Abstract
Wheat (Triticum aestivum L.) yield is directly proportional to physio-morphological traits. A high-density genetic map consisting of 2575 markers was used for mapping QTL controlling stay-green and agronomic traits in wheat grown under four diverse water regimes. A total of 108 additive QTL were identified in target traits. Among them, 28 QTL for chlorophyll content (CC) were detected on 11 chromosomes, 43 for normalized difference vegetation index (NDVI) on all chromosomes except 5B, 5D, and 7D, five for spikes per plant (NSP) on different chromosomes, nine for plant height (PH) on four chromosomes, and 23 for thousand-kernel weight (TKW) on 11 chromosomes. Considering all traits, the phenotypic variation explained (PVE) ranged from 3.61 to 41.62%. A major QTL, QNDVI.cgb-5A.7, for NDVI with a maximum PVE of 20.21%, was located on chromosome 5A. A stable and major PH QTL was observed on chromosome 4D with a PVE close to 40%. Most distances between QTL and corresponding flanking markers were less than 1 cM, and approximately one-third of the QTL coincided with markers. Each of 16 QTL clusters on 10 chromosomes controlled more than one trait and therefore could be regarded as pleiotropic regions in response to different water regimes. Forty-one epistatic QTL were identified for all traits having PVE of 6.00 to 25.07%. Validated QTL closely linked to flanking markers will be beneficial for marker-assisted selection in improving drought-tolerance in wheat.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat is one of the most important for mankind; however, its production is affected by environmental stresses such as drought, heat, salinity, and nutrient deficiency. In recent years, drought has become the major abiotic stress factor leading to lower wheat yields and has become a challenge to agricultural scientists in general and to plant breeders in particular (Langridge and Reynolds 2015). Therefore, breeding for higher yield under drought stress has become a target research area for crop breeders coping with global food security.
Quantitative trait loci (QTL) mapping has become a useful tool for dissecting the genetic basis of crop performance under stress by associating genomic information with phenotypic measurements. It may help breeders accelerate the development of abiotic stress tolerant cultivars (Fleury et al. 2010; Langridge and Reynolds 2015). Previous, research on QTL for drought tolerance mostly targeted agronomic traits. Knowledge regarding QTL linked with physiological traits conferring drought tolerance is still lacking. Although a number of QTL mapping studies for drought tolerance have been carried out, very few QTL conferring drought tolerance have been fine mapped or cloned (Ashraf 2010). Some success in improving drought tolerance by molecular marker-assisted selection (MAS) has been achieved. However, under field conditions, difficulties arise in identification and accurate measurement of vital physiological attributes that contribute to yield under drought stress (Maccaferri et al. 2009). In addition, the available low-density linkage maps mean that genes or QTL for target traits are far from the flanking markers, leading to problems in achieving successful outcomes.
Stay-green (sometimes called staygreen) refers to a heritable, delayed foliar senescence character in model and crop plant species (Thomas and Ougham 2014). In crop plants species, stay-green enables a plant to maintain greenness and continue photosynthesising even under stress (Borrell et al. 2014). Stay-green is associated with antioxidant status and photosystem II of the mesophyll cells (De Simone et al. 2014; Luo et al. 2013b). In many cereal crops, stay-green plants generally have higher chlorophyll contents and are more tolerant to abiotic stresses during grain filling. It has been observed in sorghum that during the grain-filling period under water stress conditions, genotypes possessing this trait maintain more photosynthetically active leaf areas than genotypes lacking it (Borrell et al. 2000b; Rosenow et al. 1983). Stay-green also plays an important role during the grain filling stage in wheat when assimilation is limited due to water stress (Distelfeld et al. 2014). Along with other environmental constraints, wheat production in arid and semi-arid regions of the world is mainly limited by water stress, which causes premature senescence that results in lower grain yield and poor quality. Genotypes possessing the ability to maintain green leaf area (stay-green) throughout the grain-filling period are potentially better adapted to water stress environments (Hoang and Kobata 2009).
Normalized difference vegetation index (NDVI) is a widely used indicator of canopy greenness used as an indirect selection criterion for stay-green and higher grain yield under water stress conditions (Marti et al. 2007; Babar et al. 2006a, b). However, there are also contradictory reports regarding the relationship between NDVI measured at different growth stages and yield (Hazratkulova et al. 2012). An association between NDVI and yield during the grain-filling stage in wheat has been reported by some researchers (Hazratkulova et al. 2012; Freeman et al. 2003), while other studies showed associations of yield and NDVI at the booting, heading and grain-filling stages (Babar et al. 2006b). Identification of common chromosome regions shared by NDVI and biomass and yield should be helpful in understanding the genetic basis of the relationship between these traits, and should allow breeders to utilize the trait as an indirect selection criterion for grain yield improvement. Chlorophyll content (SPAD value) was used for assessment of stay-green in sorghum, and four stay-green QTL were identified (Harris et al. 2007). It was reported that individual alleles for stay-green enhanced grain yield in sorghum under drought by modifying canopy development and water uptake patterns (Borrell et al. 2014).
For the current study in wheat we chose a genetic population with a narrow range of heading and flowering dates. This enabled measurement of stay-green traits at the same time as a means of minimising interference from vegetative and reproductive growth differences. We considered water deficiency as an abiotic stress that affected stay-green expression. Water stress occurs at any developmental stage under rainfed conditions. Therefore, we considered NDVI at growth stages S1 (post-winter seedling recovery), S2 (stem elongation) and S3 (booting) as predictors of stay-green under different water regimes.
To meet the need for wheat cultivars better adapted to drought-prone environments the study focused on understanding the genetic basis of important agronomic and physiological traits in diverse water environments.
Materials and methods
Plant materials and field trials
A doubled haploid (DH) population consisting of 150 lines derived from a cross between elite Chinese winter wheat cultivars Hanxuan 10 (H10) and Lumai 14 (L14) was used for QTL analysis (Hao et al. 2003; Jing et al. 1999). H10 is a drought tolerant cultivar released by Shanxi Academy of Agricultural Sciences in 1966, and L14 is a high yielding cultivar adapted to abundant water and fertile conditions released by Yantai Institute of Agricultural Sciences in Shandong in 1986. H10 and L14 were significant cultivars well adapted to local environments. The parents and DH lines had similar phenologies; flowering date of H10 was one day earlier than L14, and flowering dates of the DH lines ranged over four days.
Field trials were conducted at the experimental stations of the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences at Shunyi (116°56′E; 40°23′N) and Changping (116°13′E; 40°13′N), during 2011–2012 and 2012–2013, respectively. Single replicates of the DH lines and three replicates of both parents were grown in 4-row 2 m plots with a 30 cm row spacing. Forty seeds were sown in each row. The rainfalls in the 2011–2012 and 2012–2013 growing seasons were 180 and 158 mm, respectively. Water management was separated into rain-fed (RF, drought stress) and well-watered (WW). In the WW treatment, plots were watered with 750 m3/ha (75 mm) at the pre-overwintering, booting, flowering, and grain-filling stages. Thus, there were two water treatments and two locations (viz. Shunyi in 2011–2012, RF (E1) and WW (E2), and Changping in 2012–2013, RF (E3) and WW (E4)).
Phenotyping and statistical analysis
Phenotypic data were recorded at six different stages of plant growth, viz. post-winter recovery (S1), stem elongation (S2), booting (S3), flowering (S4), grain-filling (S5) and maturity (S6). Normalized difference vegetative index (NDVI) was measured with a GreenSeeker sensor (Optical Sensor Unit 2002, Ntech Industries Inc., Ukiah, CA, USA) at the S1 (only in 2011), S2 and S3 stages. S1 was the stage of initiation of vegetative growth after the overwintering period, and at this stage, the leaves were bright green. The distance between the GreenSeeker and the canopy was maintained at approximately 90 cm. Chlorophyll content (CC) was measured from the midpoint of flag leaves of randomly selected plants at the S4 and S5 stages with a handheld portable chlorophyll meter (SPAD-502, Konica-Minolta, Tokyo). Five flag leaves were measured for each line as described by the standard protocols (Pask et al. 2012). Phenotyping of agronomic traits, i.e., plant height (PH), number of spikes per plant (NSP), thousand-kernel weight (TKW) and grain yield per plot (GY), was carried out at S6.
Regression analysis was based on a general linear model. All statistical analyses were computed by IBM SPSS Statistics version 19.0 software. Broad-sense heritability (\( h_{b}^{2} \)) was estimated as s: \( h_{b}^{2} \left( \% \right) = V_{g} /(V_{g} + V_{e} ) \, \times \, 100 \), where V g and V e are the genotypic and environmental variances (Wu et al. 2010).
Molecular marker genotyping
Genomic DNA was extracted from young leaves of seedlings using a DNA quick extraction kit (Tiangen Biotech Co. Ltd, Beijing). DNA was dissolved in water and quantified using 1% agarose gel electrophoresis with λ DNA as the standard. Primers for SSR markers polymorphic between the parents were identified from GrainGenes 2.0 (http://wheat.pw.usda.gov/GG2/index.shtml). AFLP marker genotyping was also performed on the DH lines (Hao et al. 2003). A total of 8632 SNPs were genotyped across the DH lines with an Infinium iSelect 9 K SNP beadchip array (Cavanagh et al. 2013).
Genetic map construction and QTL mapping
The genetic linkage map was constructed using MAPMAKER/EXP 3.0 (Lander et al. 1987) with a minimum LOD of 3.0 and a maximum recombination fraction of 0.5. The Kosambi mapping function was used to convert recombination frequencies into centiMorgans (cM). The genetic linkage maps were drawn with MapChart version 2.2 (Voorrips 2002). Inclusive composite interval mapping (ICIM) was applied to analyse the phenotypic and genotypic data to detect QTL responsible for the studied physio-morphological traits by using QTL IciMapping version 4.0 (http://www.isbreeding.net/). An additive QTL was declared at an LOD threshold value of 2.5, while an epistatic QTL was declared at an LOD threshold value of 5.0. QTL were named using Q + trait name abbreviation + research department + chromosome (Zhou et al. 2005).
Results
Genetic map construction
The genetic map included 2205 SNPs, 251 SSR, and 119 AFLP markers. SNP accounted for 86% of the total markers (Table 1). Fifty five linkage groups were identified (LG, Supplementary File S1). The entire genetic map spanned 5008.3 cM, with an average distance of 1.9 cM between adjacent markers. The genetic distances between adjacent markers ranged from 1.1 to 4.4 cM. The numbers of markers on each chromosome were considerably different, ranging from eight on 4D to 326 on 5B. The number of markers mapped to the B genome (1250) was greater than that for the A genome (1157). The D genome had 168 markers and a marker density of 3.3 cM. Among the seven homoeologous groups, marker numbers ranged from 215 to 500. Homoeologous groups II and VII had the maximum and minimum marker numbers, respectively (Table 1).
There were 842 loci on our genetic map. SNP clustering was observed, and 2026 SNP markers (78.7%) were involved in 295 (35.0%) clusters (no recombination between markers) (Supplementary Table S1). The largest SNP cluster at the site of 106 cM on LG35-5B contained 144 SNP markers. A total of 1084 SNP markers showed segregation distortion (SD); 326 (12.7%) and 758 markers (29.4%) showed SD at P < 0.05 and P < 0.01, respectively (Supplementary Tables S1 and S2). A total of 816 markers (75.3%) were skewed to parent H10, and 268 (24.7%) were skewed to parent L14. Markers at 310 loci (36.8%) exhibited SD (Supplementary Table S1). Twenty-seven SD regions (SDR, 3 SD loci) were distributed on all chromosomes except for 1D, 2A, 3D, 4A, 4D and 7B, showing that 22 SDR were skewed in favour of H10 alleles and five in favour of L14 alleles (Supplementary File S1).
Phenotypic analysis
The mean phenotypic values of the doubled haploid lines (DHLs) and parents in the four environments are presented in Table 2. Parental lines H10 and L14 exhibited significant differences in NDVI at E2-S3 and E3-S2 (Table 2). The NDVI values of the DHLs depicted an increasing trend from S1 to S3 under the different water regimes. The NDVI values were higher under RF compared to WW conditions (P < 0.01). CC values were significantly lower for H10 than for L14 in E1 to E4. Significant differences were also observed between H10 and L14 for NSP and PH in E1 to E4. High phenotypic variability was observed in the population, with coefficients of variation (CVs) ranging from 6.8 to 21.9%. The five traits showed continuous variation, and transgressive segregation occurred at all stages in all environments. Most Skew and Kurt values were less than 1.0, suggestive of a quantitative trait. \( h_{b}^{2} \) for the four traits ranged from 55.3 to 98.8%.
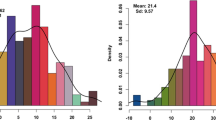
Soil moisture level affected stay-green expression. Regression analysis revealed that soil moisture explained NDVI differences among the DHLs at each developmental stage (Fig. 1a, R 2 > 0.4). The R 2 for chlorophyll contents between WW and RF ranged from 0.16 to 0.44 (Fig. 1b), suggesting significant effects of soil moisture on stay-green as indicated by the chlorophyll contents of flag leaves.
Associations among traits
Correlation analyses among all measured traits at different growth stages, water regimes and years are given in Table 3. Highly positive and significant correlations existed between the NDVI values at S1, S2 and S3 in all four environments (P < 0.01). Under WW conditions, highly negative and significant correlations were observed between NDVI and CC only in 2011.
NDVI at S1, S2, and S3 was positively and significantly correlationed with NSP under both water regimes in 2011. NDVI at S2 also showed significantly negative associations with TKW under WW conditions in both years. CC had a significantly negative correlation with PH under WW conditions (P < 0.01). Except for NSP no significantly consistent correlation between stay-green and agronomic traits was observed under RF in both years.
Based on the results of the correlation analysis, a regression analysis of NDVI (S2) with TKW under WW conditions was carried out. NDVI did not always explain TKW differences among the DH lines (Fig. 2, R 2 = 0.05–0.11), and correlation coefficients were low between NDVI and TKW (Table 3). However, CC, as a good indicator of photosynthetic capacity (Borrell et al. 2000a, b; Lopes and Reynolds 2012) was negatively correlated with PH in both years (Table 3), with an R 2 of 0.07–0.35.
GY was significantly correlated with NDVI under WW conditions in 2011, but there was no correlation in 2012. Conversely, GY was significantly correlated with CC in 2012, but not in 2011. Under RF conditions, GY was significantly correlated with NDVI in both 2011 and 2012, but no significant correlation was detected between GY and CC. This showed that significant correlations between GY and NDVI were not observed in all four environments. TKW is a yield component, but among the four environments, only one significant correlation between GY and TKW was identified and that was under WW conditions in 2012 (Table 3).
Additive QTL mapping
A total of 108 additive QTL for CC, NDVI, NSP, PH, and TKW were detected in the four environments (two water regimes by two years) on almost all chromosomes, except for 5B, 5D, and 7D. Chromosome 5A harboured a maximum number (24) of QTL linked to stay-green traits, except for one that was associated with TKW. Most of the QTL were linked with flanking markers at distances of less than 1 cM, and approximately one-third of the QTL coincided with markers (Supplementary Table S3).
Twenty-eight additive QTL were detected for CC on chromosomes 1B, 2A, 2B, 2D, 3A, 3B, 4B, 4D, 5A, 6B, 6D, and 7A, with PVE ranging from 4.60 to 21.40%. Fourteen QTL were detected in RF conditions (E1 and E3), and 14 were detected in WW conditions (E2 and E4). Twenty-four of these additive QTL showed negative effects, meaning that L14 alleles had positive effects. Only four QTL, on 1B, 5A, and 6B, had positive effects, and were therefore inherited from H10. QCC.cgb-3B.1 and QCC.cgb-3B.2 sharing an adjacent region were identified in E2-S4 and E4-S4, respectively (Fig. 3, cluster f). QCC.cgb-4D.4 in E4-S5 had the maximum PVE (21.40%) and was co-localized with QCC.cgb-4D.1 ~ 3 for CC in E1-S5, E3-S5 and E4-S4 (cluster h). All four of these QTL had negative additive effects, and the favourable alleles were inherited from L14. Another three co-localized QTL for CC were observed at positions 75 cM and 158 cM in LG32 on chromosome 5A (cluster j, k) and at position 0 cM in LG48 on chromosome 6D (cluster n). All QTL at the above three positions had negative effects, and the favourable alleles were from L14.
QTL analysis of NDVI showed 43 additive QTL on all chromosomes, expect for 5B, 5D, and 7D. Among them, 24 and 19 QTL were detected in RF and WW conditions, respectively. The PVE of these QTL ranged from 3.97 to 20.21%. QNDVI.cgb-1B.1 and QNDVI.cgb-1B.2 were repeatedly identified in E2-S2 and E3-S3, respectively (cluster a). Similarly, QNDVI.cgb-1B.4 in E1-S2 was closely linked with QNDVI.cgb-1B.5 in E2-S1 (cluster b). QNDVI.cgb-2A.1 for NDVI at S1 and QNDVI.cgb-2A.2 at S3 in E2 were co-located at position 37 cM in LG8 on chromosome 2A (cluster c). QNDVI.cgb-5A.1 and QNDVI.cgb-5A.2 for NDVI at S3 in E1 and E2 were co-located at position 12 cM in LG32 (cluster i). QNDVI.cgb-5A.6 ~ 11 sharing an adjacent region were identified in E1-E3 (cluster l). The major QTL QNDVI.cgb-5A.7, with a maximum PVE of 20.21%, was located in LG33 on chromosome 5A. All the aforementioned six QTL for NDVI were co-located on chromosome 5A and had positive additive effects. Co-localizations were observed among QNDVI.cgb-5A.12 ~ 14 in E3-S2, E3-S3 and E4-S3, respectively.
Five QTL for NSP were on chromosomes 1B, 2B, 2D, 3B, and 7A, with PVE ranging from 7.32 to 13.53%. No common QTL were identified in multiple environments. QNSP.cgb-2D had a maximum PVE of 13.53%, and the favourable allele was from L14.
Nine additive QTL controlling PH were detected on chromosomes 3D, 4D, 7A, and 7B, explaining 4.53–41.62% of the phenotypic variation under the two water regimes. QTL on 4D and 7A were expressed in more than one environment, indicating that these QTL were relatively stable. QPH.cgb-4D.1 ~ 4, co-located in LG31, were identified at SNP locus wsnp_7344 in four environments and had a relatively high PVE (38.66–41.62%), but other QTL on 3D, 7A, and 7B had lower PVE. Hence, the QTL marked by wsnp_7344 was regarded as the most stable QTL, with considerable effects on reduced plant height from the allele in L14. Three QTL for PH, QPH.cgb-7A.1 ~ 3 in E2-E4, shared marker Xcwm48.2 on LG49 of 7A. QPH.cgb-3D and QPH.cgb-7B had negative effects, which were expressed in E1, and the reduced plant height alleles were inherited from H10.
Twenty three additive QTL were detected for TKW on chromosomes 1B, 2D, 3A, 3B, 3D, 4D, 5A, 6A, 6B, 7A, and 7B. Among them, 12 and 11 were detected under RF and WW conditions, respectively. The PVE of these QTL ranged from 3.61 to 19.25%. Among all QTL, 12 and 11 favorable alleles were inherited from H10 and L14, respectively. QTKW.cgb-3A.1 and QTKW.cgb-3A.2 shared the common marker wsnp_5969 and were identified in E4 and E1, respectively. QTKW.cgb-3D.2 and QTKW.cgb-3D.3 were identified under RF conditions (E1 and E3) and shared marker wsnp_5469 in LG25 of 3D. QTKW.cgb-3D.1 and QTKW.cgb-3D.4 were also identified close to this region in E4 and E2, respectively. QTKW.cgb-4D.1 and QTKW.cgb-4D.2 were co-localized on LG31 of 4D. QTKW.cgb-6A.2 in E2 and QTKW.cgb-6A.3 in E1 mapped to adjacent regions. QTKW.cgb-7A.2 in E4 and QTKW.cgb-7A.3 in E3 were also identified in adjacent regions.
QTL mapping showed eight additive QTL clusters simultaneously controlling different stay-green and agronomic traits on chromosome 2D (cluster d), 3A (cluster e), 3D (cluster g), 4D (cluster h), 5A (cluster j, l) and 7A (cluster o, p) (Fig. 3, Supplementary Table S3). The QTL cluster (h) on chromosome 4D had QTL associated with CC, PH, NDVI and TKW. In this cluster region, the additive effect of QTL for stay-green traits (CC and NDVI) was negative, having a PVE of 5.89–21.40% under three of the four environments; however, the additive effect of QTL controlling agronomic traits was positive, having PVE of 38.66–41.62% (PH) and 5.24–19.25% (TKW). QTKW.cgb-4D.2 showed the maximum PVE in this population, and increased TKW by 2.80 g. QTL controlling CC at the grain-filling stage (S5) were detected in this region in E1, E3 and E4, QTL associated with PH were detected in all four environments (E1, E2, E3 and E4), and QTL linked with TKW were detected in E1 and E4. However, only one QTL for NDVI was identified in this cluster in E4.
Another QTL cluster (j) associated with two stay-green traits was located on 5A (LG32). In this cluster, QTL for CC and NDVI had reverse additive effects, and favorable alleles were inherited from L14 and H10, respectively. The third QTL cluster (l) on 5A (LG33) included QTL for CC, NDVI and TKW (Fig. 3). The QTL for CC and NDVI in this cluster, also had reverse additive effects, and the favorable alleles were inherited from H10 and L14, respectively. However, the QTL for TKW had negative effects and the favorable allele was from L14.
Epistatic QTL
A total of 41 pairs of epistatic QTL were identified (Supplementary Table S4). Among them, 17 pairs were associated with stay-green traits CC and NDVI. For CC, four epistatic QTL pairs were mapped and explained 9.21–12.16% of the phenotypic variation. Three of the four pairs had negative effects, and one had a positive effect. NDVI was linked with 13 pairs of epistatic QTL that were identified in E1, E2 and E4. These epistatic QTL explained 8.92–25.07% of the phenotypic variation. Three pairs of epistatic QTL were mapped for NSP; the PVE values ranging from 19.49 to 24.36% were higher than those of the corresponding additive QTL. There were 18 epistatic QTL for PH, with PVE in the range of 6.00–10.35%. Three epistatic QTL for TKW had PVE ranging from 8.78 to 14.53%.
Discussion
High-density genetic map and SNP clustering
High-density genetic maps are a powerful tool for QTL mapping and subsequent gene mining (Fleury et al. 2010). Wheat possesses a huge and complex genome and most QTL locations were based on low-density genetic maps containing only hundreds of markers (RFLP, AFLP and SSR). This resulted in a low degree of QTL resolution, especially for minor QTL. In addition, genotyping of those markers was laborious and time-consuming. Compared to earlier genetic maps based on SSR and AFLP markers (Groos et al. 2003; Hao et al. 2003; Sun et al. 2010; Xue et al. 2008; Zanetti et al. 2001; Huang et al. 2006) the present map contained mainly SNP markers (86%) and was obtained by using an Infinium iSelect 9 K SNP beadchip array. It comprised 2575 markers spanning 5008.3 cM, with an average distance of 1.9 cM between adjacent markers. The B genome contained the maximum number of markers, followed by the A and D genomes, in accordance with earlier studies (Hao et al. 2003; Xue et al. 2008; Wang et al. 2014; Wu et al. 2015).
As in previous reports (Allen et al. 2011, 2013; Wu et al. 2015) the SNP clusters were integrated with SSR and AFLP markers. This clustering phenomenon might be related to the origins of the SNP, which were developed from the transcriptomes of only 26 hexaploid wheat accessions with most SNP being in gene regions (Cavanagh et al. 2013). This suggested that lack of recombination prevented dispersion of the clusters due to the limited size of the population. Moreover, segregation distortion of markers demonstrated a consistent skew direction and formed 27 segregation distortion regions, most of which were skewed in favour of alleles from H10, and indicating the presence of segregation distortion loci in these regions (Vogl and Xu 2000). However, 547 loci (65.0%) were mapped to unique positions, providing favorable overall coverage of the whole genome with few gaps and closer average spacing than earlier maps.
Deficiency of markers in the D genome
The numbers of molecular markers in the present map were substantially higher in the A and B genomes (93.5, 94.5%, respectively) than in the D genome (6.5%). This lower number in the D genome was consistent with previous studies (Allen et al. 2011, 2013; Cavanagh et al. 2013; Wu et al. 2015). This might be due to the higher diversity of the A and B genomes compared to the D genome, which has lower diversity (Ling et al. 2013; Jia et al. 2013; Brenchley et al. 2012). The lower marker coverage of the D genome compared to the A and B genomes may be responsible for the lower number of QTL in this sub-genome. During the domestication and evolution of common wheat, the gene flow between the A and B genomes was continuous due to a weak barrier between wild emmer to tetraploid cultivated wheat and hexaploid wheat; however, gene flow between the D genome of common wheat and its D genome progenitor A. tauschii was limited. This situation allowed abundant gene interflow between the A and B genomes whereas gene flow between the D genome in common wheat and Ae. tauschii was far less due to reproductive isolation. Because hybridization of tetraploid wheat and Ae. tauschii occurred very few times the D genome in common wheat has much lower genetic diversity, a consequence of which is less molecular marker polymorphism in the D genome of common wheat (Wang et al. 2013; Luo et al. 2013a; Dubcovsky and Dvorak 2007; Salamini et al. 2002). Keeping in view that many QTL for yield and physiological traits are located in the D genome, consideration should be given to increasing the number and density of markers in the D genome through technologies such as next generation sequencing. The improvement of marker density in the D genome would promote the separation of many essential genomic regions/QTL/genes, especially those related to diverse abiotic stresses (Jia et al. 2013).
Relationship between stay-green and agronomic traits
Although there are many reports on stay-green (Gregersen et al. 2008; Verma et al. 2004; Xie et al. 2016; Pinto et al. 2016; Montazeaud et al. 2016), the underlying genetic mechanisms of stay-green and the relationship between stay-green and agronomic traits are not well understood. This might be due to fewer known genes and QTL. Until now, the isopentenyl transferase (IPT) gene (Gan and Amasino 1995) has been the most successful application of improving stay-green traits and delayed senescence for increasing productivity in specific crops (Rivero et al. 2007; Swartzberg et al. 2006), but it does not affect grain yield in wheat (Sykorova et al. 2008).
In the present research, NDVI and CC were measured to evaluate stay-green expression; however, significant correlations were observed between the two stay-green traits only in E2. Although chlorophyll retention behaves as a good indicator of photosynthetic capacity and senescence (Lopes and Reynolds 2012; Harris et al. 2007; Verma et al. 2004; Haussmann et al. 2002; Borrell et al. 2000a, b), both NDVI and CC were highly sensitive to water regime differences (Fig. 1), and water deficiency decreased CC and increased NDVI (Table 2). One reason may be that in winter wheat, moderate vegetative growth is expected in seedlings surviving a harsh winter. However, in spring wheat rapid growth at the seedling stage is important for high yield. The differential NDVI between RF and WW was likely caused by soil temperature, which is affected by water supply. That is, well-watered soil has higher moisture and lower temperature, and thus contains smaller biomass than rainfed soil in early spring. Therefore, optimal NDVI values under different water regimes should be selected as the diagnostic indicators of seedling biomass for winter wheat.
Previous studies reported that NDVI as a spectral reflectance index could predict biomass and grain yield (Marti et al. 2007; Babar et al. 2006a, b). In the present study, the NDVI at each stage under rain-fed conditions was higher than that under well-watered conditions. This might be caused by faster development under rain-fed conditions compared to well-watered conditions for winter wheat during S1–S3 due to higher soil temperature. To determine the relationship between stay-green and agronomic traits under the varying water regimes, NDVI was measured at post-wintering (S1), stem elongation (S2), and booting (S3) stages due to the ability of NDVI to predict grain yield (Marti et al. 2007; Royo et al. 2003; Ma et al. 2001). Since a positive contribution of NDVI to the grain yield of wheat had been reported (Lopes et al. 2014; Lopes and Reynolds 2012), the association between stay-green traits and TKW was investigated. Our results indicated that NDVI and CC were significantly and negatively correlated with TKW only under well-watered conditions (Table 3). Negative correlation of CC and TKW suggested that lower CC would reduce assimilation by the plant and result in reduced grain yield due to water stress. Under WW conditions GY was significantly correlated with NDVI in 2011, but not in 2012. Conversely, GY was significantly correlated with CC in 2012, but not in 2011. Under RF conditions, GY was significantly correlated with NDVI in both years, but no significant correlation was detected between GY and CC. The results showed that significant correlations between GY and NDVI were not observed under all four environments. TKW is a yield component, but among the four environments, the only significant correlation between GY and TKW was identified under WW in 2012 (Table 3).
NDVI at S3 was significantly and positively correlated with PH in E1 and E2 (Table 3). A similar result was observed in wheat under rain-fed conditions (Lopes et al. 2014) and in maize that was tolerant to low phosphorus (Zhang et al. 2015b). CC in the flag leaf had a significant negative correlation with PH, as also reported in rice (Wang et al. 2015). H10 was the taller, lower CC cultivar. The DH lines showed a significant negative correlation between CC and PH implying that these two traits might be linked, a possibility supported by a common location in LG31 on chromosome 4D (Fig. 3). A significant correlation was also observed between NDVI and NSP in the present study. Similar findings suggesting that NDVI could be used to predict tiller density at an early growth stage were reported by others (Wu et al. 2011).
Correlation between stay-green and agronomic traits depends on the drought status at different developmental stages (Tardieu 2012). Judging from the correlation analysis, measurement of NDVI and CC as criteria of stay-green for prediction of the yield-related traits was not successful in the current experiments. For the two winter wheat cultivars moderate vegetative growth is expected for seedlings surviving a harsh winter. Generally, lower correlations were observed between the assessed traits, and these were apt to be affected by environmental conditions such as water availability.
QTL mapping for trait responses to changing water regimes
NDVI was previously only used as an integrated screening indicator to evaluate the performances of different materials under drought stress (Lopes et al. 2014; Lopes and Reynolds 2012; Marti et al. 2007). Studies in rice indicated that a combination of yield-related QTL could improve NDVI under drought stress (Henry et al. 2015), suggesting that direct screening and genetic improvement of NDVI through marker-assisted selection could contribute to improvement in yield-related traits. The present study revealed a complicated genetic architecture of stay-green and agronomic traits through linkage analysis in response to changing water regimes. The high-density linkage map enabled us to accurately identify QTL underlying alleles controlling the expression of target traits by tightly linked markers. First, 43 additive QTL controlling expression of NDVI were characterized in our wheat population. Among them, 16 were located on chromosome 5A (Supplementary Table S3). This suggested that chromosome 5A was the most important chromosome for stay-green at early growth stages in our wheat population. Stable or consistently expressed QTL on 5A under both rain-fed and well-watered conditions could be utilized after validation in wheat improvement programs for drought tolerance. Most of the QTL on 5A were associated with stay-green traits. We therefore hypothesized that hotspot QTL regions on 5A are important for stay-green in wheat. In addition, there were three NDVI QTL clusters on chromosomes 1B (clusters a, b) and 2A (cluster c) controlling NDVI during different growth stages and in multiple environments (Fig. 3). Twenty-eight additive QTL for CC were identified; seven of which were novel loci located on chromosome 5A (Supplementary Table S3). QCC.cgb-5A.7 reappeared and shared the same Xgwm 291-Xwmc 74 interval, in accordance with our previous studies (Yang et al. 2007).
We characterized five additive QTL controlling NSP in specific environments, and near the QNSP.cgb-7A ortholog of Moc1 that controlled tiller development in rice. TaMoc1-A was mapped based on common markers. However, TaMoc1-A has a different function and is involved in wheat spikelet development (Zhang et al. 2015a). A QTL for plant height was identified on chromosome 4D in all four environments, pinpointed by the SNP marker wsnp_7344 (wsnp_Ku_c9140_15390166) (Supplementary Table S3). This QTL for PH located on 4D was likely Rht-D1 (Fig. 4) (Wheat, Composite, 2004 at GrainGenes 2.0). Additionally, near the QTL for PH on 3D and 7A, TaGA20ox2-D1 an ortholog of Sd-1 in rice had been mapped on 3D, and the reduced height wheat gene Rht22 is present on 7D (Fig. 5). Twenty three additive QTL were identified for TKW on chromosomes 1B, 2D, 3A, 3B, 3D, 4D, 5A, 6A, 6B, 7A and 7B (Supplementary Table S3). A region involving three QTL at position 33–36 cM on 3D detected in E1, E3 and E4 (Fig. 6) co-incided with previous studies (McCartney et al. 2005; Somers et al. 2004). Two QTL for TKW on 7A shared the same marker interval with QTL reported in previous studies, suggesting the existence of a locus for TKW in this region (Fig. 6) (Somers et al. 2004; Cuthbert et al. 2008).
Partial linkage maps of chromosomes 3D and 7A harboring QTL for plant height. They show marker collinearity between regions containing QPH.cgb-3D and TaGA20ox2-D1 on 3D (Wu et al. 2009; Somers et al. 2004) and QPH.cgb-7A.1-3 and Rht22 on 7A (Peng et al. 2011; Somers et al. 2004) in red. Common markers and mapped regions are indicated in pink and cyan, respectively. Map distances are in cM
No epistatic QTL co-located with multiple traits. Epistatic QTL involved only single traits, such as NDVI and PH. While these epistatic QTL might be difficult to apply in molecular breeding they will be valuable for understanding the genetic mechanisms of target traits.
Using the Hanxuan 10 × Lumai 14 DH population two sets of molecular linkage maps have been constructed. The previous set (Hao et al. 2003) was constructed using SSR and AFLP markers, and the present research added SNP markers to that map. Using the two linkage maps, several QTL were mapped to common chromosome intervals, such as QCC.cgb-5A.7 in the marker interval Xgwm 291–Xwmc 74 (Yang et al. 2007). These results suggested that the genetic map in our research was reliable and effective for QTL mapping.
QTL clustering and pleiotropy
In the present study there was significant clustering of QTL. A cluster for PH and TKW on 4D (Fig. 3) was reported in other studies (McCartney et al. 2005; Huang et al. 2006; Wang et al. 2009). There were one QTL for NDVI and four QTL for CC at this locus, demonstrating that it was pleiotropic for PH, TKW, NDVI and CC. QTL for CC and PH were tightly linked on 4D (cluster h) and 7A (cluster o), and had reverse additive effects, explained by the fact that H10 was taller and had lower CC, demonstrating that the traits were negatively correlated. QTL controlling CC at the grain-filling stage and PH at maturity were insensitive to water regime and were expressed stably in this DH population.
Similar clusters were located on chromosome 5A for CC and NDVI and 7A for PH, CC, and TKW. The cluster identified on chromosome 5A, where a VRN locus is located, suggests confounding by heading date even though there was variation in heading date of only 4 days. Considering that the QTL cluster on 5A could be confounded by VRN alleles, we checked the locations of VRN-A1 on 5A and the QTL identified in our research (Fig. 7). One of the 5A maps in Fig. 7 was constructed using a Nanda 2419 × Wangshuibai population (Xue et al. 2008), in which VRN-A1 locus was mapped on 5AL. Four markers, Xwmc410, Xgwm595, Xwmc524 and Xgwm291, were distal to the VRN-A1 locus. In the present map the QTL for CC and NDVI were located in the interval Xgwm595–Xgwm291 in 5A-LG33. Comparing the chromosome intervals of 5A with these four common markers, it is suggested that VRN-A1 is located at some distance from the QTL for CC and NDVI and hence were not the same locus. The pleiotropic loci and the candidate markers could be valuable for marker-assisted selection and improvement of multiple traits.
Marker maps of chromosome 5A. The left map was constructed using the Nanda2419 × Wangshuibai population (5A) (Xue et al. 2008), in which the VRN-A1 gene on 5AL was mapped. The right map is a partial linkage map of 5A (LG33) from the current research, constructed using data from the (H10 × Lumai14) population
Conclusions
We constructed a high-density genetic map consisting of 2575 molecular markers using a doubled haploid population derived from a cross of two Chinese winter wheat cultivars. Water regime significantly affected the expression of stay-green traits. Under well-watered conditions the stay-green traits were significantly correlated with agronomic traits, such as NDVI with TKW and GY, but the optimal NDVI value for winter wheat should be used to diagnose higher grain yield. QTL were generally closely flanked by the nearest markers at distances of less than 1 cM, and approximately one-third of the QTL coincided with the markers. Several important QTL regions associated with stay-green and/or agronomic traits identified in multiple environments were mapped on chromosomes 1B, 3D, 4D, 5A and 7A. Comparative mapping was performed for several QTL, such as those for plant height on 4D and 7A and those for TKW on 3D and 7A. Marker numbers were enriched in the candidate QTL regions and should assist map-based cloning and molecular marker-assisted selection. Although the genetic mechanisms controlling stay-green and yield-associated traits are very complex, use of QTL mapped on the present high-density linkage map should enable exploration of novel favorable alleles. The identified QTL for stay-green and agronomic traits will be beneficial for improving drought tolerance by molecular approaches.
References
Allen AM, Barker GL, Berry ST, Coghill JA, Gwilliam R et al (2011) Transcript-specific, single-nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.). Plant Biotechnol J 9:1086–1099. doi:10.1111/j.1467-7652.2011.00628.x
Allen AM, Barker GL, Wilkinson P, Burridge A, Winfield M et al (2013) Discovery and development of exome-based, co-dominant single nucleotide polymorphism markers in hexaploid wheat (Triticum aestivum L.). Plant Biotechnol J 11:279–295. doi:10.1111/pbi.12009
Ashraf M (2010) Inducing drought tolerance in plants: recent advances. Biotechnol Adv 28:169–183. doi:10.1016/j.biotechadv.2009.11.005
Babar MA, Reynolds MP, van Ginkel M, Klatt AR, Raun WR et al (2006a) Spectral reflectance indices as a potential indirect selection criteria for wheat yield under irrigation. Crop Sci 46:578–588. doi:10.2135/cropsci2005.0059
Babar MA, Reynolds MP, van Ginkel M, Klatt AR, Raun WR et al (2006b) Spectral reflectance to estimate genetic variation for in-season biomass, leaf chlorophyll, and canopy temperature in wheat. Crop Sci 46:1046–1057. doi:10.2135/cropsci2005.0211
Borrell AK, Hammer GL, Douglas ACL (2000a) Does maintaining green leaf area in sorghum improve yield under drought? I. Leaf growth and senescence. Crop Sci 40:1026–1037
Borrell AK, Hammer GL, Henzell RG (2000b) Does maintaining green leaf area in sorghum improve yield under drought? II. Dry matter production and yield. Crop Sci 40:1037–1048
Borrell AK, van Oosterom EJ, Mullet JE, George-Jaeggli B, Jordan DR et al (2014) Stay-green alleles individually enhance grain yield in sorghum under drought by modifying canopy development and water uptake patterns. New Phytol 203:817–830. doi:10.1111/nph.12869
Brenchley R, Spannagl M, Pfeifer M, Barker GL, D’Amore R et al (2012) Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491:705–710. doi:10.1038/nature11650
Cavanagh CR, Chao S, Wang S, Huang BE, Stephen S et al (2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Natl Acad Sci USA 110:8057–8062. doi:10.1073/pnas.1217133110
Cuthbert J, Somers D, Brûlé-Babel A, Brown PD, Crow G (2008) Molecular mapping of quantitative trait loci for yield and yield components in spring wheat (Triticum aestivum L.). Theor Appl Genet 117:595–608. doi:10.1007/s00122-008-0804-5
De Simone V, Soccio M, Borrelli GM, Pastore D, Trono D (2014) Stay-green trait-antioxidant status interrelationship in durum wheat (Triticum durum) flag leaf during post-flowering. J Plant Res 127:159–171. doi:10.1007/s10265-013-0584-0
Distelfeld A, Avni R, Fischer AM (2014) Senescence, nutrient remobilization, and yield in wheat and barley. J Exp Bot 65:3783–3798. doi:10.1093/jxb/ert477
Dubcovsky J, Dvorak J (2007) Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316:1862–1866. doi:10.1126/science.1143986
Fleury D, Jefferies S, Kuchel H, Langridge P (2010) Genetic and genomic tools to improve drought tolerance in wheat. J Exp Bot 61:3211–3222. doi:10.1093/jxb/erq152
Freeman KW, Raun WR, Johnson GV, Mullen RW, Stone ML et al (2003) Late-season prediction of wheat grain yield and grain protein. Commun Soil Sci Plant Anal 34:1837–1852. doi:10.1081/Css-120023219
Gan S, Amasino RM (1995) Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270:1986–1988
Gregersen PL, Holm PB, Krupinska K (2008) Leaf senescence and nutrient remobilisation in barley and wheat. Plant Biol (Stuttg) 10:37–49. doi:10.1111/j.1438-8677.2008.00114.x
Groos C, Robert N, Bervas E, Charmet G (2003) Genetic analysis of grain protein-content, grain yield and thousand-kernel weight in bread wheat. Theor Appl Genet 106:1032–1040. doi:10.1007/s00122-002-1111-1
Hao Z, Chang X, Guo X, Jing R, Li R et al (2003) QTL mapping for drought tolerance at stages of germination and seedling in wheat (Triticum aestivum L.) using a DH population. Agric Sci China 2:943–949
Harris K, Subudhi PK, Borrell A, Jordan D, Rosenow D et al (2007) Sorghum stay-green QTL individually reduce post-flowering drought-induced leaf senescence. J Exp Bot 58:327–338. doi:10.1093/jxb/erl225
Haussmann BIG, Mahalakshmi V, Reddy BVS, Seetharama N, Hash CT et al (2002) QTL mapping of stay-green in two sorghum recombinant inbred populations. Theor Appl Genet 106:133–142. doi:10.1007/s00122-002-1012-3
Hazratkulova S, Sharma RC, Alikulov S, Islomov S, Yuldashev T et al (2012) Analysis of genotypic variation for normalized difference vegetation index and its relationship with grain yield in winter wheat under terminal heat stress. Plant Breed 131:716–721. doi:10.1111/pbr.12003
Henry A, Swamy BPM, Dixit S, Torres RD, Batoto TC et al (2015) Physiological mechanisms contributing to the QTL-combination effects on improved performance of IR64 rice NILs under drought. J Exp Bot 66:1787–1799. doi:10.1093/jxb/eru506
Hoang TB, Kobata T (2009) Stay-green in rice (Oryza sativa L.) of drought-prone areas in desiccated soils. Plant Prod Sci 12:397–408
Huang XQ, Cloutier S, Lycar L, Radovanovic N, Humphreys DG et al (2006) Molecular detection of QTLs for agronomic and quality traits in a doubled haploid population derived from two Canadian wheats (Triticum aestivum L.). Theor Appl Genet 113:753–766. doi:10.1007/s00122-006-0346-7
Jia J, Zhao S, Kong X, Li Y, Zhao G et al (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496:91–95. doi:10.1038/nature12028
Jing R, Chang X, Jia J, Hu R (1999) Establishing wheat doubled haploid population for genetic mapping by anther culture. Biotechnology 9:4–8
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ et al (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Langridge P, Reynolds MP (2015) Genomic tools to assist breeding for drought tolerance. Curr Opin Biotechnol 32:130–135. doi:10.1016/j.copbio.2014.11.027
Ling HQ, Zhao S, Liu D, Wang J, Sun H et al (2013) Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496:87–90. doi:10.1038/nature11997
Lopes MS, Reynolds MP (2012) Stay-green in spring wheat can be determined by spectral reflectance measurements (normalized difference vegetation index) independently from phenology. J Exp Bot 63:3789–3798. doi:10.1093/jxb/ers071
Lopes MS, Saglam D, Ozdogan M, Reynolds M (2014) Traits associated with winter wheat grain yield in Central and West Asia. J Integr Plant Biol 56:673–683. doi:10.1111/jipb.12172
Luo MC, Gu YQ, You FM, Deal KR, Ma YQ et al (2013a) A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc Natl Acad Sci USA 110:7940–7945. doi:10.1073/pnas.1219082110
Luo PG, Deng KJ, Hu XY, Li LQ, Li X et al (2013b) Chloroplast ultrastructure regeneration with protection of photosystem II is responsible for the functional stay-green trait in wheat. Plant Cell Environ 36:683–696. doi:10.1111/pce.12006
Ma BL, Dwyer LM, Costa C, Cober ER, Morrison MJ (2001) Early prediction of soybean yield from canopy reflectance measurements. Agron J 93:1227–1234
Maccaferri M, Sanguineti M, Giuliani S, Tuberosa R (2009) Genomics of tolerance to abiotic stress in the Triticeae. In: Muehlbauer GJ, Feuillet C (eds) Genetics and genomics of the Triticeae, vol 7. Plant genetics and genomics: crops and models. Springer, New York, pp 481–558. doi:10.1007/978-0-387-77489-3_18
Marti J, Bort J, Slafer GA, Araus JL (2007) Can wheat yield be assessed by early measurements of normalized difference vegetation index? Ann Appl Biol 150:253–257. doi:10.1111/j.1744-7348.2007.00126.x
McCartney CA, Somers DJ, Humphreys DG, Lukow O, Ames N et al (2005) Mapping quantitative trait loci controlling agronomic traits in the spring wheat cross RL4452x’AC Domain’. Genome 48:870–883. doi:10.1139/g05-055
Montazeaud G, Karatoğma H, Özturk I, Roumet P, Ecarnot M et al (2016) Predicting wheat maturity and stay–green parameters by modeling spectral reflectance measurements and their contribution to grain yield under rainfed conditions. Field Crop Res 196:191–198
Pask AJD, Pietragalla J, Mullan DM, Reynolds MP (2012) Physiological breeding II: a field guide to wheat phenotyping. CIMMYT, Mexico
Peng ZS, Li X, Yang ZJ, Liao ML (2011) A new reduced height gene found in the tetraploid semi-dwarf wheat landrace Aiganfanmai. Genet Mol Res 10:2349–2357
Pinto RS, Lopes MS, Collins NC, Reynolds MP (2016) Modelling and genetic dissection of staygreen under heat stress. Theoretical Appl Genetics 129:1–20
Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R et al (2007) Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc Natl Acad Sci USA 104:19631–19636. doi:10.1073/pnas.0709453104
Rosenow DT, Quisenberry JE, Wendt CW, Clark LE (1983) Drought tolerant sorghum and cotton germplasm. Agric Water Manag 7:207–222
Royo C, Aparicio N, Villegas D, Casadesus J, Monneveux P et al (2003) Usefulness of spectral reflectance indices as durum wheat yield predictors under contrasting Mediterranean conditions. Int J Remote Sens 24:4403–4419. doi:10.1080/0143116031000150059
Salamini F, Ozkan H, Brandolini A, Schafer-Pregl R, Martin W (2002) Genetics and geography of wild cereal domestication in the near east. Nat Rev Genet 3:429–441. doi:10.1038/nrg817
Somers D, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114. doi:10.1007/s00122-004-1740-7
Sun X, Marza F, Ma H, Carver B, Bai G (2010) Mapping quantitative trait loci for quality factors in an inter-class cross of US and Chinese wheat. Theor Appl Genet 120:1041–1051. doi:10.1007/s00122-009-1232-x
Swartzberg D, Dai N, Gan S, Amasino R, Granot D (2006) Effects of cytokinin production under two SAG promoters on senescence and development of tomato plants. Plant Biol (Stuttg) 8:579–586. doi:10.1055/s-2006-924240
Sykorova B, Kuresova G, Daskalova S, Trckova M, Hoyerova K et al (2008) Senescence-induced ectopic expression of the A-tumefaciens ipt gene in wheat delays leaf senescence, increases cytokinin content, nitrate influx, and nitrate reductase activity, but does not affect grain yield. J Exp Bot 59:377–387. doi:10.1093/Jxb/Erm319
Tardieu F (2012) Any trait or trait-related allele can confer drought tolerance: just design the right drought scenario. J Exp Bot 63:25–31. doi:10.1093/jxb/err269
Thomas H, Ougham H (2014) The stay-green trait. J Exp Bot 65:3889–3900. doi:10.1093/jxb/eru037
Verma V, Foulkes MJ, Worland AJ, Sylvester-Bradley R, Caligari PDS et al (2004) Mapping quantitative trait loci for flag leaf senescence as a yield determinant in winter wheat under optimal and drought-stressed environments. Euphytica 135:255–263
Vogl C, Xu SZ (2000) Multipoint mapping of viability and segregation distorting loci using molecular markers. Genetics 155:1439–1447
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang RX, Hai L, Zhang XY, You GX, Yan CS et al (2009) QTL mapping for grain filling rate and yield-related traits in RILs of the Chinese winter wheat population Heshangmai × Yu8679. Theor Appl Genet 118:313–325. doi:10.1007/s00122-008-0901-5
Wang JR, Luo MC, Chen ZX, You FM, Wei YM et al (2013) Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol 198:925–937. doi:10.1111/Nph.12164
Wang S, Wong D, Forrest K, Allen A, Chao S et al (2014) Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796. doi:10.1111/pbi.12183
Wang Q, Xie W, Xing H, Yan J, Meng X et al (2015) Genetic architecture of natural variation in rice chlorophyll content revealed by a genome-wide association study. Mol Plant 8:946–957. doi:10.1016/j.molp.2015.02.014
Wu J, Kong XY, Gao LF, Ren ZL, Jia JZ (2009) Isolation and analysis of TaGA20ox2 genes in wheat. Sci Agric Sin 42:3405–3412
Wu X, Wang Z, Chang X, Jing R (2010) Genetic dissection of the developmental behaviours of plant height in wheat under diverse water regimes. J Exp Bot 61:2923–2937. doi:10.1093/jxb/erq117
Wu JH, Yue SC, Hou P, Meng QF, Cui ZL et al (2011) Monitoring winter wheat population dynamics using an active crop sensor. Spectrosc Spect Anal 31:535–538
Wu XS, Chang XP, Jing RL (2012) Genetic insight into yield-associated traits of wheat grown in multiple rain-fed environments. PLoS ONE 7:e31249
Wu QH, Chen YX, Zhou SH, Fu L, Chen JJ et al (2015) High-density genetic linkage map construction and QTL mapping of grain shape and size in the wheat population Yanda 1817 × Beinong6. PLoS ONE 10:e0118144. doi:10.1371/journal.pone.0118144
Xie Q, Mayes S, Sparkes DL (2016) Early anthesis and delayed but fast leaf senescence contribute to individual grain dry matter and water accumulation in wheat. Field Crop Res 187:24–34. doi:10.1016/j.fcr.2015.12.009
Xue S, Zhang Z, Lin F, Kong Z, Cao Y et al (2008) A high-density intervarietal map of the wheat genome enriched with markers derived from expressed sequence tags. Theor Appl Genet 117:181–189. doi:10.1007/s00122-008-0764-9
Yang DL, Jing RL, Chang XP, Li W (2007) Quantitative trait loci mapping for chlorophyll fluorescence and associated traits in wheat (Triticum aestivum). J Integr Plant Biol 49:646–654
Zanetti S, Winzeler M, Feuillet C, Keller B, Messmer M (2001) Genetic analysis of bread-making quality in wheat and spelt. Plant Breed 120:13–19. doi:10.1046/j.1439-0523.2001.00552.x
Zhang B, Liu X, Xu W, Chang J, Li A et al (2015a) Novel function of a putative MOC1 ortholog associated with spikelet number per spike in common wheat. Sci Rep 5:12211. doi:10.1038/srep12211
Zhang H, Xu R, Xie C, Huang C, Liao H et al (2015b) Large-scale evaluation of maize germplasm for low-phosphorus tolerance. PLoS ONE 10:e0124212. doi:10.1371/journal.pone.0124212
Zhou XG, Jing RL, Hao ZF, Chang XP, Zhang ZB (2005) Mapping QTL for seedling root traits in common wheat. Sci Agric Sin 38:1951–1957
Acknowledgements
We are thankful to the National Natural Science Foundation (31461143024 and 31271720) and the National Key Technologies R&D Program of China (2016ZX08010005) for financial support.
Author information
Authors and Affiliations
Contributions
RJ and BL conceived and designed research. SS and XC conducted experiments. HL contributed analytical tools and partial data analysis. SS analysed the data. SS and RJ wrote the manuscript. FA participated in partial phenotyping in the field and revised the first draft of the manuscript. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Shi, S., Azam, F.I., Li, H. et al. Mapping QTL for stay-green and agronomic traits in wheat under diverse water regimes. Euphytica 213, 246 (2017). https://doi.org/10.1007/s10681-017-2002-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-017-2002-5