanthrene, pyrene, guaiacol, azure B, and 2,6-dimethoxy phenol (2,6-DMP) were purchased from Himedia, India. All other chemicals and solvents (ethyl acetate, acetonitrile) used were of analytical grade purchased from Himedia, India.
Microorganism and culture condition
A white rot fungi strain CCG1, found on the decayed wood surface, was collected from the Janjgir Champa district of (Latitude 21°58′13.9908ʺ, Longitude 82°28′30.9936ʺ and Elevation 258.7 m) Chhattisgarh, India, fungi were isolated from the fruiting body by the spore-drop method according to Choi et al. (1999). A piece of cap tissue was cut from the fruiting body and transferred in the top of a Petri dish containing sabouraud dextrose agar (SDA) media composition (g/L): dextrose (40), peptone (10), agar (15) and streptomycin (500 mg/L) antibiotic to inhibit the bacterial growth). After the pure culture was obtained, culture was maintained in SDA media and stored at 4 °C.
Ligninolytic enzyme production
To investigate the enzyme production, 8 mm of fungal mycelium disc was transferred in the 20 mL of mineral salt broth (composition g/L: glucose—10, KH2PO4—2, MgSO4·7H2O—0.5, CaCl2·2H2O—0.1, ammonium tartrate—0.2 and trace element solution—10 (mL) with 20 mg/L phenanthrene and 20 mg/L pyrene separately (0.2 µm membrane filter sterilized). A trace element solution comprised of (in mg/L) FeSO4·7H2O (12), MnSO4·7H2O (3), ZnSO4·7H2O (3), CoSO4·7H2O (1), (NH4)6Mo7O24·4H2O (1) (Hadibarata and Kristanti 2012). Then, incubate the culture at 27 °C in a rotatory shaker incubator for 2, 5, 10, 15, 20, 25 and 30 days. The ligninolytic enzyme activity was investigated in the presence of phenanthrene and pyrene separately. Control experiment (containing 20 mL of mineral salt broth with 20 mg/L phenanthrene and pyrene separately), was done at the same time when sample experiment was done. All the experiments were performed in triplicates.
The production of ligninolytic enzyme was investigated as the following procedure:
Laccase assay
To investigate the laccase enzyme production, take 3 mL of reaction mixture containing 0.5 mL of the enzyme extract, 1.5 mL of sodium acetate buffer (10 mM, pH 5.0) and 1 mL of guaiacol (2 mM), then incubated for 2 h and absorbance read at 450 nm. The laccase enzyme activity has been expressed in international units per liter of enzyme extract (U/L) (Sandhu and Arora 1985).
Lignin peroxidase assay
For the lignin peroxidase enzyme production, take 0.5 mL of the culture filtrate, 1 mL of 125 mM sodium tartrate buffer (pH 3.0), 0.5 mL of 0.16 mM azure B, then add 0.5 mL of 2 mM hydrogen peroxide, after addition of hydrogen peroxide, reaction was initiated. One unit of enzyme activity was expressed as an O.D. decrease at 651 nm of 0.1 units per minute per liter of the culture filtrate (Archibald 1992).
Manganese peroxidase assay
According to de Jong et al. (1992) manganese peroxidase activity was accessed by the oxidation of 2,6-DMP at 468 nm. Take total 3 mL of reaction mixture containing 0.5 mL of culture filtrate, 1 mL of sodium tartrate buffer (50 mM, pH 4.0) and 1 mL of 2 mM 2,6-DMP. The reaction was started by the addition of 0.5 mL of 0.4 mM hydrogen peroxide.
Degradation of PAHs (phenanthrene and pyrene)
To investigate the phenanthrene and pyrene degradation mineral salt broth containing 20 mg/L phenanthrene and pyrene were taken separately. Then, 8 mm diameter, one culture mycelium disc was transferred in the broth media, control set containing mineral salt broth with phenanthrene and pyrene separately. Incubate the culture at 27 °C in a rotatory shaker incubator for 2, 5, 10, 15, 20, 30 days.
Extraction of degraded PAHs and analyzed by HPLC
After the incubation period degraded phenanthrene and pyrene were extracted from the medium and mycelium using ethyl acetate, equal volumes of ethyl acetate were mixed with incubated mineral broth and kept 160 rpm in a rotatory shaker for overnight, centrifuge for 10 min in 12,000g, fungal body was separated from liquid media by filtration. Separated filtrate mycelium was dried in oven at 50 °C; dry weight of mycelium was analyzed. The remaining liquid medium was shaken vigorously for 10 min and then ethyl acetate was separated by separator funnel. Extraction procedure was repeated three times with ethyl acetate (Bezalel et al. 1996). The extracted ethyl acetate was dried and evaporated with the help of sodium sulfate using vacuum evaporator. The residue obtained was redissolved in acetonitrile for further HPLC analysis. Biodegradation efficiency (%) was calculated by as following formula given by Bishnoi et al. (2008).
where C0 initial concentration of PAHs (mg/L), Ce equilibrium concentration of PAHs (mg/L).
HPLC analysis
The PAH degradation efficiency was assessed by HPLC (UFLC, Shimadzu prominence) using Luna5u, C18, 100 Å column (250 × 4.6 nm), pump (LC—20AT), detector (PDA) was set at 254 nm. A mixture of acetonitrile and water (70:30) used as mobile phase with 1.0 mL/min flow rate for 20 min in case of phenanthrene analysis and 30 min for pyrene analysis. Concentration of phenanthrene and pyrene was calculated by the comparison of peak area of sample chromatogram and peak area of standard chromatogram (Bishnoi et al. 2005).
where APAH Peak area of chromatogram of sample, ASTD Peak area of standard PAHs compound, CSTD Concentration of standard PAH compounds (mg/L).
Morphological and molecular characterization
Morphological observation of strain CCG1 was performed and molecular identification was conducted through 18S rRNA. 18S rRNA sequencing was carried out by the service provider Chromous Biotech Pvt. Ltd., Bangalore, India. They amplified the extracted DNA genome of the fungal strain CCG1 through polymerase chain reaction (PCR). The process of PCR amplification contains DNA template, primer (FP: GTAGTCATAKGCTNGTCTS, RP: GARACCTDGTTAVGACTY), taq DNA polymerase enzyme, dNTPs, buffer. PCR amplification was performed with following reactions: Started with 1st cycle at 94 °C for 5 min, 35 cycles at 940 °C for 30 s, 55 °C for 30 s, 72 °C for 2 min and ended with 1 cycle at 72 °C for 7 min. After that, sequencing reaction was performed using ABI 3500XL Genetic Analyzer sequencing machine. Later, closely related sequence was obtained from the National Center for Biotechnology Information (NCBI) database. Multiple alignments of all sequences were performed using CLUSTAL W. The neighbor-joining tree was constructed by Mega 7.0 software programs using bootstrapping at 1000 bootstraps (Tamura et al. 2007).
Data analysis
One-way ANOVA (analysis of variance) test was performed to analysis of the difference in ligninolytic enzyme production in the presence of phenanthrene and pyrene in mineral salt broth as compared to control; by applying Dunnett’s multiple comparison test with the help of statistical program Graph Pad Prism 7.
Results and discussion
Isolation and identification of collected fungal strain
Fungal strain CCG1 was collected from the hardwood stump of Janjgir Champa district of Chhattisgarh, India during the rainy season. A collected CCG1 fungus was isolated from their fruiting body. Further strain CCG1 was used for the PAHs degradation investigation on the basis of their ligninolytic enzyme activity (Agrawal et al. 2017).
Morphological and molecular characterization
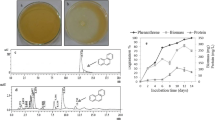
The fruiting body (Fig. 1a) of fungal stain CCG1 was annual, umbrella shaped with a semicircular reniform pileus (8–10 cm wide and 1–2 cm thick). Epicutis dull yellowish brown colour, lustrous, gills on the underside of pileus are white with tubules containing numerous pores. Stipe lateral 5–7 cm long, 1–2 cm diameter, reddish brown, lustrous.
On the basis of morphological and molecular characterization through 18S rRNA gene sequencing fungal strain CCG1 was identified as G. lucidum (Accession no. KY464926). A phylogenetic tree was created with the gene sequence of strain CCG1 and with their similar sequences of reference strain which are obtained from NCBI database. On the basis of phylogenetic tree (Fig. 1b) strain, CCG1 (Accession no. KY464926) was more similar with G. lucidum strain GI-16 (Accession no. FJ379279.1) and another 99% similar strains are Ganoderma australe strain Wu 9302-56 (Accession no. AY336763.1), Amauroderma sp. MUCL40278 (Accession no. AF255199.1), Lentinus sp. WR2 (Acccession No. GQ899200.1), Lentinus tigrinus (Accession no. AY946269.1), Hexagonia hirta strain Wu 9906-35 (Accession no. AY336759.1), Ganoderma tsugae isolate AFTOL-ID 771 (Accession no. AY705969.1), Ganoderma boninense (Accession no. AF255198.1), Coriolopsis byrsina strain CRM-46 (Accession no. AY336773.1), Earliella scabrosa strain Wu 9704-83 (Accession no. AY336766.1), Hexagonia hirta strain CBS 515.96 (Accession no. AY336760.1), Hexagonia pobeguinii strain CBS 510.96 (Accession no. AY336758.1), Ganoderma australe (Accession no. AF026629.1), Diplomitoporus crustulinus strain FP 101824 (Accession no. AY336770.1), Diplomitoporus lindbladii strain HHB 5629 (Accession no. AY336768.1), Microporus xanthopus strain Wu 9705-42 (Accession no. AY336756.1), Lignosus rhinocerus strain CH2 (Accession no. FJ899144.1), and Coriolopsis polyzona strain OH-184-SP (Accession no. AY336771.1).
Ligninolytic enzyme production
Ligninolytic enzyme production was investigated in the fungal strain CCG1 after 2, 5, 10, 15, 20, 25 and 30 days of incubation in mineral salt broth with PAHs compound. The production of ligninolytic enzyme in fungal strain CCG1 increased with the incubation period and maximum laccase, LiP and MnP activity 10,788.00, 3283.00 and 47,444.00 U/L were found in 15, 15 and 10 days of incubation period, respectively, in the phenanthrene media (Table 1). In the presence of pyrene, 10,166.00, 3613.00 and 50,977.00 U/L laccase, LiP and MnP activity was investigated after 30, 25 and 15 days, respectively (Table 1). Strain CCG1 produced a significant amount of ligninolytic enzyme in the phenanthrene- and pyrene-containing mineral salt broth as compared to without phenanthrene and pyrene up to p < 0.0001 significant level. Ting et al. (2011) also reported that the production of laccase by G. lucidum was enhanced, when the PAH is present in the media. The production of ligninolytic enzymes activated during secondary metabolic stage and increased in the carbon-, sulfur- and nitrogen-limiting condition in media (Heinzkill et al. 1998). Zhuo et al. (2011) also found that during the dye decolourization process, laccase activity increased by Ganoderma sp. En3. Ting et al. (2011) found that G. lucidum BCRC 36021 showed maximum 670.00 U/L laccase activity in the presence of 20 mg/L PAHs (phenanthrene and pyrene), but G. lucidum strain CCG1 produced maximum 10,788.00 U/L laccase, because due to the laccase gene-encoding promoter regions contain various xenobiotic and heavy metal-specific recognition site (Sannia et al. 2001; Faraco et al. 2002). The production of high amount of enzyme also depends on the cultivation media condition (Heinzkill et al. 1998).
Degradation of phenanthrene
The phenanthrene degradation efficiency was accessed by the HPLC chromatogram. In the control experiment, one single peak of phenanthrene was eluted at the retention time 12.322 (Fig. 2a). The peak area of phenanthrene was decreased continuously with the incubation period increase in the sample and other small molecule compound peaks were eluted (Fig. 2b). After 30 days of incubation, maximum 99.65% phenanthrene degradation (Fig. 2c) was investigated on 6 pH at 27 °C. As the degradation of phenanthrene increases, the concentration of biomass and protein of G. lucidum increases, maximum biomass 255.00 mg and protein 129.00 mg/L were found in the phenanthrene media, respectively, (Fig. 2c). Previously, degradation of phenanthrene was investigated by Phanerochaete chrysosporium (Sutherland et al. 1991), Pleurotus ostreatus (Bezalel et al. 1996), Trametes versicolor (Sack et al. 1997), Kuehneromyces mutabilis (Sack et al. 1997), Trametes versicolor 951022 (Han et al. 2004), Polyporus sp. S133 (Hadibarata and Tachibana 2010), G. lucidum BCRC 36021 (Ting et al. 2011), Pleurotus ostreatus sp. ATCC38540 (Tirado-Torres et al. 2016), Pleurotus eryngii (Wu et al. 2016). Ting et al. (2011) observed that G. lucidum BCRC 36021 degraded 98.70% of phenanthrene in basal medium at 30 °C but in this study, phenanthrene degradation was investigated in mineral salt broth, in our knowledge, no one study has been found on the degradation of phenanthrene by G. lucidum strain CCG1 in mineral salt broth.
According to Bezalel et al. (1996), phenanthrene contains K-region and bay region in their structure. Phenanthrene was oxidized in K-region and formed 9,10-dihydroxy phenanthrene. Then, further the action of oxidation and ortho-ring cleavage reaction 2,2-diphenic acid was formed, and then it converted into phthalic acid and finally carbon dioxide (Bezalel et al. 1996, 1997; Hadibarata and Tachibana 2010). Hammel et al. (1992, 1995a) reported that the 2,2-diphenic acid produced by the action of MnP and LiP enzyme in Phanerochaete chrysosporium. Then, formed phthalic acid enters into basal metabolism pathway suggested by Pozdnyakova et al. (2010). According to Cerniglia et al. (1992) and Hammel (1995b) the characteristic feature of ligninolytic fungi is open and mineralize aromatic ring structure compounds.
Degradation of pyrene
The degradation efficiency of pyrene by strain CCG1 was investigated by the HPLC chromatogram. In the control experiment, one single peak of pyrene was eluted at the retention time 15.835 (Fig. 3a). It was found that as the peak area of pyrene decreases with the incubation period increase in the sample and other small peaks were eluted (Fig. 3b). After 30 days of incubation, maximum 99.58% degradation of pyrene was investigated (Fig. 3c) was investigated on 6 pH at 27 °C. Previously, degradation of pyrene was investigated by Crinipellis stipitaria JK364 (Lange et al. 1994), Trametes versicolor (Sack et al. 1997), Kuehneromyces mutabilis (Sack et al. 1997), Irpex lacteus (Song 1999), Trametes versicolor KR11W (Song 1999), Phanerochaete chrysosporium (Song 1999), Pleurotus ostreatus (Song 1999), Pleurotus ostreatus HP-1 (Hardik et al. 2010), G. lucidum BCRC 36021 (Ting et al. 2011), Pseudotrametes gibbosa (Wen et al. 2011), Armillaria sp. F022 (Hadibarata and Kristanti 2013), Pleurotus eryngii F032 (Teh and Hadibarata 2014), Rhizoctonia zeae SOL3 (Khudhair et al. 2015), Pycnoporus sanguineus H1 (Zhang et al. 2015), Trametes polyzona RYNF13 (Teerapatsakul et al. 2016), Coriolopsis byrsina strain APC5 (Agrawal and Shahi 2017). G. lucidum BCRC 36021 degraded 88.80% of pyrene in basal medium at 30 °C (Ting et al. 2011), but in this research phenanthrene degradation was investigated in mineral salt broth, in our knowledge, no one study has been found of the degradation of pyrene by G. lucidum strain CCG1 in mineral salt broth.
As the pyrene degradation increases, the biomass and protein concentration of G. lucidum increases, maximum 257.00 mg biomass and 140.00 mg/L protein concentration were found in the pyrene-containing media (Fig. 3c).
Pyrene was oxidized at the 4,5 bond (K-region) and formed 4,5 dihydroxy pyrene in the ligninolytic condition, it may be converted from pyrene trans 4,5-dihydrodiol (Sack et al. 1997; Agrawal and Shahi 2017). Further, due to the oxidation process 4,5 dihydroxy pyrene converted into phenanthrene, phthalic acid and benzoic acid by the action laccase, LiP and MnP enzyme (Hadibarata and Kristanti 2013; Khudhair et al. 2015; Agrawal and Shahi 2017). According to Pozdnyakova et al. (2010) phthalic acid can be involved in basal metabolism. Bogan and Lamar (1996) and Field et al. (1992) also reported that laccase-producing fungi are capable to mineralize PAHs to CO2 and H2O.
Conclusions
PAHs are recalcitrant organic pollutants in the environment; their occurrence in the environment caused an adverse effect on the human and other animals. In this study, ligninolytic enzyme producing white rot fungi G. lucidum was used for the degradation of PAHs compound (phenanthrene and pyrene). G. lucidum degrade 99.65, 99.58% of phenanthrene and pyrene, respectively, in mineral salt broth. G. lucidum produced a significant amount of ligninolytic enzyme (p < 0.0001) in the presence of phenanthrene and pyrene containing media. Further, after successful pot and field trial G. lucidum strain CCG1 can be used as potent PAH degrader from the environment.
References
Adenipekun CO, Ejoh OE, Ogunjobi AA (2012) Bioremediation of cutting fluids contaminated soil by Pleurotus tuber-regium Singer. Environmentalist 32:11–18
Agrawal N, Shahi SK (2017) Degradation of polycyclic aromatic hydrocarbon (pyrene) using novel fungal strain Coriolopsis byrsina strain APC5. Int Biodeterior Biodegrad 122:69–81
Agrawal N, Verma P, Singh RS, Shahi SK (2017) Ganoderma lucidum: a potent source of ligninolytic enzyme production. Int J Adv Res 5(5):1977–1981
Aitken MD, Stringfellow WT, Nagel RD, Kazunga C, Chen SH (1998) Characteristics of phenanthrene-degrading bacteria isolated from soils contaminated with polycyclic aromatic hydrocarbons. Can J Microbiol 44(8):743–752
Archibald FS (1992) A new assay for lignin type peroxidase employing the dye Azure B. Appl Environ Microbiol 58:3110–3116
Balesteros MR, de Sa LRV, Pereira PM, da Silva M, de Oliveira MAL, Ferreira-Leitao VS (2014) Monitoring of atrazine biodegradation by Pleurotus ostreatus INCQS 40310 through the simultaneous analysis of atrazine and its derivatives by HPLC. Biocatal Biotransformation 32:23–33
Bezalel L, Hadar Y, Fu PP, Freeman JP, Cerniglia CE (1996) Metabolism of phenanthrene by the white rot fungus Pleurotus ostreatus. Appl Environ Microbiol 62(7):2547–2553
Bezalel L, Hadar Y, Cerniglia CE (1997) Enzymatic mechanisms involved in phenanthrene degradation by the white rot fungus Pleurotus ostreatus. Appl Environ Microbiol 63(7):2495–2501
Bishnoi NR, Mehta U, Sain U, Pandit GG (2005) Quantification of polycyclic aromatic hydrocarbons in tea and coffee samples of Mumbai city (India) by high performance liquid chromatography. Environ Monit Assess 107:399–406
Bishnoi K, Kumar R, Bishnoi NR (2008) Biodegradation of polycyclic aromatic hydrocarbons by white rot fungi Phanerochaete chrysosporium in sterile and unsterile soil. J Sci Ind Res 67:538–542
Bogan B, Lamar RT (1996) Polycyclic aromatic hydrocarbon degrading capabilities of Phanerochaete laevis HHB-1625 and its extracellular ligninolytic enzymes. Appl Environ Microbiol 62:1597–1603
Boopathy R (2000) Factors limiting bioremediation technologies. Bioresour Technol 74:63–67
Casas N, Parella T, Vicent T, Caminal G, Sarra M (2009) Metabolites from the biodegradation of triphenylmethane dyes by Trametes versicolor or laccase. Chemosphere 75:1344–1349
Casillas RP, Crow SA, Heinze TM, Deck J, Cerniglia CE (1996) Initial oxidative and subsequent conjugative metabolites produced during the metabolism of phenanthrene by fungi. J Ind Microbiol Biotechnol 16(4):205–215
Cerniglia CE, Sutherland JB, Crow SA (1992) Fungal metabolism of aromatic hydrocarbons. In: Winkelmann G (ed) Microbial degradation of natural products. VCH Press, Weinheim, pp 193–217
Choi YW, Hyde KD, Ho WH (1999) Single spore isolation of fungi. Fungal Divers 3:29–38
de Jong ED, Field JA, de Bont JA (1992) Evidence for a new extracellular peroxidase manganese inhibited peroxidase from the white rot fungus Bjerkandera sp. BOS 55. FEBS Lett 299:107–110
Di Toro DM, McGrath JA, Hansen DJ (2000) Technical basis for narcotic chemicals and polycyclic aromatic hydrocarbon criteria. I. Water and tissue. Environ Toxicol Chem 19(8):1951–1970
Dominguez A, Couto SR, Sanroman MA (2005) Dye decolorization by Trametes hirsuta immobilized into alginate beads. World J Microbiol Biotechnol 21(4):405–409
Faraco V, Giardina P, Palmieri G, Sannia G (2002) Metal activated laccase promoters. Progress Biotechnol Amst 21:105–112
Field JA, de Jong E, Costa GF, de Bont JAM (1992) Biodegradation of polycyclic aromatic hydrocarbons by new isolates of white rot fungi. Appl Environ Microbiol 58:2219–2226
Hadibarata T, Kristanti RA (2012) Fate and cometabolic degradation of benzo[a]pyrene by white rot fungus Armillaria sp. F022. Bioresour Technol 107:314–318
Hadibarata T, Kristanti RA (2013) Biodegradation and metabolite transformation of pyrene by basidiomycetes fungal isolate Armillaria sp. F022. Bioprocess Biosyst Eng 36(4):461–468
Hadibarata T, Tachibana S (2010) Characterization of phenanthrene degradation by strain Polyporus sp. S133. J Environ Sci 22(1):142–149
Hammel KE (1995a) Mechanisms for polycyclic aromatic hydrocarbon degradation by ligninolytic fungi. Environ Health Perspect 103(5):41
Hammel KE (1995b) Organopollutant degradation by ligninolytic fungi. In: Young LY, Cerniglia CE (eds) Microbial transformation and degradation of toxic organic chemicals. Wiley, New York, pp 331–346
Hammel KE, Gai WZ, Green B, Moen MA (1992) Oxidative degradation of phenanthrene by the ligninolytic fungus Phanerochaete chrysosporium. Appl Environ Microbiol 58(6):1832–1838
Han MJ, Choi HT, Song HG (2004) Degradation of phenanthrene by Trametes versicolor and its laccase. J Microbiol 42(2):94–98
Hardik P, Akshaya G, Shilpa G (2010) Biodegradation of pyrene by Pleurotus ostreatus HP-1 and its correlation with extracellular ligninolytic enzyme production. Int J Environ Agric Res 3(3):313–319
Haritash AK, Kaushik CP (2009) Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J Hazard Mater 169:1–15
Heinzkill M, Bech L, Halkier T, Schneider P, Anke T (1998) Characterization of laccases and peroxidases from wood rotting fungi (family Coprinaceae). Appl Environ Microbiol 64(5):1601–1606
Ijoma GN, Tekere M (2017) Potential microbial applications of co-cultures involving ligninolytic fungi in the bioremediation of recalcitrant xenobiotic compounds. Int J Environ Sci Technol. https://doi.org/10.1007/s13762-017-1269-3
Jin D, Jiang X, Jing X, Ou Z (2007) Effects of concentration, head group and structure of surfactants on the degradation of phenanthrene. J Hazard Mat 144:215–221
Juhasz AL, Naidu R (2000) Bioremediation of high molecular weight polycyclic aromatic hydrocarbons: a review of the microbial degradation of benzo[a]pyrene. Int Biodeterior Biodegrad 45:57–88
Kadri T, Rouissi T, Brar SK, Cledon M, Sarma S, Verma M (2017) Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by fungal enzymes: a review. J Environ Sci 51:52–74
Khudhair AB, Hadibarata T, Yusoff ARM, Teh ZC, Adnan LA, Kamya H (2015) Pyrene metabolism by new species isolated from soil Rhizoctonia zeae SOL3. Water Air Soil Pollut 226:186
Kitamura S, Suzuki T, Sanoh S, Kohta R, Jinno N, Sugihara K, Yoshihara S, Fujimoto N, Watanabe H, Ohta S (2005) Comparative study of the endocrine-disrupting activity of bisphenol A and 19 related compounds. Toxicol Sci 84:249–259
Kumar AK, Sharma S (2017) Recent updates on different methods of pretreatment of lignocellulosic feedstocks: a review. Bioresour Bioprocess 4(1):7
Lange B, Kremer S, Sterner O, Anke H (1994) Pyrene metabolism in Crinipellis stipitaria: identification of trans-4, 5-dihydro-4,5-dihydroxypyrene and 1-pyrenylsulfate in strain JK364. Appl Environ Microbiol 60(10):3602–3607
Lawton JH, Jones CG (1995) Linking species and ecosystems: organisms as ecosystem engineers. In: Linking species and ecosystems. Springer, Boston, MA, pp 141–150.
Lee SM, Koo BW, Lee SS, Kim MK, Choi DH, Hong EJ, Jeung EB, Choi IG (2004) Biodegradation of dibutyl phthalate by white rot fungi and evaluation on its estrogenic activity. Enzyme Microb Technol 35:417–423
Makkar RS, Rockne KJ (2003) Comparison of synthetic surfactants and biosurfactants in enhancing biodegradation of polycyclic aromatic hydrocarbons. Environ Toxicol Chem 22:2280–2292
Mrozik A, Piotrowska-Seget Z (2010) Bioaugmentation as a strategy for cleaning up of soils contaminated with aromatic compounds. Microbiol Res 165:363–375
Pointing SB (2001) Feasibility of bioremediation by white rot fungi. Appl Microbiol Biotechnol 57:20–33
Pothuluri JV, Selby A, Evans FE, Freeman JP, Cerniglia CE (1995) Transformation of chrysene and other polycyclic aromatic hydrocarbon mixtures by the fungus Cunninghamella elegans. Can J Microbiol 73(S1):1025–1033
Pozdnyakova NN, Nikiforova SV, Makarov OE, Chernyshova MP, Pankin KE, Turkovskaya OV (2010) Influence of cultivation conditions on pyrene degradation by the fungus Pleurotus ostreatus D1. World J Microbiol Biotechnol 26:205–211
Puglisi E, Cappa F, Fragoulis G, Trevisan M, Del Re AA (2007) Bioavailability and degradation of phenanthrene in compost amended soils. Chemosphere 67:548–556
Rengarajan T, Rajendran P, Nandakumar N, Lokeshkumar B, Rajendran P, Nishigaki I (2015) Exposure to polycyclic aromatic hydrocarbons with special focus on cancer. Asian Pac J Trop Biomed 5(3):182–189
Rigas F, Papadopoulou K, Philippoussis A, Papadopoulou M, Chatzipavlidis J (2009) Bioremediation of lindane contaminated soil by Pleurotus ostreatus in non-sterile conditions using multilevel factorial design. Water Air Soil Pollut 197:121–129
Rostami I, Juhasz AL (2011) Assessment of persistent organic pollutant (POP) bioavailability and bioaccessibility for human health exposure assessment: a critical review. Crit Rev Environ Sci Technol 41(7):623–656
Sack U, Heinze TM, Deck J, Cerniglia CE, Martens R, Zadrazil F, Fritsche W (1997) Comparison of phenanthrene and pyrene degradation by different wood-decaying fungi. Appl Environ Microbiol 63(10):3919–3925
Sandhu DK, Arora DS (1985) Laccase production by Polyporus sanguineus under different nutritional and environmental conditions. Experientia 41:355–356
Sannia G, Faraco V, Giardina P, Palmieri G (2001) Metal-activated laccase promoters. In: Proceedings of the 8th international conference on biotechnology in the pulp and paper industry, Helsinki, Finland
Singh H (2006) Mycoremediation: fungal bioremediation. Wiley, New Jersey
Song HG (1999) Comparison of pyrene biodegradation by white rot fungi. World J Microbiol Biotechnol 5:669–672
Sutherland JB, Selby AL, Freeman JP, Evans FE, Cerniglia CE (1991) Metabolism of phenanthrene by Phanerochaete chrysosporium. Appl Environ Microbiol 57(11):3310–3316
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Teerapatsakul C, Pothiratana C, Chitradon L, Thachepan S (2016) Biodegradation of polycyclic aromatic hydrocarbons by a thermotolerant white rot fungus Trametes polyzona RYNF13. J Gen Appl Microbiol. https://doi.org/10.2323/jgam.2016.06.001
Teh ZC, Hadibarata T (2014) Enhanced degradation of pyrene and metabolite identification by Pleurotus eryngii F032. Water Air Soil Pollut 225(5):1909
Ting WTE, Yuan SY, Wu SD, Chang BV (2011) Biodegradation of phenanthrene and pyrene by Ganoderma lucidum. Int Biodeterior Biodegrad 65:238–242
Tirado-Torres D, Gayosso-Canales M, Marmolejo-Santillan Y, Romo-Gomez C, Acevedo-Sandoval O (2016) Removal of polycyclic aromatic hydrocarbons by Pleurotus ostreatus sp. ATCC38540 in liquid medium. Acad J Sci Res 4(10):376–379
Tisma M, Sudar M, Vasic-Racki D, Zelic B (2010) Mathematical model for Trametes versicolor growth in submerged cultivation. Bioprocess Biosyst Eng 33:749–758
Wang C, Sun H, Li Y, Zhang Q (2009) Enzyme activities during degradation of polycyclic aromatic hydrocarbons by white rot fungus Phanerochaete chrysosporium in soils. Chemosphere 77:733–738
Wen J, Gao D, Zhang B, Liang H (2011) Co-metabolic degradation of pyrene by indigenous white-rot fungus Pseudotrametes gibbosa from the northeast China. Int Biodeterior Biodegrad 65(4):600–604
Wong DWS (2009) Structure and action mechanism of ligninolytic enzymes. Appl Biochem Biotechnol 157:174–209
Wu M, Xu Y, Ding W, Li Y, Xu H (2016) Mycoremediation of manganese and phenanthrene by Pleurotus eryngii mycelium enhanced by Tween 80 and saponin. Appl Microbiol Biotechnol 100:7249–7261
Wunder T, Kremer S, Sterner O, Anke H (1994) Metabolism of the polycyclic aromatic hydrocarbon pyrene by Aspergillus niger SK 9317. Appl Microbiol Biotechnol 42(4):636–641
Zhang S, Ning Y, Zhang X, Zhao Y, Yang X, Wu K, Yang S, La G, Sun X, Li X (2015) Contrasting characteristics of anthracene and pyrene degradation by wood rot fungus Pycnoporus sanguineus H1. Int Biodeterior Biodegrad 105:228–232
Zhuo R, Ma L, Fan F, Gong Y, Wan X, Jiang M, Zhang X, Yang Y (2011) Decolorization of different dyes by a newly isolated white-rot fungi strain Ganoderma sp. En3 and cloning and functional analysis of its laccase gene. J Hazard Mater 192(2):855–873
Authors’ contributions
NA performed the research experiments and wrote the manuscript. PV helped in the experiments and manuscript writing. SKS guided both author during the experiments and manuscript preparation. All authors read and approved the final manuscript.
Acknowledgements
The authors are thankful to Mr. Anand Barapatre, post doc fellow, JNARDDC Campus, Nagpur for their support in the statistical analysis and to improve my research work. The authors are also thankful to Mr. Swaroop Biswas, JLA, CIF-PD Lab Bose Institute, Kolkata for HPLC analysis. The authors are thankful to Head, Department of Botany, Guru Ghasidas Vishwavidyalaya, providing infrastructural facilities. In addition, thanks to Guru Ghasidas Vishwavidyalaya, Bilaspur (C.G.) for providing financial assistance.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All the authors have agreed to provide the data and materials.
Consent for publication
The authors approved the consent for publishing the manuscript.
Ethics approval and consent to participate
All the authors have read and agreed the ethics for publishing the manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Agrawal, N., Verma, P. & Shahi, S.K. Degradation of polycyclic aromatic hydrocarbons (phenanthrene and pyrene) by the ligninolytic fungi Ganoderma lucidum isolated from the hardwood stump. Bioresour. Bioprocess. 5, 11 (2018). https://doi.org/10.1186/s40643-018-0197-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40643-018-0197-5