Abstract
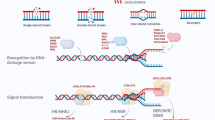
The ascomycetous yeast Yarrowia lipolytica has been established as model system for studies of several research topics as well as for biotechnological processes in the last two decades. However, frequency of heterologous recombination is high in this yeast species, and so knockouts of genes are laborious to achieve. Therefore, the aim of this study was to check whether a reduction of non-homologous end-joining (NHEJ) of double strand breaks (DSB) results in a strong increase of proportion of homologous recombinants. The Ku70–Ku80 heterodimer is known as an essential protein complex of the NHEJ. We show that deletion of YlKU70 and/or YlKU80 results in an increase of the rate of transformants with homologous recombination (HR) up to 85 % in each case. However, it never reaches near 100 % of HR in any case as described for some other yeast. Furthermore, we demonstrated that growth of Δylku strains was similar to that of the wild-type strain. In addition, no differences were detected between the Δylku strains and the parent strain in respect to sensitivity to the mutagenic agent EMS as well as to the antibiotics hygromycin, bleomycin and nourseothricin. However, Δylku70 and Δylku80 strain showed a slightly higher sensitivity against UV rays. Thus, the new constructed Δylku strains are attractive recipient strains for homologous integration of DNA fragments and a valuable tool for directed knockouts of genes. Nevertheless, our data suggest the existence of another system of non-homologous recombination what may be subject of further investigation.


Similar content being viewed by others
References
Abdel-Banat BM, Nonklang S, Hoshida H, Akada R (2010) Random and targeted gene integrations through the control of non-homologous end joining in the yeast Kluyveromyces marxianus. Yeast 27:29–39
Barth G, Gaillardin C (1996) Yarrowia lipolytica. In: Wolf K (ed) Nonconventional yeasts in biotechnology, pp 313–388
Barth G, Gaillardin C (1997) Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol Rev 19:219–237
Barth G, Weber H (1982) Genetic studies on the yeast Saccharomycopsis lipolytica: inactivation and mutagenesis. Z Allg Mikrobiol. 23(3):147–157
Barth G, Beckerich JM, Dominguez A, Kerscher S, Ogrydziak DM, Titorenko VI, Gaillardin C (2003) Functional genetics of Yarrowia lipolytica. In: de Winde JH (ed) Functional genetics of industrial yeasts, vol 2, pp 227–271
Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C (1993) A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res 21:3329–3330
Boulton SJ, Jackson SP (1996a) Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res 24:4639–4648
Boulton SJ, Jackson SP (1996b) Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J 15:5093–5103
Casaregola S, Neuveglise C, Lepingle A, Bon E, Feynerol C, Artiguenave F, Wincker P, Gaillardin C (2000) Genomic exploration of the hemiascomycetous yeasts: 17 Yarrowia lipolytica. FEBS Lett 487:95–100
Chang PK (2008) A highly efficient gene-targeting system for Aspergillus parasiticus. Lett Appl Microbiol 46:587–592
Cheon SA, Han EJ, Kang HA, Ogrydziak DM, Kim JY (2003) Isolation and characterization of the TRP1 gene from the yeast Yarrowia lipolytica and multiple gene disruption using a TRP blaster. Yeast 20:677–685
Daley JM, Palmbos PL, Wu D, Wilson TE (2005) Nonhomologous end joining in yeast. Annu Rev Genet 39:431–451
Decottignies A (2007) Microhomology-mediated end joining in fission yeast is repressed by pku70 and relies on genes involved in homologous recombination. Genetics 176:1403–1415
Dower WJ, Miller JF, Ragsdale CW (1988) High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res 16:6127–6145
Dujon B, Sherman D, Fischer G et al (2004) Genome evolution in yeasts. Nature 430:35–44
Duquesne S, Bordes F, Fudalej F, Nicaud JM, Marty A (2012) The yeast Yarrowia lipolytica as a generic tool for molecular evolution of enzymes. Methods Mol Biol 861:301–312
Fickers P, Le Dall MT, Gaillardin C, Thonart P, Nicaud JM (2003) New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica. J Microbiol Methods 55:727–737
Fickers P, Fudalej F, Nicaud JM, Destain J, Thonart P (2005) Selection of new over-producing derivatives for the improvement of extracellular lipase production by the non-conventional yeast Yarrowia lipolytica. J Biotechnol 115:379–386
Förster A, Aurich A, Mauersberger S, Barth G (2007a) Citric acid production from sucrose using a recombinant strain of the yeast Yarrowia lipolytica. Appl Microbiol Biotechnol 75:1409–1417
Förster A, Jacobs K, Juretzek T, Mauersberger S, Barth G (2007b) Overexpression of the ICL1 gene changes the product ratio of citric acid production by Yarrowia lipolytica. Appl Microbiol Biotechnol 77:861–869
Friedberg EC, Bardwell AJ, Bardwell L, Feaver WJ, Kornberg RD, Svejstrup JQ, Tomkinson AE, Wang Z (1995) Nucleotide excision repair in the yeast Saccharomyces cerevisiae: its relationship to specialized mitotic recombination and RNA polymerase II basal transcription. Philos Trans R Soc Lond B Biol Sci 347:63–68
Goins CL, Gerik KJ, Lodge JK (2006) Improvements to gene deletion in the fungal pathogen Cryptococcus neoformans: absence of Ku proteins increases homologous recombination, and co-transformation of independent DNA molecules allows rapid complementation of deletion phenotypes. Fungal Genet Biol 43:531–544
Hoffman CS, Winston F (1987) A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267–272
Holz M, Förster A, Mauersberger S, Barth G (2009) Aconitase overexpression changes the product ratio of citric acid production by Yarrowia lipolytica. Appl Microbiol Biotechnol 81:1087–1096
Holz M, Otto C, Kretzschmar A, Yovkova V, Aurich A, Pötter M, Marx A, Barth G (2011) Overexpression of alpha-ketoglutarate dehydrogenase in Yarrowia lipolytica and its effect on production of organic acids. Appl Microbiol Biotechnol 89:1519–1526
Kanaar R, Hoeijmakers JH, van Gent DC (1998) Molecular mechanisms of DNA double strand break repair. Trends Cell Biol 8:483–489
Kato A, Akamatsu Y, Sakuraba Y, Inoue H (2004) The Neurospora crassa mus-19 gene is identical to the qde-3 gene, which encodes a RecQ homologue and is involved in recombination repair and postreplication repair. Curr Genet 45:37–44
Kooistra R, Hooykaas PJ, Steensma HY (2004) Efficient gene targeting in Kluyveromyces lactis. Yeast 21:781–792
Lindegren G, Hwang YL, Oshima Y, Lindegren CC (1965) Genetical mutants induced by ethyl methanesulfonate in Saccharomyces. Can J Genet Cytol 7:491–499
Ma JL, Kim EM, Haber JE, Lee SE (2003) Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol Cell Biol 23:8820–8828
Maassen N, Freese S, Schruff B, Passoth V, Klinner U (2008) Nonhomologous end joining and homologous recombination DNA repair pathways in integration mutagenesis in the xylose-fermenting yeast Pichia stipitis. FEMS Yeast Res 8:735–743
Madzak C, Gaillardin C, Beckerich JM (2004) Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: a review. J Biotechnol 109:63–81
Madzak C, Otterbein L, Chamkha M, Moukha S, Asther M, Gaillardin C, Beckerich JM (2005) Heterologous production of a laccase from the basidiomycete Pycnoporus cinnabarinus in the dimorphic yeast Yarrowia lipolytica. FEMS Yeast Res 5:635–646
Mauersberger S, Wang HJ, Gaillardin C, Barth G, Nicaud JM (2001) Insertional mutagenesis in the n-alkane-assimilating yeast Yarrowia lipolytica: generation of tagged mutations in genes involved in hydrophobic substrate utilization. J Bacteriol 183:5102–5109
Mauersberger S, Kruse K, Barth G (2003) Chapter 63. Induction of citric acid/isocitric acid and α-ketoglutaric acid production in the yeast Yarrowia lipolytica. In: Wolf K, Breunig K, Barth G (eds.) Non-conventional yeasts in genetics, biochemistry and biotechnology. Practical protocols. Springer, Berlin, pp 393–400
Meyer V, Arentshorst M, El-Ghezal A, Drews AC, Kooistra R, van den Hondel C, Rahm A (2007) Highly efficient gene targeting in the Aspergillus niger kusA mutant. J Biotechnol 128:770–775
Naatsaari L, Mistlberger B, Ruth C, Hajek T, Hartner FS, Glieder A (2012) Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS One 7:e39720. doi:10.1371/journal.pone.0039720
Ninomiya Y, Suzuki K, Ishii C, Inoue H (2004) Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci USA 101:12248–12253
Otto C, Yovkova V, Barth G (2011) Overproduction and secretion of α-ketoglutaric acid by microorganisms Mini-review. Appl Microbiol Biotechnol 92:689–695
Pastink A, Lohman PH (1999) Repair and consequences of double-strand breaks in DNA. Mutat Res 428:141–156
Pastink A, Eeken JC, Lohman PH (2001) Genomic integrity and the repair of double-strand DNA breaks. Mutat Res 480–481:37–50
Pöggeler S, Kück U (2006) Highly efficient generation of signal transduction knockout mutants using a fungal strain deficient in the mammalian ku70 ortholog. Gene 378:1–10
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning. A laboratory manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sauer B, Whealy M, Robbins A, Enquist L (1987) Site-specific insertion of DNA into a pseudorabies virus vector. Proc Natl Acad Sci USA 84:9108–9112
Shinohara A, Ogawa T (1995) Homologous recombination and the roles of double-strand breaks. Trends Biochem Sci 20:387–391
Sperrhake M (2008) Entwicklung neuer Wirts-Vektor-Systeme zur Selektion von Transformaden der Hefe Yarrowia lipolytica. TU Dresden, Diploma thesis
Stottmeister U, Behrens U, Weissbrodt E, Barth G, Franke-Rinker D, Schulze E (1982) Utilization of paraffins and other noncarbohydrate carbon sources for microbial citric acid synthesis. Z Allg Mikrobiol 22:399–424
Symington LS (2002) Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev 66:630–670
Ueno K, Uno J, Nakayama H, Sasamoto K, Mikami Y, Chibana H (2007) Development of a highly efficient gene targeting system induced by transient repression of YKU80 expression in Candida glabrata. Eukaryot Cell 6:1239–1247
Uhlig T (2009) Herstellung neuer Rezipientenstämme der Hefe Yarrowia lipolytica. TU Dresden, Bachelor thesis
Verbeke J, Beopoulos A, Nicaud JM (2012) Efficient homologous recombination with short length flanking fragments in Ku70 deficient Yarrowia lipolytica strains. Biotechnol Lett. doi:10.1007/s10529-012-1107-0
Wang TT, Lee CF, Lee BH (1999) The molecular biology of Schwanniomyces occidentalis klocker. Crit Rev Biotechnol 19:113–143
Acknowledgments
We would like to thank Maria Sperrhake and Thomas Uhlig for supplying the vectors pTRPPTU and pUC-HIS3-DK as well as Annett Buchwald for her technical help during this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Gaillardin.
Rights and permissions
About this article
Cite this article
Kretzschmar, A., Otto, C., Holz, M. et al. Increased homologous integration frequency in Yarrowia lipolytica strains defective in non-homologous end-joining. Curr Genet 59, 63–72 (2013). https://doi.org/10.1007/s00294-013-0389-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-013-0389-7