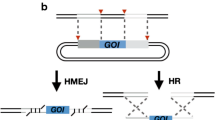
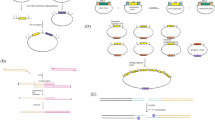
Abstract
Yarrowia lipolytica is an oleaginous saccharomycetous yeast with a long history of industrial use. It aroused interest several decades ago as host for heterologous protein production. Thanks to the development of numerous molecular and genetic tools, Y. lipolytica is now a recognized system for expressing heterologous genes and secreting the corresponding proteins of interest. As genomic and transcriptomic tools increased our basic knowledge on this yeast, we can now envision engineering its metabolic pathways for use as whole-cell factory in various bioconversion processes. Y. lipolytica is currently being developed as a workhorse for biotechnology, notably for single-cell oil production and upgrading of industrial wastes into valuable products. As it becomes more and more difficult to keep up with an ever-increasing literature on Y. lipolytica engineering technology, this article aims to provide basic and actualized knowledge on this research area. The most useful reviews on Y. lipolytica biology, use, and safety will be evoked, together with a resume of the engineering tools available in this yeast. This mini-review will then focus on recently developed tools and engineering strategies, with a particular emphasis on promoter tuning, metabolic pathways assembly, and genome editing technologies.

Similar content being viewed by others
References
Groenewald, M., Boekhout, T., Neuvéglise, C., Gaillardin, C., van Dijck, P. W., & Wyss, M. (2014). Yarrowia lipolytica: Safety assessment of an oleaginous yeast with a great industrial potential. Critical Reviews in Microbiology, 40(3), 187–206.
Sibirny, A., Madzak, C., & Fickers, P. (2014). Genetic engineering of non-conventional yeast for the production of valuable compounds. In F.D. Harzevili, H. Chen (Eds.), Microbial biotechnology: Progress and trends (pp. 63–111). Boca Raton: CRC Press.
Bankar, A. V., Kumar, A. R., & Zinjarde, S. S. (2009). Environmental and industrial applications of Yarrowia lipolytica. Applied Microbiology and Biotechnology, 84(5), 847–865.
Zinjarde, S. S. (2014). Food-related applications of Yarrowia lipolytica. Food Chemistry, 152, 1–10.
Madzak, C., Gaillardin, C., & Beckerich, J. M. (2004). Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: A review. Journal of Biotechnology, 109(1–2), 63–81.
Madzak, C., & Beckerich, J. M. (2013). Heterologous protein expression and secretion, in Yarrowia lipolytica. In G. Barth (Ed.), Yarrowia lipolytica: Biotechnological applications (Vol. 25, pp. 1–76). Heidelberg: Springer.
Madzak, C. (2015). Yarrowia lipolytica: Recent achievements in heterologous protein expression and pathway engineering. Applied Microbiology and Biotechnology, 99(11), 4559–4577.
Barth, G. (2013). Microbiology monographs, Vol. 24: Yarrowia lipolytica: Genetics, genomics, and physiology. Heidelberg: Springer.
Barth, G. (2013). Microbiology monographs, Vol. 25: Yarrowia lipolytica: Biotechnological applications. Heidelberg: Springer.
Nicaud, J. M., Fabre, E., & Gaillardin, C. (1989). Expression of invertase activity in Yarrowia lipolytica and its use as a selective marker. Current Genetics, 16(4), 253–260.
Lazar, Z., Rossignol, T., Verbeke, J., Crutz-Le Coq, A. M., Nicaud, J. M., & Robak, M. (2013). Optimized invertase expression and secretion cassette for improving Yarrowia lipolytica growth on sucrose for industrial applications. Journal of Industrial Microbiology and Biotechnology, 40(11), 1273–1283.
Le Dall, M. T., Nicaud, J. M., & Gaillardin, C. (1994). Multiple-copy integration in the yeast Yarrowia lipolytica. Current Genetics, 26(1), 38–44.
Fournier, P., Abbas, A., Chasles, M., Kudla, B., Ogrydziak, D. M., et al. (1993) Colocalization of centromeric and replicative functions on autonomously replicating sequences isolated from the yeast Yarrowia lipolytica. Proceedings of the National Academy of Sciences of the United States of America, 90(11), 4912–4916.
Chen, D. C., Beckerich, J. M., & Gaillardin, C. (1997). One-step transformation of the dimorphic yeast Yarrowia lipolytica. Applied Microbiology and Biotechnology, 48(2), 232–235.
Juretzek, T., Le Dall, M. T., Mauersberger, S., Gaillardin, C., Barth, G., & Nicaud, J. M. (2001). Vectors for gene expression and amplification in the yeast Yarrowia lipolytica. Yeast, 18(2), 97–113.
Neuvéglise, C., Nicaud, J. M., Ross-Macdonald, P., & Gaillardin, C. (1998). A shuttle mutagenesis system for tagging genes in the yeast Yarrowia lipolytica. Gene, 213(1–2), 37–46.
Müller, S., Sandal, T., Kamp-Hansen, P., & Dalbøge, H. (1998). Comparison of expression systems in the yeasts Saccharomyces cerevisiae, Hansenula polymorpha, Klyveromyces lactis, Schizosaccharomyces pombe and Yarrowia lipolytica. Cloning of two novel promoters from Yarrowia lipolytica. Yeast, 14(14), 1267–1283.
Madzak, C., Tréton, B., & Blanchin-Roland, S. (2000). Strong hybrid promoters and integrative expression/secretion vectors for quasi-constitutive expression of heterologous proteins in the yeast Yarrowia lipolytica. Journal of Molecular Microbiology and Biotechnology, 2(2), 207–216.
Madzak, C. (2003). New tools for heterologous protein production in the yeast Yarrowia lipolytica. In S. G. Pandalai (Ed.), Recent research developments in microbiology (Vol. 7, pp. 453–479). Trivandrum: Research Signpost.
Nicaud, J. M., Madzak, C., van den Broek, P., Gysler, C., Duboc, P., et al. (2002). Protein expression and secretion in the yeast Yarrowia lipolytica. FEMS Yeast Research, 2(3), 371–379.
Fickers, P., Le Dall, M. T., Gaillardin, C., Thonart, P., & Nicaud, J. M. (2003). New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica. Journal of Microbiological Methods, 55(3), 727–737.
Dujon, B., Sherman, D., Fischer, G., Durrens, P., Casaregola, S., et al. (2004). Genome evolution in yeasts. Nature., 430(6995), 35–44.
Madzak, C., Mimmi, M. C., Caminade, E., Brault, A., Baumberger, S., et al. (2006). Shifting the optimal pH of activity for a laccase from the fungus Trametes versicolor by structure-based mutagenesis. Protein Engineering, Design & Selection, 19(2), 77–84.
Bordes, F., Cambon, E., Dossat-Létisse, V., André, I., Croux, C., et al. (2009). Improvement of Yarrowia lipolytica lipase enantioselectivity by using mutagenesis targeted to the substrate binding site. Chembiochem, 10(10), 1705–1713.
Galli, C., Gentili, P., Jolivalt, C., Madzak, C., & Vadalà, R. (2011). How is the reactivity of laccase affected by single-point mutations? Engineering laccase for improved activity towards sterically demanding substrates. Applied Microbiology and Biotechnology, 91(1), 123–131.
Pignède, G., Wang, H. J., Fudalej, F., Seman, M., Gaillardin, C., & Nicaud, J. M. (2000). Autocloning and amplification of LIP2 in Yarrowia lipolytica. Applied and Environment Microbiology, 66(8), 3283–3289.
Park, J. N., Song, Y., Cheon, S. A., Kwon, O., Oh, D. B., et al. (2011). Essential role of YlMPO1, a novel Yarrowia lipolytica homologue of Saccharomyces cerevisiae MNN4, in mannosylphosphorylation of N- and O-linked glycans. Applied and Environment Microbiology, 77(4), 1187–1195.
De Pourcq, K., Vervecken, W., Dewerte, I., Valevska, A., Van Hecke, A., & Callewaert, N. (2012). Engineering the yeast Yarrowia lipolytica for the production of therapeutic proteins homogeneously glycosylated with Man8GlcNAc2 and Man5GlcNAc2. Microbial Cell Factories, 11, 53.
De Pourcq, K., Tiels, P., Van Hecke, A., Geysens, S., Vervecken, W., & Callewaert, N. (2012) Engineering Yarrowia lipolytica to produce glycoproteins homogeneously modified with the universal Man3GlcNAc2 N-glycan core. PLoS ONE, 7(6), e39976.
Moon, H. Y., Van, T. L., Cheon, S. A., Choo, J., Kim, J. Y., & Kang, H. A. (2013). Cell-surface expression of Aspergillus saitoi-derived functional α-1,2-mannosidase on Yarrowia lipolytica for glycan remodeling. Journal of Microbiology, 51(4), 506–514.
Bordes, F., Fudalej, F., Dossat, V., Nicaud, J. M., & Marty, A. (2007). A new recombinant protein expression system for high-throughput screening in the yeast Yarrowia lipolytica. Journal of Microbiological Methods, 70(3), 493–502.
Leplat, C., Nicaud, J. M., & Rossignol, T. (2015) High-throughput transformation method for Yarrowia lipolytica mutant library screening. FEMS Yeast Research, 15(6), fov052.
Xue, Z., Sharpe, P. L., Hong, S. P., Yadav, N. S., Xie, D., et al. (2013). Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nature Biotechnology, 31(8), 734–740.
Xie, D., Jackson, E. N., & Zhu, Q. (2015). Sustainable source of omega-3 eicosapentaenoic acid from metabolically engineered Yarrowia lipolytica: From fundamental research to commercial production. Applied Microbiology and Biotechnology, 99(4), 1599–1610.
Yue, L., Chi, Z., Wang, L., Liu, J., Madzak, C., Li, J., & Wang, X. (2008) Construction of a new plasmid for surface display on cells of Yarrowia lipolytica. Journal of Microbiological Methods, 72(2), 116–123
Yang, X. S., Jiang, Z. B., Song, H. T., Jiang, S. J., Madzak, C., & Ma, L. X. (2009). Cell-surface display of the active mannanase in Yarrowia lipolytica with a novel surface-display system. Biotechnology and Applied Biochemistry, 54(3), 171–176.
Duquesne, S., Bozonnet, S., Bordes, F., Dumon, C., Nicaud, J. M., & Marty, A. (2014) Construction of a highly active xylanase displaying oleaginous yeast: Comparison of anchoring systems. PLoS ONE, 9(4), e95128.
Yuzbasheva, E. Y., Yuzbashev, T. V., Laptev, I. A., Konstantinova, T. K., & Sineoky, S. P. (2011). Efficient cell surface display of Lip2 lipase using C-domains of glycosylphosphatidylinositol-anchored cell wall proteins of Yarrowia lipolytica. Applied Microbiology and Biotechnology, 91(3), 645–654.
Beopoulos, A., Chardot, T., & Nicaud, J. M. (2009). Yarrowia lipolytica: A model and a tool to understand the mechanisms implicated in lipid accumulation. Biochimie, 91(6), 692–696.
Dulermo, T., & Nicaud, J. M. (2011). Involvement of the G3P shuttle and β-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metabolic Engineering, 13(5), 482–491.
Blazeck, J., Hill, A., Liu, L., Knight, R., Miller, J., et al. (2014). Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nature Communications, 5, 3131.
Qiao, K., Abidi, I., Liu, S. H., Zhang, H., Chakraborty, H., S. et al (2015). Engineering lipid overproduction in the oleaginous yeast Yarrowia lipolytica. Metabolic Engineering, 29, 56–65.
Chuang, L. T., Chen, D. C., Nicaud, J. M., Madzak, C., Chen, Y. H., & Huang, Y. S. (2010). Co-expression of heterologous desaturase genes in Yarrowia lipolytica. New Biotechnology, 27(4), 277–282.
Celińska, E., & Grajek, W. (2013). A novel multigene expression construct for modification of glycerol metabolism in Yarrowia lipolytica. Microbial Cell Factories, 12, 102.
Blazeck, J., Liu, L., Redden, H., & Alper, H. (2011). Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Applied and Environment Microbiology, 77(22), 7905–7914.
Blazeck, J., Reed, B., Garg, R., Gerstner, R., Pan, A., et al. (2013). Generalizing a hybrid synthetic promoter approach in Yarrowia lipolytica. Applied Microbiology and Biotechnology, 97(7), 3037–3052.
Dulermo, R., Brunel, F., Dulermo, T., Ledesma-Amaro, R., Vion, J., et al. (2017). Using a vector pool containing variable-strength promoters to optimize protein production in Yarrowia lipolytica. Microbial Cell Factories, 16(1), 31.
Shabbir-Hussain, M., Wheeldon, I., & Blenner, M. A. (2017). A strong hybrid fatty acid inducible transcriptional sensor built from Yarrowia lipolytica upstream activating and regulatory sequences. Biotechnology Journal, 12(10), 1700248.
Trassaert, M., Vandermies, M., Carly, F., Denies, O., Thomas, S., et al. (2017). New inducible promoter for gene expression and synthetic biology in Yarrowia lipolytica. Microbial Cell Factories, 16(1), 141.
Loira, N., Dulermo, T., Nicaud, J. M., & Sherman, D. J. (2012). A genome-scale metabolic model of the lipid-accumulating yeast Yarrowia lipolytica. BMC Systems Biology, 6, 35.
Pan, P., & Hua, Q. (2012) Reconstruction and in silico analysis of metabolic network for an oleaginous yeast, Yarrowia lipolytica. PLoS ONE 7(12), e51535.
Kavšček, M., Bhutada, G., Madl, T., & Natter, K. (2015). Optimization of lipid production with a genome-scale model of Yarrowia lipolytica. BMC Systems Biology, 9, 72.
Trébulle, P., Nicaud, J. M., Leplat, C., & Elati, M. (2017). Inference and interrogation of a coregulatory network in the context of lipid accumulation in Yarrowia lipolytica. NPJ Systems Biology and Applications, 3, 21.
Tiels, P., Baranova, E., Piens, K., De Visscher, C., Pynaert, G., et al. (2012). A bacterial glycosidase enables mannose-6-phosphate modification and improved cellular uptake of yeast-produced recombinant human lysosomal enzymes. Nature Biotechnology, 30(12), 1225–1231.
Verbeke, J., Beopoulos, A., & Nicaud, J. M. (2013). Efficient homologous recombination with short length flanking fragments in Ku70 deficient Yarrowia lipolytica strains. Biotechnology Letters, 35(4), 571–576.
Kretzschmar, A., Otto, C., Holz, M., Werner, S., Hübner, L., & Barth, G. (2013). Increased homologous integration frequency in Yarrowia lipolytica strains defective in non-homologous end-joining. Current Genetics, 59(1–2), 63–72.
Han, Z., Madzak, C., & Su, W. W. (2013). Tunable nano-oleosomes derived from engineered Yarrowia lipolytica. Biotechnology and Bioengineering, 110(3), 702–710.
Liu, L., & Alper, H. S. (2014) Draft genome sequence of the oleaginous yeast Yarrowia lipolytica PO1f, a commonly used metabolic engineering host. Genome Announcements, 2(4), e00652–e006614.
Pomraning, K. R., & Baker, S. E. (2015) Draft genome sequence of the dimorphic yeast Yarrowia lipolytica strain W29. Genome Announcements, 3(6), e01211–e01215.
Magnan, C., Yu, J., Chang, I., Jahn, E., Kanomata, Y., et al. (2016) Sequence assembly of Yarrowia lipolytica strain W29/CLIB89 shows transposable element diversity. PLoS ONE, 11(9), e0162363.
Gao, S., Han, L., Zhu, L., Ge, M., Yang, S., et al. (2014). One-step integration of multiple genes into the oleaginous yeast Yarrowia lipolytica. Biotechnology Letters, 36(12), 2523–2528.
Gao, S., Tong, Y., Zhu, L., Ge, M., Jiang, Y., et al. (2017). Production of β-carotene by expressing a heterologous multifunctional carotene synthase in Yarrowia lipolytica. Biotechnology Letters, 39(6), 921–927.
Liu, H. H., Madzak, C., Sun, M. L., Ren, L., Song, P., et al. (2017). Engineering Yarrowia lipolytica for arachidonic acid production through rapid assembly of metabolic pathway. Biochemical Engineering Journal, 119, 52–58.
Schwartz, C. M., Hussain, M. S., Blenner, M., & Wheeldon, I. (2016). Synthetic RNA polymerase III promoters facilitate high-efficiency CRISPR-Cas9-mediated genome editing in Yarrowia lipolytica. ACS Synthetic Biology, 5(4), 356–359.
Gao, S., Tong, Y., Wen, Z., Zhu, L., Ge, M., et al. (2016). Multiplex gene editing of the Yarrowia lipolytica genome using the CRISPR-Cas9 system. Journal of Industrial Microbiology and Biotechnology, 43(8), 1085–1093.
Schwartz, C., Shabbir-Hussain, M., Frogue, K., Blenner, M., & Wheeldon, I. (2017). Standardized markerless gene integration for pathway engineering in Yarrowia lipolytica. ACS Synthetic Biology, 6(3), 402–409.
Morse, N. J., Wagner, J. M., Reed, K. B., Gopal, M. R., Lauffer, L. H., & Alper, H. S. (2018). T7 polymerase expression of guide RNAs in vivo allows exportable CRISPR-Cas9 editing in multiple yeast hosts. ACS Synthetic Biology, 7(4), 1075–1084.
Bredeweg, E. L., Pomraning, K. R., Dai, Z., Nielsen, J., Kerkhoven, E. J., & Baker, S. E. (2017). A molecular genetic toolbox for Yarrowia lipolytica. Biotechnology for Biofuels, 10, 2.
Celińska, E., Ledesma-Amaro, R., Larroude, M., Rossignol, T., Pauthenier, C., & Nicaud, J. M. (2017). Golden Gate Assembly system dedicated to complex pathway manipulation in Yarrowia lipolytica. Microbial Biotechnology, 10(2), 450–455.
Larroude, M., Celinska, E., Back, A., Thomas, S., Nicaud, J. M., & Ledesma-Amaro, R. (2018). A synthetic biology approach to transform Yarrowia lipolytica into a competitive biotechnological producer of β-carotene. Biotechnology and Bioengineering, 115(2), 464–472.
Rigouin, C., Gueroult, M., Croux, C., Dubois, G., Borsenberger, V., et al. (2017). Production of medium chain fatty acids by Yarrowia lipolytica: Combining molecular design and TALEN to engineer the fatty acid synthase. ACS Synthetic Biology, 6(10), 1870–1879.
Wong, L., Engel, J., Jin, E., Holdridge, B., & Xu, P. (2017). YaliBricks, a versatile genetic toolkit for streamlined and rapid pathway engineering in Yarrowia lipolytica. Metabolic Engineering Communications, 5, 68–77.
Schwartz, C., Frogue, K., Ramesh, A., Misa, J., & Wheeldon, I. (2017). CRISPRi repression of nonhomologous end-joining for enhanced genome engineering via homologous recombination in Yarrowia lipolytica. Biotechnology and Bioengineering, 114(12), 2896–2906.
Holkenbrink, C., Dam, M. I., Kildegaard, K. R., Beder, J., Dahlin, J., et al. (2018) EasyCloneYALI: CRISPR/Cas9-based synthetic toolbox for engineering of the yeast Yarrowia lipolytica. Biotechnology Journal. https://doi.org/10.1002/biot.201700543.
Wagner, J. M., Williams, E. V., & Alper, H. S. (2018). Developing a piggyBac transposon system and compatible selection markers for insertional mutagenesis and genome engineering in Yarrowia lipolytica. Biotechnology Journal, 13(5), e1800022.
Jang, I. S., Yu, B. J., Jang, J. Y., Jegal, J., & Lee, J. Y. (2018) Improving the efficiency of homologous recombination by chemical and biological approaches in Yarrowia lipolytica. PLoS ONE 13(3), e0194954.
Juretzek, T., Wang, H. J., Nicaud, J. M., Mauersberger, S., & Barth, G. (2000). Comparison of promoters suitable for regulated overexpression of β-galactosidase in the alkane-utilizing yeast Yarrowia lipolytica. Biotechnology and Bioprocess Engineering, 5, 320–326.
Tai, M., & Stephanopoulos, G. (2013). Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metabolic Engineering, 15, 1–9.
Kopecný, D., Pethe, C., Sebela, M., Houba-Hérin, N., Madzak, C., et al. (2005). High-level expression and characterization of Zea mays cytokinin oxidase/dehydrogenase in Yarrowia lipolytica. Biochimie, 87(11), 1011–1022.
Curran, K. A., Morse, N. J., Markham, K. A., Wagman, A. M., Gupta, A., & Alper, H. S. (2015). Short synthetic terminators for improved heterologous gene expression in yeast. ACS Synthetic Biology, 4(7), 824–832.
Barth, G., & Gaillardin, C. (1996) Yarrowia lipolytica. In K. Wolf (Ed.), Nonconventional yeasts in biotechnology: A handbook. Springer, Heidelberg, pp. 313–388.
Yan, J., Han, B., Gui, X., Wang, G., Xu, L., et al. (2018). Engineering Yarrowia lipolytica to simultaneously produce lipase and single cell protein from agro-industrial wastes for feed. Science Report, 8(1), 758.
Guo, H., Su, S., Madzak, C., Zhou, J., Chen, H., & Chen, G. (2016). Applying pathway engineering to enhance production of alpha-ketoglutarate in Yarrowia lipolytica. Applied Microbiology and Biotechnology, 100(23), 9875–9884.
Yovkova, V., Otto, C., Aurich, A., Mauersberger, S., & Barth, G. (2014). Engineering the α-ketoglutarate overproduction from raw glycerol by overexpression of the genes encoding NADP+-dependent isocitrate dehydrogenase and pyruvate carboxylase in Yarrowia lipolytica. Applied Microbiology and Biotechnology, 98, 2003–2013.
Förster, A., Aurich, A., Mauersberger, S., & Barth, G. (2007). Citric acid production from sucrose using a recombinant strain of the yeast Yarrowia lipolytica. Applied Microbiology and Biotechnology, 75(6), 1409–1417.
Jost, B., Holz, M., Aurich, A., Barth, G., Bley, T., & Müller, R. A. (2015). The influence of oxygen limitation for the production of succinic acid with recombinant strains of Yarrowia lipolytica. Applied Microbiology and Biotechnology, 99(4), 1675–1686.
Kerkhoven, E. J., Kim, Y. M., Wei, S., Nicora, C. D., Fillmore, T. L., et al. (2017). Leucine biosynthesis is involved in regulating high lipid accumulation in Yarrowia lipolytica. MBio, 8(3), e00857–e00817.
Schmid-Berger, N., Schmid, B., & Barth, G. (1994). Ylt1, a highly repetitive retrotransposon in the genome of the dimorphic fungus Yarrowia lipolytica. Journal of Bacteriology, 176, 2477–2482.
Tsakraklides, V., Brevnova, E., Stephanopoulos, G., & Shaw, A. J. (2015) Improved gene targeting through cell cycle synchronization. PLoS ONE 10(7), e0133434.
Liu, L., Otoupal, P., Pan, A., & Alper, H. S. (2014). Increasing expression level and copy number of a Yarrowia lipolytica plasmid through regulated centromere function. FEMS Yeast Research, 14(7), 1124–1127.
Liu, H. H., Ji, X. J., & Huang, H. (2015). Biotechnological applications of Yarrowia lipolytica: Past, present and future. Biotechnology Advances, 33(8), 1522–1546.
Xie, D. (2017). Integrating cellular and bioprocess engineering in the non-conventional yeast Yarrowia lipolytica for biodiesel production: A review. Frontiers in Bioengineering and Biotechnology, 5, 65.
Yan, J., Yan, Y., Madzak, C., & Han, B. (2017). Harnessing biodiesel-producing microbes: From genetic engineering of lipase to metabolic engineering of fatty acid biosynthetic pathway. Critical Reviews in Biotechnology, 37(1), 26–36.
Ledesma-Amaro, R., & Nicaud, J. M. (2016). Yarrowia lipolytica as a biotechnological chassis to produce usual and unusual fatty acids. Progress in Lipid Research, 61, 40–50.
Abghari, A., Madzak, C., & Chen, S. (2016). Combinatorial engineering Yarrowia lipolytica as a promising cell biorefinery platform for the production of multi-purpose long chain dicarboxylic acid building blocks from glycerol. Fermentation, 3(3), 40.
Gao, C., Qi, Q., Madzak, C., & Lin, C. S. (2015). Exploring medium-chain-length polyhydroxyalkanoates production in the engineered yeast Yarrowia lipolytica. Journal of Industrial Microbiology and Biotechnology, 42(9), 1255–1262.
Markham, K. A., Palmer, C. M., Chwatko, M., Wagner, J. M., Murray, C., et al. (2018) Rewiring Yarrowia lipolytica toward triacetic acid lactone for materials generation. Proceedings of the National Academy of Sciences of the United States of America, 115(9), 2096–2101.
Wei, H., Wang, W., Alahuhta, M., Wall, V., Baker, T., Taylor, J.O. et al. (2014). Engineering towards a complete heterologous cellulase secretome in Yarrowia lipolytica reveals its potential for consolidated bioprocessing. Biotechnology for Biofuels, 7(1), 148.
Guo, Z., Duquesne, S., Bozonnet, S., Cioci, G., Nicaud, J. M., et al. (2015). Development of cellobiose-degrading ability in Yarrowia lipolytica strain by overexpression of endogenous genes. Biotechnology for Biofuels, 8, 109.
Guo, Z. P., Robin, J., Duquesne, S., O’Donohue, M. J., Marty, A., & Bordes, F. (2018). Developing cellulolytic Yarrowia lipolytica as a platform for the production of valuable products in consolidated bioprocessing of cellulose. Biotechnology for Biofuels, 11, 141.
Ledesma-Amaro, R., & Nicaud, J. M. (2016). Metabolic engineering for expanding the substrate range of Yarrowia lipolytica. Trends in Biotechnology, 34(10), 798–809.
Morin, N., Cescut, J., Beopoulos, A., Lelandais, G., Le Berre, V., et al. (2011) Transcriptomic analyses during the transition from biomass production to lipid accumulation in the oleaginous yeast Yarrowia lipolytica. PLoS ONE. 6(11), e27966.
Pomraning, K. R., Wei, S., Karagiosis, S. A., Kim, Y. M., Dohnalkova, A. C., et al. (2015) Comprehensive metabolomic, lipidomic and microscopic profiling of Yarrowia lipolytica during lipid accumulation identifies targets for increased lipogenesis. PLoS ONE. 10(4), e0123188.
Sabra, W., Bommareddy, R. R., Maheshwari, G., Papanikolaou, S., & Zeng, A. P. (2017). Substrates and oxygen dependent citric acid production by Yarrowia lipolytica: Insights through transcriptome and fluxome analyses. Microbial Cell Factories, 16(1), 78.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Madzak, C. Engineering Yarrowia lipolytica for Use in Biotechnological Applications: A Review of Major Achievements and Recent Innovations. Mol Biotechnol 60, 621–635 (2018). https://doi.org/10.1007/s12033-018-0093-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-018-0093-4