Abstract
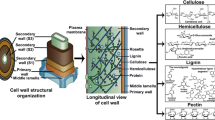
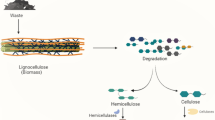
Lignin is a polyphenolic biopolymer synthesized by plants, for providing strength and rigidity to the plant cellular structure. It is closely associated with other plant polysaccharides in the cell wall such as cellulose and hemicellulose, constituting the most abundant biopolymer on the earth’s surface. However, the complete utilization of it is being explored for the past few years. Various research groups around the world are trying to replace conventional fuels with the second-generation biofuels from lignocellulose. Several physical, chemical, and biological conversion methods have been developed for the separation and utilization of this biomass, as a result of which biological methods for lignocellulose conversion are considered to be cheap and environment-friendly. Microorganisms, especially fungi and bacteria, have been able to degrade the lignocellulose network and convert it to commercially important biofuels by secreting several intra- and extra-cellular enzymes. In the past few years, research has been conducted to isolate efficient lignin-degrading microorganisms, as separation of lignin from cellulosic biomass is considered as a major hurdle in biofuel and pulping industries. In this article, we extensively discuss different small- and large-scale methods developed for the isolation and characterization of lignin-degrading microorganisms. We have also comprehensively discussed about the qualitative and quantitative methods for the identification and characterization of the lignin-degrading and lignin-degrading auxiliary enzymes by comparing different methods based on their efficiency. This review can be used as a primer for understanding and selecting the most efficient method for isolation and characterization of lignin-degrading microorganisms and their enzymes.



Similar content being viewed by others
References
Sattler SE, Funnell-Harris DL (2013) Modifying lignin to improve bioenergy feedstocks: strengthening the barrier against pathogens. Front Plant Sci 4:70. doi:10.3389/fpls.2013.00070
Kumar P, Barrett DM, Delwiche MJ, Stroeve P (2009) Methods for pretreatment of lignocellulosic biomass for efficient hydrolysis and biofuel production. Ind Eng Chem Res 48(8):3713–3729
Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M (2014) Lignin valorization: improving lignin processing in the biorefinery. Science 344(6185):1246843
Rinaldi R, Jastrzebski R, Clough MT, Ralph J, Kennema M, Bruijnincx PC, Weckhuysen BM (2016) Paving the way for lignin valorisation: recent advances in bioengineering, biorefining and catalysis. Angew Chem Int Ed Engl 55(29):8164–8215
Bugg TD, Ahmad M, Hardiman EM, Singh R (2011) The emerging role for bacteria in lignin degradation and bio-product formation. Curr Opin Biotechnol 22(3):394–400
Pollegioni L, Tonin F, Rosini E (2015) Lignin‐degrading enzymes. FEBS J 282(7):1190–1213
Kirk TK, Farrell RL (1987) Enzymatic “combustion”: the microbial degradation of lignin. Annu Rev Microbiol 41(1):465–501
Levasseur A, Piumi F, Coutinho PM, Rancurel C, Asther M, Delattre M, Henrissat B, Pontarotti P, Asther M, Record E (2008) FOLy: an integrated database for the classification and functional annotation of fungal oxidoreductases potentially involved in the degradation of lignin and related aromatic compounds. Fungal Genet Biol 45(5):638–645
Morii H, Nakamiya K, Kinoshita S (1995) Isolation of a lignin-decolorizing bacterium. J Ferment Bioeng 80(3):296–299
Pfennig N, Lippert KD (1966) Über das vitamin B12-Bedürfnis phototropher Schwefelbakterien. Arch Mikrobiol 55(3):245–256
Raj A, Reddy MK, Chandra R, Purohit HJ, Kapley A (2007) Biodegradation of kraft-lignin by Bacillus sp. isolated from sludge of pulp and paper mill. Biodegradation 18(6):783–792
Haq I, Kumar S, Kumari V, Singh SK, Raj A (2016) Evaluation of bioremediation potentiality of ligninolytic Serratia liquefaciens for detoxification of pulp and paper mill effluent. J Hazard Mater 305:190–199
Chakar FS, Ragauskas AJ (2004) Review of current and future softwood kraft lignin process chemistry. Ind Crop Prod 20(2):131–141
Pointing S (2001) Feasibility of bioremediation by white-rot fungi. Appl Microbiol Biotechnol 57(1–2):20–33
Cripps C, Bumpus JA, Aust S (1990) Biodegradation of azo and heterocyclic dyes by Phanerochaete chrysosporium. Appl Environ Microbiol 56(4):1114–1118
Rodriguez E, Pickard MA, Vazquez-Duhalt R (1999) Industrial dye decolorization by laccases from ligninolytic fungi. Curr Microbiol 38(1):27–32
Abadulla E, Tzanov T, Costa S, Robra K-H, Cavaco-Paulo A, Gübitz GM (2000) Decolorization and detoxification of textile dyes with a laccase from Trametes hirsuta. Appl Environ Microbiol 66(8):3357–3362
Podgornik H, Poljanšek I, Perdih A (2001) Transformation of Indigo carmine by Phanerochaete chrysosporium ligninolytic enzymes. Enzym Microb Technol 29(2):166–172
Spadaro JT, Gold MH, Renganathan V (1992) Degradation of azo dyes by the lignin-degrading fungus Phanerochaete chrysosporium. Appl Environ Microbiol 58(8):2397–2401
Heinfling A, Bergbauer M, Szewzyk U (1997) Biodegradation of azo and phthalocyanine dyes by Trametes versicolor and Bjerkandera adusta. Appl Microbiol Biotechnol 48(2):261–266
Levin L, Papinutti L, Forchiassin F (2004) Evaluation of Argentinean white rot fungi for their ability to produce lignin-modifying enzymes and decolorize industrial dyes. Bioresour Technol 94(2):169–176
Bandounas L, Wierckx NJ, de Winde JH, Ruijssenaars HJ (2011) Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential. BMC Biotechnol 11(1):94
Pointing SB (1999) Qualitative methods for the determination of lignocellulolytic enzyme production by tropical fungi. Fungal Diversity 2:17–33
Sundman V, Nase L (1971) A simple plate test for direct visualization of biological lignin degradation. Paperi ja puu 2:67–71
Tekere M, Mswaka A, Zvauya R, Read J (2001) Growth, dye degradation and ligninolytic activity studies on Zimbabwean white rot fungi. Enzym Microb Technol 28(4):420–426
Bavendamm W (1928) Über das Vorkommen und den Nachweis von Oxydasen bei holzzerstörenden Pilzen. Z Pflanzenkr Pflanzenschutz 38(9/10):257–276
Temp U, Eggert C, Eriksson K-EL (1998) A small-scale method for screening of lignin-degrading microorganisms. Appl Environ Microbiol 64(4):1548–1549
Eriksson K-EL, Blanchette R, Ander P (2012) Microbial and enzymatic degradation of wood and wood components. Springer Science & Business Media, Berlin Heidelberg
Haider K, Trojanowski J (1975) Decomposition of specifically 14C-labelled phenols and dehydropolymers of coniferyl alcohol as models for lignin degradation by soft and white rot fungi. Arch Microbiol 105(1):33–41
Rajan JS, Srinivasan V (1992) A colorimetric assay for lignin based on its reaction with diazotized sulfanilic acid and its use in studies on lignin biodegradation by bacteria. Biotechnol Tech 6(3):219–222
Ahmad M, Taylor CR, Pink D, Burton K, Eastwood D, Bending GD, Bugg TD (2010) Development of novel assays for lignin degradation: comparative analysis of bacterial and fungal lignin degraders. Mol Biosyst 6(5):815–821
Zimmermann W, Paterson A, Broda P (1988) Conventional and high-performance size-exclusion chromatography of graminaceous lignin–carbohydrate complexes. Methods Enzymol 161:191–199
McAuliffe L, Ellis RJ, Lawes JR, Ayling RD, Nicholas RA (2005) 16S rDNA PCR and denaturing gradient gel electrophoresis; a single generic test for detecting and differentiating Mycoplasma species. J Med Microbiol 54(8):731–739
Muyzer G, Smalla K (1998) Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek 73(1):127–141
Muyzer G, De Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59(3):695–700
Sheffield VC, Cox DR, Lerman LS, Myers RM (1989) Attachment of a 40-base-pair G+ C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci 86(1):232–236
Niku-Paavola M, Raaska L, Itävaara M (1990) Detection of white-rot fungi by a non-toxic stain. Mycol Res 94(1):27–31
Thurston CF (1994) The structure and function of fungal laccases. Microbiology 140(1):19–26
Soden D, O’callaghan J, Dobson A (2002) Molecular cloning of a laccase isozyme gene from Pleurotus sajor-caju and expression in the heterologous Pichia pastoris host. Microbiology 148(12):4003–4014
Harkin JM, Larsen MJ, Obst JR (1974) Use of syringaldazine for detection of laccase in sporophores of wood rotting fungi. Mycologia 66:469–476
Duffey S, Aldrich J, Blum M (1977) Biosynthesis of phenol and guaiacol by the hemipteran Leptoglossus phyllopus. Comp Biochem Physiol B 56(2):101–102
Dorfner R, Ferge T, Kettrup A, Zimmermann R, Yeretzian C (2003) Real-time monitoring of 4-vinylguaiacol, guaiacol, and phenol during coffee roasting by resonant laser ionization time-of-flight mass spectrometry. J Agric Food Chem 51(19):5768–5773
Sheikhi F, Ardakani MR, Enayatizamir N, Rodriguez-Couto S (2012) The determination of assay for laccase of Bacillus subtilis WPI with two classes of chemical compounds as substrates. Indian J Microbiol 52(4):701–707
Kiiskinen LL, Rättö M, Kruus K (2004) Screening for novel laccase‐producing microbes. J Appl Microbiol 97(3):640–646
López M, Guisado G, Vargas-García M, Suárez-Estrella F, Moreno J (2006) Decolorization of industrial dyes by ligninolytic microorganisms isolated from composting environment. Enzym Microb Technol 40(1):42–45
Childs RE, Bardsley WG (1975) The steady-state kinetics of peroxidase with 2, 2′-azino-di-(3-ethyl-benzthiazoline-6-sulphonic acid) as chromogen. Biochem J 145:93–103
Johannes C, Majcherczyk A (2000) Laccase activity tests and laccase inhibitors. J Biotechnol 78(2):193–199
Heinzkill M, Bech L, Halkier T, Schneider P, Anke T (1998) Characterization of laccases and peroxidases from wood-rotting fungi (family Coprinaceae). Appl Environ Microbiol 64(5):1601–1606
Edens WA, Goins TQ, Dooley D, Henson JM (1999) Purification and characterization of a secreted laccase of Gaeumannomyces graminis var. tritici. Appl Environ Microbiol 65(7):3071–3074
Prillinger H, Esser K (1977) The phenoloxidases of the ascomycete Podospora anserina. Mol Gen Genet 156(3):333–345
Harkin JM, Obst JR (1973) Syringaldazine, an effective reagent for detecting laccase and peroxidase in fungi. Experientia 29(4):381–387
Dedeyan B, Klonowska A, Tagger S, Tron T, Iacazio G, Gil G, Le Petit J (2000) Biochemical and molecular characterization of a laccase from Marasmius quercophilus. Appl Environ Microbiol 66(3):925–929
Collins PJ, Kotterman M, Field JA, Dobson A (1996) Oxidation of Anthracene and Benzo[a]pyrene by Laccases from Trametes versicolor. Appl Environ Microbiol 62(12):4563–4567
Johannes C, Majcherczyk A, Hüttermann A (1996) Degradation of anthracene by laccase of Trametes versicolor in the presence of different mediator compounds. Appl Microbiol Biotechnol 46(3):313–317
Majcherczyk A, Johannes C, Hüttermann A (1998) Oxidation of polycyclic aromatic hydrocarbons (PAH) by laccase of Trametes versicolor. Enzym Microb Technol 22(5):335–341
Johannes C, Majcherczyk A, Hüttermann A (1998) Oxidation of acenaphthene and acenaphthylene by laccase of Trametes versicolor in a laccase-mediator system. J Biotechnol 61(2):151–156
Pickard MA, Roman R, Tinoco R, Vazquez-Duhalt R (1999) Polycyclic aromatic hydrocarbon metabolism by white rot fungi and oxidation by Coriolopsis gallica UAMH 8260 laccase. Appl Environ Microbiol 65(9):3805–3809
Alcalde M, Bulter T (2003) Colorimetric assays for screening laccases. In: Arnold FH, Giorgiu G (eds.) Directed enzyme evolution. Springer, Berlin Heidelberg, pp 193–201
Glenn JK, Gold MH (1983) Decolorization of several polymeric dyes by the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol 45(6):1741–1747
Gold MH, Glenn JK, Alic M (1988). Use of polymericdyes in lignin biodegradation assays. Methods Enzymol161:74–78
Xu F (1996) Catalysis of novel enzymatic iodide oxidation by fungal laccase. Appl Biochem Biotechnol 59(3):221–230
Gabler M, Hensel M, Fischer L (2000) Detection and substrate selectivity of new microbial D-amino acid oxidases. Enzym Microb Technol 27(8):605–611
Lonergan G, Baker W (1995) Comparative study of substrates of fungal laccase. Lett Appl Microbiol 21(1):31–33
Boominathan K, Reddy C, Arora D, Elander R, Mukerji K (1992) Handbook of applied mycology. Marcel Dekker, New York
Pointing S, Vrijmoed L, Jones E (1999) Laccase is produced as the sole lignin-modifying enzyme in submerged liquid culture by the white-rot fungus Pycnoporus sanguineus L. Mycologia 91:345–349
Pointing S, Vrijmoed L, Jones E (1998) A qualitative assessment of lignocellulose degrading enzyme activity in marine fungi. Bot Mar 41(1–6):293–298
Archibald FS (1992) A new assay for lignin-type peroxidases employing the dye azure B. Appl Environ Microbiol 58(9):3110–3116
Magalhães DB, de Carvalho MEA, Bon E, Neto JSA, Kling SH (1996) Colorimetric assay for lignin peroxidase activity determination using methylene blue as substrate. Biotechnol Tech 10(4):273–276
Brown JA, Li D, Alic M, Gold MH (1993) Heat shock induction of manganese peroxidase gene transcription in Phanerochaete chrysosporium. Appl Environ Microbiol 59(12):4295–4299
Arora DS, Gill PK (2001) Comparison of two assay procedures for lignin peroxidase. Enzym Microb Technol 28(7):602–605
Tariq V-N, Irvine W (1995) Ligninolytic enzyme activity: a comparison of the veratryl alcohol and Azure B assay systems. Biotechnol Tech 9(3):197–202
Paszczyński A, Crawford RL, Huynh V-B (1988) Manganese peroxidase of Phanerochaete chrysosporium: purification. Methods Enzymol 161C:264–270
Vares T, Kalsi M, Hatakka A (1995) Lignin peroxidases, manganese peroxidases, and other ligninolytic enzymes produced by phlebia radiata during solid-state fermentation of wheat straw. Appl Environ Microbiol 61(10):3515–3520
Orth AB, Royse D, Tien M (1993) Ubiquity of lignin-degrading peroxidases among various wood-degrading fungi. Appl Environ Microbiol 59(12):4017–4023
Paszczyński A, Huynh V-B, Crawford R (1985) Enzymatic activities of an extracellular, manganese-dependent peroxidase from Phanerochaete chrysosporium. FEMS Microbiol Lett 29(1–2):37–41
Martinez MJ, Ruiz-Duenas FJ, Guillen F, Martinez AT (1996) Purification and catalytic properties of two manganese peroxidase isoenzymes from Pleurotus eryngii. Eur J Biochem 237(2):424–432
Vasilchenko LG, Ludwig R, Yershevich OP, Haltrich D, Rabinovich ML (2012) High‐throughput screening for cellobiose dehydrogenases by Prussian Blue in situ formation. Biotechnol J 7(7):919–930
Kremer SM, Wood PM (1992) Evidence that cellobiose oxidase from Phanerochaete chrysosporium is primarily an Fe (III) reductas. Eur J Biochem 205(1):133–138
Baminger U, Subramaniam SS, Renganathan V, Haltrich D (2001) Purification and characterization of cellobiose dehydrogenase from the plant pathogen Sclerotium (Athelia) rolfsii. Appl Environ Microbiol 67(4):1766–1774
Baminger U, Nidetzky B, Kulbe KD, Haltrich D (1999) A simple assay for measuring cellobiose dehydrogenase activity in the presence of laccase. J Microbiol Methods 35(3):253–259
Henriksson G, Polk V, Eriksson K-E (1997) Assay for cellobiose dehydrogenase in the presence of laccase. Biotechnol Tech 11(10):743–745
Guillen F, Martinez AT, Martinez MJ (1992) Substrate specificity and properties of the aryl‐alcohol oxidase from the ligninolytic fungus Pleurotus eryngii. Eur J Biochem 209(2):603–611
Gutierrez A, Caramelo L, Prieto A, Martínez MJ, Martinez AT (1994) Anisaldehyde production and aryl-alcohol oxidase and dehydrogenase activities in ligninolytic fungi of the genus Pleurotus. Appl Environ Microbiol 60(6):1783–1788
Ruiz-Dueñas FJ, Ferreira P, Martínez MJ, Martínez AT (2006) In vitro activation, purification, and characterization of Escherichia coli expressed aryl-alcohol oxidase, a unique H 2 O 2-producing enzyme. Protein Expr Purif 45(1):191–199
Ferreira P, Hernández-Ortega A, Herguedas B, Rencoret J, Gutiérrez A, Martinez M, Jiménez-Barbero J, Medina M (2010) Kinetic and chemical characterization of aldehyde oxidation by fungal aryl-alcohol oxidase. Biochem J 425:585–593
Tamboli DP, Telke AA, Dawkar VV, Jadhav SB, Govindwar SP (2011) Purification and characterization of bacterial aryl alcohol oxidase from Sphingobacterium sp. ATM and its uses in textile dye decolorization. Biotechnol Bioprocess Eng 16(4):661–668
Feldman D, Kowbel DJ, Glass NL, Yarden O, Hadar Y (2015) Detoxification of 5-hydroxymethylfurfural by the Pleurotus ostreatus lignolytic enzymes aryl alcohol oxidase and dehydrogenase. Biotechnol Biofuels 8(1):63
Ferreira P, Medina M, Guillén F, Martinez M, Van Berkel W, Martinez A (2005) Spectral and catalytic properties of aryl-alcohol oxidase, a fungal flavoenzyme acting on polyunsaturated alcohols. Biochem J 389:731–738
Danneel HJ, Rossner E, Zeeck A, Giffhorn F (1993) Purification and characterization of a pyranose oxidase from the basidiomycete Peniophora gigantea and chemical analyses of its reaction products. Eur J Biochem 214(3):795–802
Furukawa H, Wieser M, Morita H, Sugio T, Nagasawa T (1999) Purification and characterization of vanillyl-alcohol oxidase from Byssochlamys fulva V107. J Biosci Bioeng 87(3):285–290
Fraaije MW, Veeger C, van Berkel WJ (1995) Substrate specificity of flavin-dependent vanillyl-alcohol oxidase from Penicillium simplicissimum. Eur J Biochem 234:271–277
Jong E, Berkel WJ, Zwan RP, Bont JA (1992) Purification and characterization of vanillyl‐alcohol oxidase from Penicillium simplicissimum. Eur J Biochem 208(3):651–657
Fraaije MW, Pikkemaat M, Van Berkel W (1997) Enigmatic gratuitous induction of the covalent flavoprotein vanillyl-alcohol oxidase in Penicillium simplicissimum. Appl Environ Microbiol 63(2):435–439
van den Heuvel RH, van den Berg WA, Rovida S, van Berkel WJ (2004) Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin. J Biol Chem 279(32):33492–33500
Kersten PJ, Kirk TK (1987) Involvement of a new enzyme, glyoxal oxidase, in extracellular H2O2 production by Phanerochaete chrysosporium. J Bacteriol 169(5):2195–2201
Kersten PJ (1990) Glyoxal oxidase of Phanerochaete chrysosporium: its characterization and activation by lignin peroxidase. Proc Natl Acad Sci 87(8):2936–2940
Brugger D, Krondorfer I, Shelswell C, Huber-Dittes B, Haltrich D, Peterbauer CK (2014) Engineering pyranose 2-oxidase for modified oxygen reactivity. PLoS One 8(9):e109242
Baron AJ, Stevens C, Wilmot C, Seneviratne KD, Blakeley V, Dooley DM, Phillips S, Knowles PF, McPherson MJ (1994) Structure and mechanism of galactose oxidase. The free radical site. J Biol Chem 269(40):25095–25105
Raba J, Mottola HA (1995) Glucose oxidase as an analytical reagent. Crit Rev Anal Chem 25(1):1–42
Dobrick LA (1958) Screening method for glucose of blood serum utilizing glucose oxidase and an indophenol indicator. J Biol Chem 231(1):403–409
Salomon L, Johnson JE (1959) Enzymatic microdetermination of glucose in blood and urine. Anal Chem 31(3):453–456
Morley G, Dawson A, Marks V (1968) Manual and autoanalyzer methods for measuring blood glucose using guaiacum and glucose oxidase. Assoc Clin Biochem Proc 5(2):42–45
Trinder P (1969) Determination of blood glucose using an oxidase-peroxidase system with a non-carcinogenic chromogen. J Clin Pathol 22(2):158–161
Werner W, Rey H-G, Wielinger H (1970) Über die Eigenschaften eines neuen Chromogens für die Blutzuckerbestimmung nach der GOD/POD-Methode. Fresenius’ Z Anal Chem 252(2–3):224–228
Kabasakalian P, Kalliney S, Westcott A (1974) Enzymatic blood glucose determination by colorimetry of N, N-diethylaniline-4-aminoantipyrine. Clin Chem 20(5):606–607
Clapp PA, Evans DF (1991) Spectrophotometric determination of hydrogen peroxide with leuco Patent Blue Violet. Anal Chim Acta 243:217–220
Bateman RC Jr, Evans JA (1995) Using the glucose oxidase/peroxidase system in enzyme kinetics. J Chem Educ 72(12):A240
Betancor L, López-Gallego F, Hidalgo A, Alonso-Morales N, Dellamora-Ortiz G, Guisán JM, Fernández-Lafuente R (2006) Preparation of a very stable immobilized biocatalyst of glucose oxidase from Aspergillus niger. J Biotechnol 121(2):284–289
Guilbault GG, Brignac PJ Jr, Zimmer M (1968) Homovanillic acid as a fluorometric substrate for oxidative enzymes. Analytical applications of the peroxidase, glucose oxidase, and xanthine oxidase systems. Anal Chem 40(1):190–196
Brock BJ, Rieble S, Gold MH (1995) Purification and characterization of a 1, 4-benzoquinone reductase from the basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol 61(8):3076–3081
Kim K (2002) Characterization of 1, 4-benzoquinone reductase from bovine liver. Biotechnol Bioprocess Eng 7(4):216–220
Constam D, Muheim A, Zimmermann W, Fiechter A (1991) Purification and partial characterization of an intracellular NADH: quinone oxidoreductase from Phanerochaete chrysosporium. J Gen Microbiol 137(9):2209–2214
Rehman AU, Thurston CF (1992) Purification of laccase I from Armillaria mellea. Microbiology 138(6):1251–1257
Yadav M, Yadav P, Yadav KDS (2009) Purification and characterization of lignin peroxidase from Loweporus lividus MTCC‐1178. Eng Life Sci 9(2):124–129
Cheng X, Jia R, Li P, Tu S, Zhu Q, Tang W, Li X (2007) Purification of a new manganese peroxidase of the white-rot fungus Schizophyllum sp. F17, and decolorization of azo dyes by the enzyme. Enzym Microb Technol 41(3):258–264
Asada Y, Watanabe A, Ohtsu Y, Kuwahara M (1995) Purification and characterization of an aryl-alcohol oxidase from the lignin-degrading basidiomycete Phanerochaete chrysosporium. Biosci Biotechnol Biochem 59(7):1339–1341
Leitner C, Volc J, Haltrich D (2001) Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor. Appl Environ Microbiol 67(8):3636–3644
Paukner R, Staudigl P, Choosri W, Haltrich D, Leitner C (2015) Expression, purification, and characterization of galactose oxidase of Fusarium sambucinum in E. coli. Protein Expr Purif 108:73–79
Swoboda BE, Massey V (1965) Purification and properties of the glucose oxidase from Aspergillus niger. J Biol Chem 240(5):2209–2215
Martinez D, Larrondo LF, Putnam N, Gelpke MDS, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F (2004) Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol 22(6):695–700
Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek CP, Ferreira P, Ruiz-Duenas FJ, Martinez AT, Kersten P (2009) Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci 106(6):1954–1959
Kameshwar AkS QW (2016) Recent developments in using advanced sequencing technologies for the genomic studies of lignin and cellulose degrading microorganisms. Int J Biol Sci 12(2):156–171
Ohm RA, Riley R, Salamov A, Min B, Choi I-G, Grigoriev IV (2014) Genomics of wood-degrading fungi. Fungal Genet Biol 72:82–90
Acknowledgments
This work was supported by NSERC-RDF fund to Wensheng Qin and Ontario Trillium Scholarship (OTS) to Ayyappa Kumar Sista Kameshwar.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kameshwar, A.K.S., Qin, W. Qualitative and Quantitative Methods for Isolation and Characterization of Lignin-Modifying Enzymes Secreted by Microorganisms. Bioenerg. Res. 10, 248–266 (2017). https://doi.org/10.1007/s12155-016-9784-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-016-9784-5