Abstract
Background
Yarrowia lipolytica is an oleaginous yeast which has emerged as an important microorganism for several biotechnological processes, such as the production of organic acids, lipases and proteases. It is also considered a good candidate for single-cell oil production. Although some of its metabolic pathways are well studied, its metabolic engineering is hindered by the lack of a genome-scale model that integrates the current knowledge about its metabolism.
Results
Combining in silico tools and expert manual curation, we have produced an accurate genome-scale metabolic model for Y. lipolytica. Using a scaffold derived from a functional metabolic model of the well-studied but phylogenetically distant yeast S. cerevisiae, we mapped conserved reactions, rewrote gene associations, added species-specific reactions and inserted specialized copies of scaffold reactions to account for species-specific expansion of protein families. We used physiological measures obtained under lab conditions to validate our predictions.
Conclusions
Y. lipolytica iNL895 represents the first well-annotated metabolic model of an oleaginous yeast, providing a base for future metabolic improvement, and a starting point for the metabolic reconstruction of other species in the Yarrowia clade and other oleaginous yeasts.
Similar content being viewed by others
Background
Even if lipid metabolism is common to all microorganisms, we call oleaginous those that can store at least 20% of their dry mass as lipids. It is possible to find oleaginous organisms among plants, algae, bacteria and yeasts. Plants and algae are technically difficult (and controversial) to modify genetically, while oleaginous bateria present a low growth rate. On the other side, oleaginous yeasts enjoy well-developed genetic tools for their improvement and grow quickly. Also, oleaginous yeasts can accumulate up to 70% of their dry mass as lipids[1], making them the best candidates for industrial lipid production such as microbial oil for biodiesel.
One of those oleaginous yeasts, Yarrowia lipolytica, normally found as a food contaminant, has been extensively studied experimentally. It is easy to modify genetically, and presents many opportunities for metabolic engineering. For example, Y. lipolytica has been used as a food supplement, given its easily modifiable lipid composition. It is also studied as a potential source of biodiesel[2–4], because lipids producedby this species are similar to vegetable oils and fats. While Y. lipolytica is a hemiascomycete yeast, it is phylogenetically distant from S. cerevisiae and other well-studied yeasts, manifesting many metabolic differences: it is an obligate aerobic yeast, that can use normal hydrocarbons and various fats as carbon sources; it secretes diverse hydrolytic enzymes (proteases, lipases, RNases); its perixosome is constitutive.
Metabolic models are an important tool for metabolic engineering. Their uses include the guidance of metabolic engineering, the contextualization of high-throughput data and helping hypothesis-driven discovery.
Genome-scale metabolic models have up to now been principally produced for bacterial species and for a few higher organisms (see[5] for a review). This focus on model organisms is in part due to the great cost of obtaining high-quality annotated complete genome sequences, which requires considerable human effort regardless of the relative low cost of obtaining the genome sequence. A further need is to produce new experimental data to verify and improve the reconstructed model. Most models are reconstructed starting from the genome annotation, assembling known reactions into connected networks[6]. This requires a lengthy and expensive period of manual curation. Software has been designed to deal with process, although most existing tools are designed for bacteria.
Y. lipolytica is an ideal species for metabolic reconstruction in eukaryotes through comparative genomics. As one of the hemiascomycetous yeasts completely sequenced in the Génolevures program, it enjoys a high quality manual annotation by a network of expert curators[7, 8]. Careful analysis of conservation and species-specific expansion and contraction of families of protein-coding genes makes it possible to identify orthologs with known genes in the clade as well as functionally important paralogous families. The conservation of core metabolism with other yeasts is enough to allow the use of existing metabolic models from S. cerevisiae as a template, into which species-specific reactions and secondary metabolism can be assembled.
In this work we present the first genome-scale functional metabolic model for Y. lipolytica, built with an iterative process of automatic reconstruction and manual curation. We started from a scaffold derived from existing S. cerevisiae models, extracting information about enzymatic reactions, molecular species, transport reactions, and compartments. With this scaffold we built an in silico draft by mapping known enzyme-encoding genes, using gene homology information obtained from Génolevures protein families[8, 9] and complemented with other in silico methods, and filled network gaps in order to make it functional (i.e. to be able to predict growth from available metabolites in the media). We performed a manual curation of the initial draft model, adding species-specific metabolic reactions, in particular those related with central carbon and fatty acid metabolism. To assess the predictive power of our model, we compared our predictions against published experimental results of growth under different media conditions and gene knockouts. This comparison shows high degree of agreement between predictions and experimental results.
Results
Properties of the model
Our functional genome-scale metabolic model for Yarrowia lipolytica iNL895 describes 2 002 reactions encoded by 895 Y. lipolytica genes, the 1 847 metabolites consumed and produced by those reactions, the 16 compartments in which those reactions take place and a biomass function which describes the metabolic requirements for growth.
From the total of reactions, 139 (7%) are transport reactions with a gene association, 286 (14.3%) transport reactions that are spontaneous or without a known gene association, 171 (8.5%) are exchanges with the media, 1 055 (52.7%) enzymatic reactions with a gene association and 351 (17.5%) without.
The 1 055 enzymatic reactions with associated genes in the curated model were distributed into 39 biological processes, based on the associated GO Slim annotation of the closest ortholog in S. cerevisiae.
Gains
For alkane degradation we have introduced the ω-oxidation pathway including cytochrome P450 oxidases (12 genes ALK1–ALK12) and the cytochrome P450 reductase (CPR) reaction from each alkane (decane, dodecane, hexadecane, etc.) to the corresponding alcohol. We added reactions from alcohols to aldehydes then aldehydes to corresponding fatty acids, following[10].
For triglyceride degradation Y. lipolytica secretes lipases that are either extracellular (Lip2p) or membrane-bound (Lip7 and Lip8). These are part of a 19-gene multi-gene family[3].
For fatty acid synthesis we have included in reaction r_2008 the effect of a new gene that codes a member of the type 1 acyl-CoA:diacylglycerol acyltransferase family (DGAT1), which has not previously been identified in yeasts, but is commonly found in mammals and plants, and proposed in[11] to give the oleaginous character.
For transport and export of hydrophobic substrates we have included reactions for binding, export (by an ABC transporter, the gene ABC1 for alkane utilization[10]), and metabolite transport. The latter transport is necessary to explain growth of the TCA cycle mutant and the malate dehydrogenase mutant..
Losses
For oleaginous character, we have taken into account the loss in Y. lipolytica of genes linked to glycerol 3 phosphate (G3P), following[12], specifically in reaction r_0528 reported inAdditional file 1: Table S1.
For galactose and sucrose substrates we verified the absence of reactions that are not present in Y. lipolytica due to missing genes (compared to S. cerevisiae), in particular Y. lipolytica cannot use sucrose as a sole carbon source due to the lack of the corresponding invertase. Note that transformation of Y. lipolytica strains is made possible through the inclusion of a selective marker built from a fusion of the S. cerevisiae SUC2 gene with the promoter and signal sequence of Y. lipolytica alkaline extracellular protease XPR2[13].
For ethanol production, unlike S. cerevisiae, Y. lipolytica uses ethanol only with difficulty and does not produce it. These losses are reflected in reactions r_0176 through r_0190 concerning acetaldehyde to ethanol transformation.
Validation of the model
The draft model was verified by experts in Y. lipolytica, and approved in terms of agreement with the literature: This model is not capable of producing ethanol, it cannot grow anaerobically, fatty acid metabolism presented expansions and contractions of protein families, and new species-specific reactions for the intake of alkanes were automatically detected.
Also, to assess the completeness of our model, we compared its phenotypic predictions in terms of growth/no growth, against published experimental results of observed growth, under several carbon sources and gene knockouts (Additional file 2: Table S2). We used flux balance analysis (FBA), and a constraint based optimization approach[14] to predict whether a phenotype was present. After defining restrictions in the intake capacity of the organism, based on a selection of experimental data, we used FBA to predict biomass production, and thus the capacity of the organism to grow under those restrictions. Gene knockouts were modeled as deletions in the reconstructed metabolic network.
Media conditions, in particular different carbon sources, were extracted from the literature (See Table 1). Alas, not all experiments were well documented in terms of molecular species present in the media, so a rich media (YPD) was assumed and modified based on the general description of the media. See[15] for a discussion about uncertainty in media conditions.
In order to facilitate comparison, quantitative results from experiments and from simulations of biomass production were simplified into binary values (growth/no growth). Corresponding binary results were obtained for 98 experiments paired with simulations, with exact agreement in 64 cases (39 true positives and 25 true negatives). The 18 false negatives we observed may be attributed to missing reactions, corresponding to Y. lipolytica genes that are still unannotated, or to gaps in understanding of redundancy in the network. These 18 cases are currently being used to target improvements in gene annotation. The remaining cases, 16 false positives, are likely the product of over-optimistic flux simulations and can be reduced through parameter tuning. Overall, using this simplified binary comparison we obtain an accuracy (geometric mean of sensitivity and specificity) of 0.65.
We stress that this qualitative validation does not substitute for quantitative comparison, but does show that each of the tested conditions is connected from uptake through to the biomass function. It thus serves to validate the completeness of the model, in particular with respect to overall network topology.
Conclusions and discussion
Combining in silico tools and expert manual curation, we produced an accurate genome-scale metabolic model of the oleaginous yeast Y. lipolytica, using a functional metabolic model of the phylogenetically related yeast S. cerevisiae as a scaffold for the reconstruction. The method developed in the present work can be used for genome-scale metabolic model reconstruction of other organisms, making it a useful tool for biotechnology and research.
We noticed that, even if the list of S. cerevisiae reactions not present in Y. lipolytica was short, there was an important number of changes in the gene associations between both organisms. Also, the loss of some phenotypes in Y. lipolytica, compared to S. cerevisiae, was characterized by a loss of a small number of genes.
Thirteen new transport reactions were added to the new model in order to connect enzymatic reactions inside the peroxisome with molecular species in the cytosol, and to import species from extracellular space to the cytosol. We could not find genes encoding for all those transports, but we expect that the eventual characterization of the 1 034 (16%) Y. lipolytica genes with unknown function, will provide evidence for some of them. The lack of accuracy at predicting some experiments could be explained by missing reactions in the model, especially regarding the transport of specific carbon sources. This gives us hints about possible ways to improve our model.
The modifications to the draft model performed by the manual curators allowed us to formalize a set of edit operations over metabolic models. This facilitated an automatic iteration process, from improvements to the reconstruction method, to improved draft models, to automatic application of curator edits, to automatic assertion of accuracy.
The present model can be used to predict growth under different media conditions and gene knock-outs. It can also be used as a general description of the state-of-the-art in Y. lipolytica metabolism. Data from high-throughput experiments, like microarrays and metabolomics, can be mapped to this model to have an overview of metabolic changes under different media conditions.
Current understanding of Y. lipolytica is constantly improving, and a number of features of its metabolism are the subject of ongoing work and consequently improvements to the model. Multigene families such as POX1–POX6 in peroxisomal β-oxidation could be modeled with better precision, since there are enzymatic specificities linked to the length of the carbon chain (e.g. Pox2 for long chains, Pox3 for short chain fatty acids, see for example[26]). This is also true for multigene families LIP1–LIP19 hydrolases of triacylglycerides, where there also exists chain-length specificity[3], although the specificities of the ALK1–ALK19 genes are not completely known. In general, lipid metabolism in Y. lipolytica is still under study and there is a lack of knowledge in several areas, such as transport between compartments, or the link between nitrogen abundance and the production of either lipid or citric acid[11].
Expansion of families of isozymes is detectable through expansion of paralogous protein families, but the method used here cannot detect these differences because FBA does not differentiate isoenzyme activities in the same reaction. Dynamic models that describe the kinetics of individual enzymes in reactions must be developed. This will require acquiring and integrating metabolic and transcriptomic data for targeted pathways, and developing models. Alvarez-Vasquez et al.[27], for example, used biochemical systems theory to develop a model of S. cerevisiae sphingolipid metabolism; more recently, Gupta et al.[28] developed a quantitative model of this pathway in mammalian cells by combining metabolite and transcriptome data in their estimation of kinetic rate constants. In general, the constraint-based FBA approach used here for validation cannot describe Y. lipolytica metabolic pathways with the same precision as dynamic differential equation models, but does have the merit of permitting a whole-genome model.
The most pressing need in further iterations of the model is refinement of alkane degradation for decane and hexadecane. Indeed the analysis of alkane growth of ANT1 and ABC1 mutants were performed on n-alkane from C10 to C16, including C11, C13, and C15, in[10]. Also, Y. lipolytica is described as growing on n-alkane paraffin (petroleum distillate) containing n-alkane oil (C12 to C18 n-akanes) and also n-paraffin wax (C20 and above, solid alkane) in[29]. This suggests that it is necessary to introduce all even and odd chain lengths including C1, since Y. lipolytica could use very long alkane chains above C20.
Methods
Scaffold-based reconstruction
Genome-scale metabolic models describe the network of enzymatic and transport reactions in an organism. The main idea of most metabolic model reconstruction algorithms is to look for the presence of enzymatic reactions in the annotated genome of the organism to be modeled, and create a network of those reactions, representing the interconnected production and consumption of metabolites[6].
The construction of metabolic models is costly and time consuming, so tools have been developed to automatically create initial, draft versions of the models, to be further improved by manual curation. Some of the current methods and platforms are Pathway Tools[30], The SEED[31], AUTOGRAPH[32], and several machine learning methods[33].
These methods are mostly designed for bacterial organisms and are not always adequate for reconstruction of yeasts models. In particular, some of them lack proper handling of compartments, rewriting of gene associations, or rely on the strong functional relations provided by operons. Also, fine tuning existing programs was not always possible, given the lack of public source code availability. To cover these shortcomings, we implemented our own automatic reconstruction method (to be published separately). SeeAdditional file 3: Figure S1 for an overview of our method.
Briefly, the method developed for the present work uses a scaffold model for the reconstruction. For each one of the genes associated to reactions described in the scaffold, we look for possible orthologs in the target organism. If certain conditions are met, the reaction is considered to be conserved, and added to the network of the target organism.
This method of projection can be applied to any pair of phylogenetically close species. Given a set of ortholog maps between two genomes, and a well-annotated metabolic model for one of them, it automatically produces a draft model for the target, providing a well-documented starting point for manual curation.
Well-curated models include information about the dependency of each reaction on proteins and genes, which is called Gene-Protein-Reaction associations (GPR). The Gene Association is the dependency of a reaction on the presence of a combination of genes, described as a logical formula between gene identifiers. For example, S. cerevisiae reaction R_0005 (“1,3-beta-glucan synthase”) can be performed by either the product of gene YLR342W (FKS1) or the product of gene YGR032W (GSC2), so its Gene Association is “(YGR032W or YLR342W)”.
During the reconstruction of the iNL895 Y. lipolytica model, we used three functional models published for S. cerevisiae: iMM904[34], iIN800[35] and the consensus model version 4.36[36]. The latter was used as a scaffold for the reconstruction of the Y. lipolytica metabolic model, and will be referenced as the ‘scaffold model’ in what follows. We used the detailed fatty acid metabolism described in iIN800[35] as a scaffold for Y. lipolytica fatty acid metabolism. From the scaffold model, we extracted the reactions predicted to be present in Y. lipolytica, the metabolites consumed and produced by them, the cellular compartments and all the non-enzymatic transport reactions. To make our model functional, we produced a list of genes that restored connectivity between the metabolites imported by the organism and the metabolic requirements of the biomass function. This list of genes provided as a starting point for the manual curation of the model.
Orthology
Orthology detection based on sequence similarity is the most used approach to predict if a biological function, encoded by genes, is conserved between two organisms[37]. Some special cases need to be treated carefully: two ortholog genes, with originally similar functions, can mutate slightly and change its function, or can suffer a duplication, so only one of the two copies will keep the same biological function. Also, a fusion or fission event can integrate or divide certain domains into different genes. All those cases need to be integrated in the study of the conservation of function between two organisms and, in our experience, none of the current methods of ortholog mapping is good at all of them.
Based on homology between the genome of the scaffold (S. cerevisiae) and the genome of the target (Y. lipolytica), we determine if the original genes that encode the protein required for the enzymatic activity are conserved. Our method determines a) if a reaction is conserved, b) if a re-written gene association formula for the reaction is necessary (Additional file 4: Figure S2).
For the reconstruction of the metabolic model of Y. lipolytica, we leveraged data provided by the Génolevures program[38], in the form of multi-species protein families and gene synteny. Protein families identify phylogenetic groups of proteins sequences that are a leading indication of functional analogy.
Génolevures protein families were further subdivided into groups with the same protein domain architecture (DOM), and synteny (SONS[38]) This initial high quality annotation allowed us to map most, but not all, of the genes used by the scaffold model, so we complemented this mapping with orthology from Inparanoid-DB[39] and OrthoMCL-DB[40].
In the cases of divergent predictions, consensus was determined by the following election procedure: From the different methods we produce a tally of the number of times each paralog group appears between all existing homolog map.
Our translator, using the rules described in Table 2, looks for the possible rewritings of the scaffold gene formulas in terms of genes of the target organism. To rewrite the new gene associations, an homolog map was built with the votes between all our available methods to detect orthologs (Additional file 4: Figure S2).
The formulas that could not be resolved where reported to manual curation, as a possible loss of function (see Table 1). The resulting formulae were normalized to conjunctive normal form, as a list of alternative ways to encode the same reaction. Some examples of formula rewriting are provided in Table 2.
A projected model
After rewriting gene associations, we kept the corresponding molecular species with its identifiers and annotations. We kept all the relevant non-enzymatic transport reactions and compartments.
A model that is able to predict growth is called a functional model. To predict growth, a measure of the molecular requirements to create a copy of the organism should be provided, in the form of a biomass function. This is usually obtained by the analysis of the molecular contents of live cells[41].
The biomass function of the S. cerevisiae model was used as a starting point for the Y. lipolytica model. Some coefficients were adjusted using the amount of DNA to be produced and the GC contents of the target organism[15]. G + C content and genome length of Y. lipolytica were obtained from the Génolevures program[38].
Automatic reconstructions may produce incomplete networks, missing the presence of some reactions that are part of an existing path of reactions. These “gaps” may lead to incorrect predictions, so they need to be fixed. We analyzed those gaps, generated lists of candidates, and included them as part of the manual curation stage. We also verified whether any of the Y. lipolytica genes were annotated with an EC code not present in the draft, adding new reactions to the model (see alsoAdditional file 5).
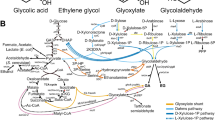
Given the importance of compartmentalization in eukaryotic organism, we built a model with 16 compartments, allowing us to map reactions and metabolites to different parts of the cell. We are interested in the oleaginous nature of Y. lipolytica, and its possible biotechnological applications, so it was critical to focus on the differences in fatty acid metabolism with respect to other yeasts. We started with the description of β-oxydation and fatty acid elongation from iIN800, projected them to Y. lipolytica, and manually modified to mirror the relevant literature (Figure1).
Projecting Fatty Acid β -oxidation from S. cerevisiaeto Y. lipolytica.
This simplified schematic view shows how the Fatty Acid β-oxidation scaffold pathway from S. cerevisiae iIN800[35] was modified to adequately describe Y. lipolytica metabolism. (a) Simplified version of fatty acid β-oxidation diagram of S. cerevisiae iIN800. (b) Fatty acid β-oxidation in the reconstructed model for Y. lipolytica, with a constitutive peroxisome compartment and cytosol ↔ peroxisome transport reactions. Species-specific transport mechanisms for long and short fatty acid chains (PXA1,2 and PEX11) are highlighted in green and blue. Long chains are activated (-CoA) before being transported to the peroxisome. Y. lipolytica can directly process Octanoic (C8), Hexanoic (C6), Butyric (C4) acid, and C18:2, so they were added to our model (in yellow). Our method predicted the family expansion of S. cerevisiae POX1/FOX1 into POX1-6, and the reduction of S. cerevisiae family FAA1-4 to FAA1 (YALI0D17864g), which modified the genome associations of most of the pathway. POX1-6 are written in order of specificity: POX2,5,4 for long chains and POX3,5,4 for short chains[42].
We used the diagram of iIN800[35] as an starting point for our own diagram of Y. lipolytica metabolism. This poster was used to discuss the draft model with the curators, who suggested changes based on their experience with the modeled species. These changes were translated to edits operations, and applied to our draft model.
The feedback obtained from the simulations of growth under different conditions (see below) and the results of gap-filling analysis were also used as part of the manual curation.
Validation
To assess the predictive power of our metabolic model, we compared growth predictions, obtained using Flux Balance Analysis (FBA)[14], against 152 experimental results extracted from the literature. The effects of media conditions on growth, and the effects of gene knockouts in the system were included as constraints to the linear programming problem solved during FBA. From the literature we manually extracted experimental evidence (a growth/no growth indicator or a growth curve in time for each condition/deletion).
When growth curves were provided, we calculated a boolean value representing growth (true) or no growth (false), where the threshold was decided based on 1/3 of the average of growth in time (OD), for all mutants studied[43, 44]. The same was done with simulated results: a threshold was used to decide between growth and no growth.
A confusion matrix and geometric mean[45] was used to measure the accuracy of our predictions versus experimental results. This approach was used to assess the quality of a model as a predictor, as it was done with the reconstruction of S. cerevisiae iIN800[35] and iLL672[46].
From the list of experimental results from the literature we produced a table of experiments, summarizing media conditions, gene knockouts, and observed growth (SeeAdditional file 2: Table S2).
The description of media conditions were not standard between different works, so we defined, to the best of our knowledge, a base condition based on YPD, where only non-carbon sources were available (nitrogen, oxygen, etc.). This was modified for each simulation, controlling the availability of different carbon sources. The name of media conditions used inAdditional file 2: Table S2, were obtained from the literature listed in Table 1, and describe the following combinations: YNBD: base + Glucose, YNBcas: YNBD + Casaminoacids, YNBO: base + Oleic acid, YNBC10: base + Decane, YNBC16: base + Hexadecane, YNBT: base + Trybutirin, YNBDptr: YNBD + Putrescine, YNBDtry: YNBD + Tryptophane.
We used FBA from COBRA Tools[47] to predict growth rate under different media conditions and gene knockouts that matched the available experimental results. From those simulations and the associated experiment, we automatically generated MATLAB tests, which generated an accuracy report of our model, consisting of False Positives/Negatives, True Positives/Negatives between the expected and predicted phenotype. The MATLAB file that simulates the 152 experiments is available asAdditional file 6. The results are also available inAdditional file 2: Table S2.
We called our reconstructed model iNL895, following the rules defined in[48]. We produced a version of our model in SBML format (Systems Biology Markup Language)[49], in order to analyze it with compatible existing tools, and share it with the community (seeAdditional file 7). An updated COBRA-compatible SBML version of our model can be retrieved from the BioModels database (http://biomodels.org), searching for the model id MODEL1111190000.
References
Li Q, Du W, Liu D: Perspectives of microbial oils for biodiesel production. Appl Microbiol Biotechnol 2008,80(5):749-756. 10.1007/s00253-008-1625-9
Papanikolaou S, Chevalot I, Komaitis M, Marc I, Aggelis G: Single cell oil production by Yarrowia lipolytica growing on an industrial derivative of animal fat in batch cultures. Appl Microbiol Biotechnol 2002,58(3):308-312. 10.1007/s00253-001-0897-0
Fickers P, Benetti PH, Waché Y, Marty A, Mauersberger S, Smit MS, Nicaud JM: Hydrophobic substrate utilisation by the yeast Yarrowia lipolytica, and its potential applications. FEMS Yeast Res 2005,5(6–7):527-543.
Beopoulos A, Cescut J, Haddouche R, Uribelarrea J, Molina-Jouve C, Nicaud JM: Yarrowia lipolytica as a model for bio-oil production. Prog Lipid Res, ; 2009.
Oberhardt MA, Palsson BO, Papin JA: Applications of genome-scale metabolic reconstructions. Mol Syst Biol 2009, 5: 320.
Thiele I, Palsson BO: A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc 2010, 5: 93-121.
Dujon B, Sherman DJ, Fischer G, Durrens P, Casaregola S, Lafontaine I, Montigny JD, Marck C, Neuvéglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich JM, Beyne E, Bleykasten C, Boisramé A, Boyer J, Cattolico L, Confanioleri F, de Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H, Groppi A, Hantraye F, Hennequin C, Jauniaux N, Joyet P, Kachouri R, Kerrest A, Koszul R, Lemaire M, Lesur I, Ma L, Muller H, Nicaud JM, Nikolski M, Oztas S, Ozier-Kalogeropoulos O, Pellenz S, Potier S, Richard GF, Straub ML, Suleau A, Swennen D, Tekaia F, Wésolowski-Louvel M, Westhof E, Wirth B, Zeniou-Meyer M, Zivanovic I, Bolotin-Fukuhara M, Thierry A, Bouchier C, Caudron B, Scarpelli C, Gaillardin C, Weissenbach J, Wincker P, Souciet JL: Genome evolution in yeasts. Nature 2004,430(6995):35-44. 10.1038/nature02579
Sherman DJ, Martin T, Nikolski M, Cayla C, Souciet JL, Durrens P: Génolevures: protein families and synteny among complete hemiascomycetous yeast proteomes and genomes. Nucleic Acids Res 2009,37(Database Issue):550-D554.
Nikolski M, Sherman DJ: Family relationships: should consensus reign?–consensus clustering for protein families. Bioinformatics 2007,23(2):e71-e76. 10.1093/bioinformatics/btl314
Thevenieau F, Dall MTL, Nthangeni B, Mauersberger S, Marchal R, Nicaud JM: Characterization of Yarrowia lipolytica mutants affected in hydrophobic substrate utilization. Fungal Genet Biol 2007,44(6):531-542. 10.1016/j.fgb.2006.09.001
Beopoulos A, Haddouche R, Kabran P, Dulermo T, Chardot T, Nicaud JM: Identification and characterization of DGA2, an acyltransferase of the DGAT1 acyl-CoA:diacylglycerol acyltransferase family in the oleaginous yeastYarrowia lipolytica. New insights into the storage lipid metabolism of oleaginous yeasts. Appl Microbiol Biotechnol 2011, 1-15. [10.1007/s00253-011-3506-x]
Dulermo T, Nicaud JM: Involvement of the G3P shuttle andβ-oxidation pathway in the control of TAG synthesis and lipid accumulation inYarrowia lipolytica. Metabolic Eng 2011,13(5):482-91. http://www.sciencedirect.com/science/article/pii/S1096717611000516 10.1016/j.ymben.2011.05.002
Nicaud JM, Fabre E, Gaillardin C: Expression of invertase activity inYarrowia lipolyticaand its use as a selective marker. Curr Genet 1989,16(4):253-260. Pmid:2627713 10.1007/BF00422111
Lee JM, Gianchandani EP, Papin JA: Flux balance analysis in the era of metabolomics. Brief Bioinformatics 2006,7(2):140-150. 10.1093/bib/bbl007
Suthers PF, Dasika MS, Kumar VS, Denisov G, Glass JI, Maranas CD: A genome-scale metabolic reconstruction of Mycoplasma genitalium, iPS189. PLoS Comput Biol 2009,5(2):e1000285. 10.1371/journal.pcbi.1000285
BioloMICS CK: CBS-KNAW Fungal Biodiversity Centre. http://bit.ly/enWFr3
van den Tempel T, Jakobsen M: The technological characteristics of Debaryomyces hansenii and Yarrowia lipolytica and their potential as starter cultures for production of Danablu. Int Dairy J 2000,10(4):263-270. 10.1016/S0958-6946(00)00053-4
Jardón R, Gancedo C, Flores CL: The gluconeogenic enzyme fructose-1,6-bisphosphatase is dispensable for growth of the yeast Yarrowia lipolytica in gluconeogenic substrates. Eukaryot Cell 2008,7(10):1742-1749. 10.1128/EC.00169-08
Flores CL, Gancedo C: Yarrowia lipolytica mutants devoid of pyruvate carboxylase activity show an unusual growth phenotype. Eukaryot Cell 2005,4(2):356-364. 10.1128/EC.4.2.356-364.2005
Yamagami S, Iida T, Nagata Y, Ohta A, Takagi M: Isolation and characterization of acetoacetyl-CoA thiolase gene essential for n-decane assimilation in yeast Yarrowia lipolytica. Biochem Biophys Res Commun 2001,282(3):832-838. 10.1006/bbrc.2001.4653
Haddouche R: Unpublished thesis (personal communication). Thesis, Laboratoire de Microbiologie et Génétique Moléculaire, INRA; 2010.
Kabran P: Etude du stockage et de la mobilisation des triglycérides chez la levure Yarrowia lipolytica. Thesis, Laboratoire de Microbiologie et Génétique Moléculaire, INRA; 2010.
Beopoulos A, Mrozova Z, Thevenieau F, Dall MTL, Hapala I, Papanikolaou S, Chardot T, Nicaud JM: Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl Environ Microbiol 2008,74(24):7779-7789. 10.1128/AEM.01412-08
Jiménez-Bremont JF, Ruiz-Herrera J, Dominguez A: Disruption of gene YlODC reveals absolute requirement of polyamines for mycelial development in Yarrowia lipolytica. FEMS Yeast Res 2001,1(3):195-204.
Cheon SA, Han EJ, Kang HA, Ogrydziak DM, Kim JY: Isolation and characterization of the TRP1 gene from the yeast Yarrowia lipolytica and multiple gene disruption using a TRP blaster. Yeast 2003,20(8):677-685. 10.1002/yea.987
Wang HJ, Dall MTL, Wach Y, Laroche C, Belin JM, Gaillardin C, Nicaud JM: Evaluation of acyl coenzyme A oxidase (Aox) isozyme function in the n-alkane-assimilating yeastYarrowia lipolytica. J Bacteriol 1999,181(17):5140-5148.
Alvarez-Vasquez F, Sims K, Cowart L, Okamoto Y, Voit E, Hannun Y: Simulation and validation of modelled sphingolipid metabolism in Saccharomyces cerevisiae. Nature 2005,433(7024):425-430. 10.1038/nature03232
Gupta S, Maurya MR, Merrill AH, Glass CK, Subramaniam S: Integration of lipidomics and transcriptomics data towards a systems biology model of sphingolipid metabolism. BMC Systems Biology 2011, 5: 26. 10.1186/1752-0509-5-26
Barth G, Gaillardin C: Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol Rev 1997,19(4):219-237. 10.1111/j.1574-6976.1997.tb00299.x
Paley SM, Karp PD: Evaluation of computational metabolic-pathway predictions for Helicobacter pylori. Bioinformatics 2002,18(5):715-724. 10.1093/bioinformatics/18.5.715
DeJongh M, Formsma K, Boillot P, Gould J, Rycenga M, Best A: Toward the automated generation of genome-scale metabolic networks in the SEED. BMC Bioinforma 2007, 8: 139. 10.1186/1471-2105-8-139
Notebaart RA, van Enckevort FHJ, Francke C, Siezen RJ, Teusink B: Accelerating the reconstruction of genome-scale metabolic networks. BMC Bioinforma 2006, 7: 296. 10.1186/1471-2105-7-296
Dale JM, Popescu L, Karp PD: Machine learning methods for metabolic pathway prediction. BMC Bioinforma 2010, 11: 15. 10.1186/1471-2105-11-15
Mo M, Palsson BO, Herrgard MJ: Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst Biol 2009, 3: 37. 10.1186/1752-0509-3-37
Nookaew I, Jewett MC, Meechai A, Thammarongtham C, Laoteng K, Cheevadhanarak S, Nielsen J, Bhumiratana S: The genome-scale metabolic model iIN800 of Saccharomyces cerevisiae and its validation: a scaffold to query lipid metabolism. BMC Syst Biol 2008, 2: 71. 10.1186/1752-0509-2-71
Herrgard MJ, Swainston N, Dobson P, Dunn WB, Arga KY, Arvas M, Blüthgen N, Borger S, Costenoble R, Heinemann M, Hucka M, Novère NL, Li P, Liebermeister W, Mo ML, Oliveira AP, Petranovic D, Pettifer S, Simeonidis E, Smallbone K, Spasić I, Weichart D, Brent R, Broomhead DS, Westerhoff HV, Kirdar B, Penttilä M, Klipp E, Palsson BO, Sauer U, Oliver SG, Mendes P, Nielsen J, Kell DB: A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat Biotechnol 2008,26(10):1155-1160. 10.1038/nbt1492
Kuzniar A, van Ham RCHJ, Pongor S, Leunissen JAM: The quest for orthologs: finding the corresponding gene across genomes. Trends Genet 2008,24(11):539-551. 10.1016/j.tig.2008.08.009
Souciet JL, Dujon B, Gaillardin C, Johnston M, Baret P, Cliften P, Sherman DJ, Weissenbach J, Westhof E, Wincker P, Jubin C, Poulain J, Barbe V, Ségurens B, Artiguenave F, Anthouard V, Vacherie B, Val M, Fulton R, Minx P, Wilson R, Durrens P, Jean G, Marck C, Martin T, Nikolski M, Rolland T, Seret M, Casaregola S, Despons L, Fairhead C, Fischer G, Lafontaine I, Leh V, Lemaire M, de Montigny J, Neuvéglise C, Thierry A, Blanc-Lenfle I, Bleykasten C, Diffels J, Fritsch E, Frangeul L, Goëffon A, Jauniaux N, Kachouri-Lafond R, Payen C, Potier S, Pribylova L, Ozanne C, Richard G, Sacerdot C: Straub M. Comparative genomics of protoploid Saccharomycetaceae. Genome Res, Talla E; 2009.
Remm M, Storm CE, Sonnhammer EL: Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol 2001,314(5):1041-1052. 10.1006/jmbi.2000.5197
Li L, Stoeckert CJ, Roos DS: OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 2003,13(9):2178-2189. 10.1101/gr.1224503
Varma A, Palsson BO: Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl Environ Microbiol 1994,60(10):3724-3731.
Beopoulos A, Chardot T, Nicaud JM: Yarrowia lipolytica: A model and a tool to understand the mechanisms implicated in lipid accumulation. Biochimie 2009.
Kumar VS, Maranas CD: GrowMatch: an automated method for reconciling in silico/in vivo growth predictions. PLoS Comput Biol 2009,5(3):e1000308. 10.1371/journal.pcbi.1000308
Joyce AR, Reed J, White A, Edwards R, Osterman A, Baba T, Mori H, Lesely SA, Palsson BO, Agarwalla S: Experimental and computational assessment of conditionally essential genes in Escherichia coli. J Bacteriol 2006,188(23):8259-8271. 10.1128/JB.00740-06
Kubat M: Holte R. Machine learning for the detection of oil spills in satellite radar images. Machine Learning, Matwin S; 1998.
Kuepfer L, Sauer U, Blank LM: Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res 2005,15(10):1421-1430. 10.1101/gr.3992505
Becker SA, Feist AM, Mo ML, Hannum G, Palsson BO, Herrgard MJ: Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protoc 2007,2(3):727-738. 10.1038/nprot.2007.99
Reed J, Vo TD, Schilling CH, Palsson BO: An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol 2003,4(9):R54. 10.1186/gb-2003-4-9-r54
Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles ED, Ginkel M, Gor V, Goryanin II, Hedley WJ, Hodgman TC, Hofmeyr JH, Hunter PJ, Juty NS, Kasberger JL, Kremling A, Kummer U, Novère NL, Loew LM, Lucio D, Mendes P, Minch E, Mjolsness ED, Nakayama Y, Nelson MR, Nielsen PF, Sakurada T, Schaff JC, Shapiro BE, Shimizu TS, Spence HD, Stelling J, Takahashi K, Tomita M, Wagner J, Wang J, Forum S: The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 2003,19(4):524-531. 10.1093/bioinformatics/btg015
Acknowledgements
We thank Dr. Thomas Desfougeres for initial discussions of Y. lipolytica fatty acid metabolism, and Intawat Nookaew for kindly providing us the source diagram of iIN800. This work was supported in part by the French GDR “Génolevures” (GDR 2354 CNRS) and the ANR “GENARISE” project (ANR-05-BLAN-0331-03). Nicolás Loira was supported by a doctoral fellowship from CONICYT-INRIA and by the Center for Mathematical Modeling (UMI-CNRS), Universidad de Chile. Thierry Dulermo was supported by the EU in the 7th Framework Program for Research and Technological Development, under ALFA-BIRD (Alternative Fuels and Biofuels for Aircraft Development) project. The authors would like to thank the anonymous reviewers for valuable advice and criticism.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
NL and DJS conceived the study and wrote the paper. NL implemented the reconstruction method and produced the model. TD and JMN provided expert knowledge in the form of manual curation. All authors read and approved the final manuscript.
Electronic supplementary material
12918_2011_920_MOESM1_ESM.doc
Additional file 1: Table S1 Manual curation of lost reactions. In many cases, orthology results fail to associate a target gene to an enzyme-coding gene in the scaffold model, suggesting that the reaction is absent. Each of these predictions were manually reviewed, where a reaction was confirmed as being absent (‘Lost’), or was upheld (‘Retained’) when empirical evidence was available. Genes for which no ortholog could be found are underlined in the gene association column. (DOC 42 KB)
12918_2011_920_MOESM2_ESM.pdf
Additional file 2 : Table S2 Validation of the iNL895 model. This table lists 152 experiments extracted from the literature, detailing media conditions, gene KOs, and observed growth (as yes/no). It also includes our simulations of the same experiments, obtained using FBA/COBRA Tools, and the comparison between observed and the simulated growth. (PDF 124 KB)
12918_2011_920_MOESM3_ESM.pdf
Additional file 3: Figure S1Projection pipeline from S. cerevisiae scaffold model to Y. lipolytica iNL895. The three main parts of our pipeline for the reconstruction of the Y. lipolytica model are: Projection, where the S. cerevisiae scaffold model and the information from different sources of orthology between S. cerevisiae and Y. lipolytica are used to produce a draft model, Curation, where the expert curators revised the candidates for gap-filling and added species-specific reactions and Validation, where experiments obtained from the literature were compared with our simulations, producing a detailed accuracy report. (PDF 238 KB)
12918_2011_920_MOESM4_ESM.pdf
Additional file 4: Figure S2 Gene Association rewrite from S. cerevisiae reactions to Y. lipolytica. Pipeline for gene-association rewriting, as part of the projection of Y. lipolytica iNL895 model. From the 4 ortholog maps provided by different methods, a map of votes of possible ortholog mappings is created. Then, from the scaffold model, we extracted gene associations for each reaction, and re-wrote them based on our map of homologs (e.g.: Reaction1: (SourceGene1 or SourceGene2) ↔ (TargetGene1)). The new reactions, this time associated with Y. lipolytica genes, constituted the base of the reconstructed model. (PDF 42 KB)
12918_2011_920_MOESM5_ESM.xls
Additional file 5 : Selected gene annotations in Y. lipolytica . This table lists Y. lipolytica genes used in the manual curation of the metabolic model. (XLS 34 KB)
12918_2011_920_MOESM6_ESM.zip
Additional file 6 : Complete validation tests for Y. lipolytica. This MATLAB file runs the validation tests of the Y. lipolytica metabolic model. It requires the COBRA Toolbox (2.0+). Each of the 152 tests is declared as a MATLAB function, in order to help the curation process. All tests can be ran in batch mode using: matlab -nodisplay -nosplash -nojvm -r “model0=runTests(‘supp_2.xml’, ‘test.results’); exit;” (ZIP 8 KB)
12918_2011_920_MOESM7_ESM.zip
Additional file 7 : Y. lipolytica iNL895 SBML model. SBML representation of the reconstructed model of Y. lipolytica. This XML file is compatible with SBML Level 2, Version 4, and has been tested with COBRA Toolbox (2.0) and CellDesigner (4.1). This model can also be retrieved from the BioModels database (http://biomodels.org), under model id MODEL1111190000. (ZIP 89 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Loira, N., Dulermo, T., Nicaud, JM. et al. A genome-scale metabolic model of the lipid-accumulating yeast Yarrowia lipolytica. BMC Syst Biol 6, 35 (2012). https://doi.org/10.1186/1752-0509-6-35
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1752-0509-6-35