Abstract
Cowpea (Vigna unguiculata (L.) Walp) is an important legume, particularly in developing countries. However, little is known about its genome or chromosome structure. We used molecular cytogenetics to characterize the structure of pachytene chromosomes to advance our knowledge of chromosome and genome organization of cowpea. Our data showed that cowpea has highly distinct chromosomal structures that are cytologically visible as brightly DAPI-stained heterochromatic regions. Analysis of the repetitive fraction of the cowpea genome present at centromeric and pericentromeric regions confirmed that two retrotransposons are major components of pericentromeric regions and that a 455-bp tandem repeat is found at seven out of 11 centromere pairs in cowpea. These repeats likely evolved after the divergence of cowpea from common bean and form chromosomal structure unique to cowpea. The integration of cowpea genetic and physical chromosome maps reveals potential regions of suppressed recombination due to condensed heterochromatin and a lack of pairing in a few chromosomal termini. This study provides fundamental knowledge on cowpea chromosome structure and molecular cytogenetics tools for further chromosome studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cowpea (Vigna unguiculata (L.) Walp) is an important agronomic crop in Africa and other developing countries and is particularly tolerant to drought and heat stress, especially as compared to other legume crops (Hall 2004). Cowpea is closely related to other economically important legumes such as soybean (Glycine max), common bean (Phaseolus vulgaris), and pigeonpea (Cajanus cajan) and has a relatively small genome size of 620 Mb, consisting of 2n = 2x = 22 chromosomes (Arumuganathan and Earle 1991). Cowpea was identified as an “orphan crop” with limited genomic resources (Naylor et al. 2004); however, since then, a consensus genetic map with high-density single-nucleotide polymorphism (SNP) markers has been developed from 11 mapping populations (Muchero et al. 2009; Lucas et al. 2011; consensus genetic linkage map version 6 in http://harvest.ucr.edu). Extensive genomic resources such as bacterial artificial chromosome (BAC) libraries, BAC end sequences (BESs), and a physical map have been constructed. Moreover, there is an ongoing genome sequencing project (https://www.integratedbreeding.net/126/communities/genomics-crop-info/agricultural-genomics/genome-sequencing/cowpea). These resources have and will facilitate marker-assisted breeding, association mapping, and comparative analyses of cowpea with other crops. In addition, whole genome sequencing projects on other important Vigna crops such as mung bean (Vigna radiata) and azuki bean (Vigna angularis) have been published (Kang et al. 2014, 2015), which will accelerate the development of genetic and genomic resources for these closely related species and allow them to leverage each other to advance our ability to genetically manipulate these crops.
Despite recent rapid advances in the establishment of genetic and genomic resources, association between genomic information and chromosomal organization of cowpea has been limited. A cowpea karyotype, chromosomal banding patterns, karyotype comparisons among wild cowpea species, chromosomal distribution of ribosomal DNA (rDNA), a centromeric repetitive DNA family, and Ty1-copia-like retrotransposable elements have been previously reported (Barone and Saccardo 1990; Pignone et al. 1990; Saccardo et al. 1992; Galasso et al. 1995, 1997; Guerra et al. 1996; Venora and Padulosi 1997). A comparative cytogenetics study between cowpea and common bean was done using common bean BAC clones mapped to cowpea mitotic chromosomes (Vasconcelos et al. 2015). Polytene chromosomes from anther tapetum cells of Vigna species have been observed and used for cytogenetic studies (reviewed in Guerra 2001). However, most of these studies were done when genomic resources were limited and do not provide a detailed genome organization associated with chromosome structures.
With the development of BAC libraries, fluorescence in situ hybridization (FISH) using BAC clones as probes became a powerful tool in modern cytogenetics. FISH using genetically anchored single- or low-copy BAC probes has been used to integrate genetic and chromosome maps in numerous plant species. This approach has helped to identify individual chromosomes, reveal recombination patterns, find discrepancies between genetic and chromosome maps, and correlate genetic markers with cytological features such as telomeres, heterochromatin, and euchromatin (Cheng et al. 2001a, b; Kulikova et al. 2001; Zhang et al. 2005; Wang et al. 2006a; Fonseca et al. 2010; Ohmido et al. 2010; Iovene et al. 2011).
FISH using repetitive BAC probes has been used to determine chromosomal distribution of the repeats (Lin et al. 2005; Fonseca et al. 2010; Xiong and Pires 2011). In higher plant species, repetitive DNA sequences represent a large fraction of most genomes. For example, in soybean, ∼59 % of the genome is made up of repetitive elements (Schmutz et al. 2010). Based on chromosomal organization in other plants, repetitive sequences are typically enriched in the pericentromeric heterochromatic regions. The pericentromeric regions of soybean contain fast-evolving tandem repeats with interspersed retroelements (Lin et al. 2005). Thus far, in cowpea, only Ty1-copia-type retrotransposons have been identified, which were dispersed relatively uniformly across all chromosomes except in centromeres and subtelomeres (Galasso et al. 1997).
Centromeric regions in plants are typically rich in satellite repeats (a type of tandem repeat) and retrotransposons (reviewed in Jiang et al. 2003). Despite the functional conservation of centromeres, centromeric satellite repeats across species are highly diverged. Of the identified satellite repeats in plant species, for example, CentO in rice, CentC in maize, pAL in Arabidopsis, CentGm in soybean, and CentPv in common bean, all have unit sizes that range between ∼90 and ∼180 bp (Martinez-Zapater et al. 1986; Murata et al. 1994; Ananiev et al. 1998; Cheng et al. 2002; Gill et al. 2009; Iwata et al. 2013). In cowpea, a 488-bp AT-rich centromeric repetitive sequence isolated from a DraI digestion of genomic DNA was previously reported (Galasso et al. 1995). However, it is unknown if this 488-bp repeat is part of satellite repeats or derived from some type of transposable element, and what other repetitive components compose centromeres in cowpea. In common bean, which diverged from cowpea ∼5 million years ago (MYA), two centromeric satellite repeats, CentPv1 and CentPv2, were identified (Iwata et al. 2013). Southern analysis showed that neither of these repeats was conserved in cowpea, indicating that other satellite repeats might dominate cowpea centromeres.
In this study, we utilized available genomic resources with the aim of advancing our knowledge of chromosome organization in cowpea. Molecular cytogenetics analysis and integration of genetic and physical chromosome maps revealed highly distinct structure of cowpea chromosomes. This study contributes to a better understanding of chromosome organization of cowpea and facilitates an understanding of chromosome/genome evolution in legumes.
Materials and methods
Plant materials
Blackeye 5 line 9405C and African accession IT97K-499-35 were grown in greenhouse and field for DNA extraction and preparations of mitotic and meiotic chromosomes.
Preparation of mitotic and meiotic chromosomes
Mitotic chromosome preparations were conducted as previously described (Gill et al. 2009 and Findley et al. 2010) with the following modifications. Root tips from potted plants were treated with pressurized nitrous oxide for 90 min, fixed in a solution composed of 3:1 ethanol and glacial acetic acid for a day at room temperature, and then stored at 4 °C. After rinsing fixed root tips in distilled water, root tips were digested with an enzyme solution containing 1 % Pectolyase (MP Biomedicals) and 2 % Cellulase (MP Biomedicals) in citric buffer (10 mM sodium citrate, 10 mM sodium EDTA, pH 5.5) for 80 min at 37 ° C. For meiotic chromosome preparation, tiny flower buds (1∼2 mm) of cowpea grown in field were collected and fixed in 3:1 ethanol and glacial acetic acid for 24 h at room temperature and then stored at 4 °C. Flower buds were dissected under dissecting microscope and anthers around 0.6 mm in length were selected for preparation of meiotic pachytene chromosome spreads. The preparation was based on the published protocol with the following modifications (Ross et al. 1996). The selected anthers were incubated in an enzyme mixture containing 1 % (w/v) Pectolyase (MP Biomedicals) and 2 % (w/v) Cellulase (MP Biomedicals) in citric buffer (10 mM sodium citrate, 10 mM sodium EDTA, pH 5.5) for 2.5 h at 37 °C. The digested anthers were macerated on glass slides in 60 % acetic acids at 50 °C with fine forceps. Subsequently, ice-cold 3:1 ethanol and glacial acetic acid were added to the slide, and the slide was dried.
Identification of highly abundant tandem repeats
To identify potential centromeric satellite repeats, we searched for highly abundant tandem repeats in the cowpea genome. First, tandem repeats were searched against the BAC end sequences (BESs) of VUH2 BAC library (NCBI GI 146506166-146551943 and GI 270244997-270250498) using tandem repeats finder (TRF; Benson 1999), and tandem repeats with a consensus size greater than 60 bp were further analyzed. In order to find highly abundant tandem repeats in the whole genome, we used a randomly selected subset of 26-bp (26mer) sequences of top 10,000 highest copy numbers corresponding to the 26-bp sequences occurring more than 271,512 times in a 60× draft genome. We performed BLAST analysis using the subset of 26-bp sequences as queries against the BES-derived tandem repeats.
Fluorescence in situ hybridization
BAC clones anchored to linkage groups were selected and used as a FISH probe (Table 1; Muchero et al. 2009). BAC DNAs were extracted using QIAGEN large construct kit. The 455- and 285-bp tandem repeats were amplified from genomic DNA of IT97K-499-35 using primer sets P1 and P2, respectively (Supplemental Table 1). The retrotransposon gag-pol region of VUH2_70J18 and the retrotransposon long terminal repeat (LTR) region of VUH2_81M23 were amplified from each BAC clone using primer sets of P3 and P4, respectively (Supplemental Table 1). The amplified PCR fragments were purified from agarose gel and used for a FISH probe. An 18S rDNA clone developed from soybean was provided by D.A. Johnson at University of Ottawa, and 5S rDNA clone was cloned from common bean. FISH was carried out according to Walling et al. 2005. BAC DNAs, rDNAs, and the purified PCR products were nick translated with either biotin dUTP or digoxigenin dUTP (Roche) and visualized with Streptavidin Alexa Fluor 488 (Invitrogen) or Anti-Digoxigenin-Rhodamine (Roche), respectively. FISH with 176-bp tandem repeat and 18S rDNA was conducted using an oligonucleotide probes (5′-AATACCATGAAAGTCTTGGTGCAC-3′) labeled with FAM (Integrated DNA Technologies) and 18S rDNA directly labeled with Cy5, according to the published protocol (Gill et al. 2009; Iwata et al. 2013). The images were taken with Zeiss Axio Imager M2 microscope, equipped with AxioCam MRm, controlled by Axio Vision 40 V4.8.2.0. The images were adjusted for publication using Adobe Photoshop CS5.1 (Adobe Systems Incorporated). The chromosome lengths were measured using Axio Vision 40. V4.8.2.0. Straightened chromosome images were obtained using ImageJ (Kocsis et al. 1991). Pachytene chromosomes were numbered according to their corresponding linkage groups (Muchero et al. 2009).
Bacterial artificial chromosome DNA sequencing and analysis
BAC DNAs of VUH2_81M23 (NCBI ID: 24466881) and VUH2_70J18 (NCBI ID: 24462974) were extracted using QIAGEN large construct kit (BAC 70J18 subsequent to sequencing was lost due to library contamination; however, sequences are available from the original colony described here). For sequencing of VUH2_81M23, shotgun clone library for BAC sequencing was constructed as previously described in SanMiguel et al. (2002) and Lin et al. (2005). VUH2_81M23 was sequenced from both directions using Big Dye Terminator chemistry (Applied Biosystems) and run on an ABI3730 sequencer. Base calling and quality assessment were done using PHRED, assembled by PHRAP, and edited with CONSED (Ewing et al. 1998; Gordon et al. 1998). FGENESH (www.softberry.com/) was used for de novo gene prediction. Predicted genes were annotated using deduced amino acid sequences by BLASTP against the NCBI nonredundant protein database with an e-value cutoff of 10−10. Predicted coding segments (CDS) were used for annotating genes using BLASTX against the NCBI nonredundant protein database with an e-value of 10−10. Repetitive sequences were annotated using de novo methods and homology searches. De novo identification of LTR retrotransposons was done using LTR-struc (McCarthy and McDonald 2003) and LTR-finder (Xu and Wang 2007). Homology searches for LTR retrotransposons and DNA transposons were conducted using BLASTX of FGENESH predicted gene models against the NCBI nonredundant protein database. An exhaustive search for repetitive sequence was performed using RepeatMasker (www.repeatmasker.org/) with a customized cowpea repeat database created by RECON analysis (Bao and Eddy 2002) from VUH2 BESs.
The DNA of VUH2_70J18 was sequenced using Roche 454 sequencing at the University of Georgia. With a mean size of 515 bp, 160,945 reads were obtained of which 91,460 nonredundant reads were used for repeat analysis. Similarity-based clustering of the reads, assembly of the reads, and classification of repeats in individual clusters were performed using RepeatExplorer (Novak et al. 2013).
Immunodetection of 5-methylcytosine
Postfixation of the slides was performed according to the published protocol (Lysak et al. 2006). Briefly, the slides were postfixed in 4 % formaldehyde in 1× phosphate-buffered saline (PBS) for 10 min at room temperature, washed twice in 1× PBS for 5 min each, and dehydrated in an ethanol series (70, 90, and 100 %). The immunodetection was performed according to Zhang et al. (2008) using mouse anti-5-methylcytosine (1:500, Eurogentec) detected with Alexa Flour 568 goat anti-mouse IgG (Life Technologies). The chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Images were taken with Zeiss Axio Imager M2 microscope, equipped with AxioCam MRm, and controlled by Axio Vision 40 V4.8.2.0. Adobe Photoshop CS5 (Adobe Systems Incorporated) was used to produce publication images.
Results
Microscopic analysis of cowpea chromosomal structure
We first analyzed the chromosomal structure of cowpea using DAPI staining (Fig. 1). Consistent with previous reports (Galasso et al. 1995; Vasconcelos et al. 2015), cowpea mitotic metaphase chromosomes were small (2–3 μm) and most were metacentric or submetacentric (Fig. 1a). The individual chromosomes were morphologically similar, and it was nearly impossible to distinguish homologous chromosomes pairs.
Cowpea chromosomes stained with 4′,6-diamidino-2-phenylindole (DAPI). a Mitotic metaphase chromosomes. Bar = 5 μm. b Meiotic pachytene chromosomes. The image was converted into black and white to enhance the visualization of cytological features along the pachytene chromosomes. Chromosomes were distinguished and identified based on centromeric positions, heterochromatin distributions, and chromosome lengths and numbered according to the corresponding linkage groups. Opened arrowheads indicate the centromere positions based on the chromatin structure. Bar = 10 μm. Arrows show large heterochromatic knobs at chromosomal termini of 4S, 6S, 7S, and 10S. There are two heterochromatic knobs at chromosome 6S due to unpaired chromosomal termini. Closed arrowheads show small heterochromatic knobs at chromosomal termini of 9S and 11L
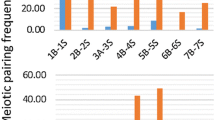
We next analyzed meiotic pachytene chromosomes derived from cowpea pollen mother cells (Fig. 1b). Pachytene chromosomes are often used for cytological studies in plant species, as they show conspicuous cytological features. As seen in other plants such as rice and Medicago (Cheng et al. 2001b; Kulikova et al. 2001), pericentromeric regions of all chromosomes were composed of highly condensed heterochromatin blocks with the chromosome arms being mostly euchromatic (Fig. 1b). There were several interesting cytological features unique to cowpea pachytene chromosomes, not even observed in other closely related legume species such as soybean and common bean (Walling et al. 2006; David et al. 2009). First, centromeres were cytologically larger (∼2.7 μm on average) as compared to other plant species (Cheng et al. 2001b; Zhang et al. 2005; Tang et al. 2009; Iovene et al. 2011), and based on DAPI staining, the centromeric chromatin looked markedly different from heterochromatic and euchromatic regions (open arrowheads in Fig. 1b). Second, some of the heterochromatin was highly distinct to form “knob-like” structures, particularly around the pericentromeres and telomeres. Pericentromeric heterochromatic knobs were found flanking all 11 centromeres. Four chromosomal termini had large heterochromatic knob structures (arrows in Fig. 1b); these regions were found to correspond to chromosomes 4S, 6S, 7S, and 10S. Two chromosomal termini (9S and 11L) possessed small knobs (filled arrowheads in Fig. 1b). Third, the chromosomal termini with large heterochromatin have euchromatic subtelomeric regions where homologous chromosomes were not always completely paired—chromosomes 4S, 6S, 7S, and 10S. These unpaired chromosomal termini were found in early to late pachytene stages. Based on the analysis of 50 pollen mother cells, the structure of these four chromosomal termini was grouped into three types: unpaired, partially paired, and fully paired (Fig. 2; Supplemental Table 2).
Five different chromosomal termini are categorized into three types of structures at chromosomal termini with big heterochromatic knobs. Only chromosomal termini were cropped and shown in the figure. Unpaired: euchromatic subtelomeric regions and heterochromatic knobs are completely unpaired. Partially paired: (1) euchromatic subtelomeric regions are paired, and heterochromatic knobs are somewhat paired, but not completely, showing a heart-like structure (arrow); (2) euchromatic subtelomeric regions are unpaired, but heterochromatic knobs are paired showing a bubble-shape structure (arrow); and (3) euchromatic subtelomeric regions are paired, but heterochromatic knobs are unpaired. Fully paired: euchromatic subtelomeric regions and heterochromatic knobs are completely paired. Bar = 10 μm
Fluorescence in situ hybridization analysis of ribosomal DNA
To further investigate chromosomal organization related to pachytene chromosome structure, 18S and 5S rDNAs were mapped to pachytene chromosomes using FISH (Supplemental Fig. 1). Strong 18S rDNA signals coincided with the large heterochromatin knobs at the four chromosomal termini (chromosome 4S, 6S, 7S, and 10S). One additional interstitial locus was detected in the middle of a chromosome (arrow in Supplemental Fig. 1; later determined to be chromosome 8S by FISH; unpublished data). Weak 18S rDNA signals were also detected at a small heterochromatic knob on chromosome 9S (Supplemental Fig. 1, arrowhead). Therefore, we concluded that 18S rDNA is the major component of heterochromatic knobs at chromosomal termini. Two 5S rDNA loci were found. One pair of chromosomes possessed both 18S rDNA and 5S rDNA on opposite arms, which is homologous to chromosome 10 of common bean (Vasconcelos et al. 2015).
Integration of linkage groups with the chromosome map
Mapping of genetically anchored cloned sequences to pachytene chromosomes has been used to investigate the position of individual genetic markers on chromosomes, identify recombination hot/cold spots, evaluate the accuracy of linkage mapping, and associate chromosomal structures with genetic markers (Cheng et al. 2001a; Kulikova et al. 2001; Zhang et al. 2005; Wang et al. 2006a; Szinay et al. 2008; Ohmido et al. 2010; Iovene et al. 2011). Twenty-two cowpea BAC clones were selected and mapped to individual chromosomes using FISH (Table 1; Fig. 3). Most BAC clones were genetically anchored near the ends of linkage groups, and as expected, their signals localized to subtelomeric regions (Table 1). The exception was H074C16, which was genetically anchored to the short arm of linkage group 7; however, the FISH signal was detected close to centromere and on the same arm as M026L23, which was genetically anchored to long arm of linkage group 7 (Fig. 4a). To obtain a better FISH marker for the short arm of chromosome 7, M051D16, anchored to short arm of linkage group 7 (3.3 cM), was also tested, but the FISH signal also localized on the long arm (Supplemental Fig. 2). This suggests that there are no or very few genetic markers on the short arm of chromosome 7.
Cowpea pachytene chromosomes probed with chromosome-specific BAC clones (listed in Table 1). Arrowheads indicate centromeric positions based on chromatin staining. Bar = 10 μm. The names of BAC clones mapped to individual chromosomes using FISH are shown next to signals in green or red corresponding to their signal colors
Comparisons between linkage (Muchero et al. 2009) and cytogenetic maps. FISH images of pachytene chromosomes straightened and lengths adjusted to those of linkage map for relative distance comparisons. Centromeres on chromosomes are indicated by arrowheads. a Chromosome 7. b Chromosome 9
Another discrepancy observed between genetic and chromosome maps was the orientation of genetic maps for chromosomes 6, 9, and 10 (example of chromosome 9 in Fig. 4b). For consistency, we refer to the cytologically defined long and short arms; thus, the genetic maps for chromosomes 6, 9, and 10 are inverted relative to the chromosome orientation.
Identification and characterization of individual pachytene chromosomes
Cowpea pachytene chromosomes are relatively long and were often entangled; thus, in a single cell, it was difficult to trace all of 11 chromosomes from end to end. Therefore, different cells were used to collect the 11 chromosome images used to measure chromosome lengths and analyze cytological features (Table 2). Chromosome lengths averaged 67.72 μm with a range of ∼127.06 μm (chromosome 3) to 45.66 μm (chromosome 5). Chromosome identification was done using chromosome-specific BAC clones for FISH, but it was also possible to distinguish individual chromosomes by cytological features including heterochromatin distribution, chromosome lengths, and arm ratios. We developed ideogram showing cytological features and BAC positions (Fig. 5).
Molecular characterization of pericentromeric heterochromatin
Microscopic observation of pachytene chromosomes revealed heterochromatin in pericentromeric regions, indicating the presence of repetitive sequences in these regions. It was shown that repetitive sequences at pericentromeric regions of soybean were not conserved in Vigna species (Lin et al. 2005). To analyze the genome organization of cowpea pericentromeric regions, we used two BAC clones selected randomly from a BAC library of VUH2 Blackeye 5 line 9405C, VUH2_70J18 and VUH2_81M23, both containing repetitive sequences.
FISH of BACs, VUH2_70J18 (green) and VUH2_81M23 (red), on mitotic metaphase chromosomes showed that both BACs were distributed across pericentromeric regions of all chromosomes (Fig. 6a–d). Since we wanted to correlate BAC signals with chromosomal structure, pachytene chromosomes were used to further investigate the chromosomal localization of these BACs (Fig. 6e–h). FISH signals from both BAC were at the pericentromeres with distinct distribution patterns. VUH2_70J18 was primarily localized at the very condensed heterochromatic knob-like structures flanking the centromeres, whereas VUH2_81M23 was more dispersed along chromosome arms than VUH2_70J18. Signals of both BACs were very weak or not detected in centromeric regions (centromeres are shown with arrowheads in Fig. 4f), indicating that the BACs consist primarily of pericentromeric repetitive sequences. Part of the signals of VUH2_70J18 and VUH2_81M23 overlapped which indicates that both BAC clones may contain some common repeat sequences or interspersed with each other.
To better understand the genomic makeup of pericentromeric regions, we sequenced BAC VUH2_81M23 using Sanger sequencing technology. The BAC DNA was sequenced to phase II as there are four gaps in the BAC sequence, even though the coverage of this BAC is high, ∼21×. Seven genes were annotated (Supplemental Table 3), three of which were duplicated, and repetitive DNA was found throughout the sequence (Supplemental Fig. 3). Gene duplications interspersed with repetitive DNA were also found in another legume (Lin et al. 2010).
Two intact gypsy LTR retrotransposons were annotated, and their insertions truncate the 5′ ends of two previous LTR retrotransposon insertions. The intact LTR retrotransposon, located at 10,901–20,911 bp, is abundant in the cowpea genome based on BLASTN against a customized cowpea repeat database. Another region with multiple hits against the customized repeat database was located from 59,080 to 59,606 bp and belongs to repeat sequence, fam26-16498. Both the LTR retrotransposons and repeat sequence fam26-16498 have abundant hits from the analysis of 500-bp windows against the repeat database, indicating that both may contribute to the dispersed chromosomal distribution signal of this BAC across pericentromere regions. In several plant genomes, LTR retrotransposons and tandem repeats are known to be major components of pericentromeric regions (Jiang et al. 2002; Feng et al. 2002; Lin et al. 2005; Chang et al. 2008). We PCR amplified the LTR region of the intact LTR retrotransposon on 10,901–20,911 bp to examine its distribution. FISH signals of the LTR showed that it is concentrated in pericentromeric heterochromatic regions (Supplemental Fig. S4a, b), confirming that it is a major constituent of pericentromeric regions of cowpea.
The other pericentromeric BAC clone VUH2_70J18 was sequenced using 454 sequencing technology (Roche) resulting in 91,460 nonredundant reads. Since the read lengths were short (441 bp on average), it was not possible to assemble the entire sequence of the BAC. Instead, we conducted repeat analysis using RepeatExplorer which was developed to use 454 short reads as input (Novak et al. 2010, 2013). Similarity-based clustering of the short reads resulted in 50 clusters and included 90,738 reads. The top 18 clusters (more than 2816 reads per individual clusters) were further analyzed using RepeatMasker with the custom transposon database of soybean (Du et al. 2010) and common bean (http://www.phytozome.net/). Eighteen percent and nearly 50 % of the reads in the cluster 5 (CL5) showed hits against gypsy retrotransposons in soybean and common bean, respectively. We further analyzed the contigs assembled from the short reads contained within CL5 by the CAP3 program as implemented in RepeatExplorer. Two (CL5Contig3 and 7) out of 12 assembled contigs were 1.8 and 1.4 kb long, and a BlastX query against GenBank indicated the presence of gag-pol polyprotein in these contigs, further confirming the presence of a LTR retrotransposon in BAC VUH2_70J18. To confirm that the LTR retrotransposon is a genomic component of the knob-like heterochromatin in pericentromeres, the gag-pol region was amplified from the BAC DNA and used as a FISH probe which targeted the heterochromatic knob-like regions, confirming that this retrotransposon is a constituent of these unusual pericentromeric knobs (Supplemental Fig. 4c, d).
Next, we determined the presence of any common repeats in the two pericentromeric BACs, VUH2_70J18 and VUH2_81M23, using BLAST. Sequences from BAC VUH2_70J18 were used with BLASTN to query against assembled VUH2_81M23 sequence with a cutoff e-value 1E−4. Only one common repeat in both BACs, Gypsy LTR retrotransposon B, was found (Supplemental Fig. 3). Alignment of Gypsy LTR retrotransposon B sequences from the two BACs was short, 27, 71, and 75 bp. Thus, even though both BACs were physically located in pericentromeres, there were few shared sequences between the BACs suggesting a either a high rate of sequence change for the gypsy LTR retrotransposon or that it is an ancient element. These results are consistent with distinct distribution patterns of the two BACs within the pericentromere regions.
Identification of a potential centromeric DNA
In cowpea, large, light-staining centromeres were observed on pachytene chromosomes which are intriguing given that plant centromeres typically consist of megabase-sized arrays of tandem repeats and retrotransposons (Jiang et al. 2003). Previously, a DraI-digested genomic DNA clone, pVuKB1, was reported to be distributed at all centromeric regions of cowpea (Galasso et al. 1995). However, the detailed structure of centromeric regions in cowpea was still undetermined. The most abundant tandem repeats in a plant genome are often centromeric (Gill et al. 2009; Melters et al. 2013). Tandem repeats finder (TRF, Benson 1999) has been used to identify potential centromeric or highly abundant tandem repeats from genome assemblies or sequences (Gill et al. 2009; Melters et al. 2013; Iwata et al. 2013). Therefore, we ran TRF against BESs derived from the VUH2 BAC library.
Since TRF results show only the copy numbers of tandem repeats within a BES, we used a set of 26-bp sequences that occurred frequently in a 60× cowpea draft genome to estimate the abundance of the tandem repeats in the whole genome. The set of 26-bp sequences were sorted according to frequency and used as a query for BLAST analysis against BES-derived tandem repeats
We found that a 176 bp (GenBank seq ID: EI903718.1) and a 445 bp (GenBank seq ID: EI938113.1) were the first and second most abundant tandem repeats in the genome. BLAST analysis revealed that part of the 176-bp tandem repeat was similar to the rDNA intergenic spacer subrepeat (Unfried et al. 1991) also confirmed by FISH showing co-localization of the 176-bp tandem repeat and 18S rDNA (Supplemental Fig. 5). We detected additional strong signals of the 176-bp tandem repeat (arrows on Supplemental Fig. 5c) at the proximal region of one pair of homologous chromosomes indicating independent amplification of the rDNA intergenic spacer subrepeat in cowpea.
Based on FISH, the 455-bp tandem repeat was found at seven of the 11 pairs of centromeres (chromosome 1, 2, 4, 5, 6, 10, and 11; closed arrowheads in Fig. 7). The 455-bp FISH signals covered the large centromeric regions of the seven centromeres and part of the flanking pericentromeric regions. Very weak FISH signals were also detected on the other four pairs of centromeres of pachytene chromosomes indicating the presence of very few or diverged sequences (open arrowheads in Fig. 7). None of these two tandem repeats showed sequence identity with pVuKB1; therefore, we used pVuKB1 sequence as a query to BLAST against the tandem repeats identified from the BESs and found a 285-bp (EI917305.1, EI917305.1, EI920835.1) tandem repeat with 80 % sequence identity to pVuKB1 (91st to 351st base). FISH using the 285-bp satellite repeat as a probe, however, showed distribution at pericentromeric regions of only one chromosome, possibly because the cowpea strain used in this study is different from the one used in Galasso et al. (1995), or our probe is missing bases 1 to 90 of the pVuKB1 clone (Supplemental Fig. 6). BLAST analysis using the first 90 bp of pVuKB1 sequence showed no hits against any identified tandem repeats of our TRF analysis. It is possible that the pVuKB1 sequence is organized independently from tandem repeats including the 285-bp tandem repeat. Another possibility is that the pVuKB1 sequence is not present at centromeres in accessions that we used as BLAST analysis using the first 90 bp of pVuKB1 sequence against BESs, and the genome assembly showed that they are present at relatively low copy numbers.
FISH analysis of potential centromeric repeats, 455-bp tandem repeat on mitotic prometaphase (a–c) and meiotic pachytene chromosomes (d, e). b Chromosomes counterstained with DAPI. c, e Signals of 455-bp tandem repeat. a, d Merged images. Closed arrowheads in d, e indicate centromeres with strong FISH signals, and open arrowheads indicate centromeres with very weak FISH signals. Bars = 5 μm (a) and 10 μm (d), respectively
Chromosomal distribution of cytosine DNA methylation in cowpea
Since cowpea pachytene chromosomes have very distinct large centromeres flanked by unusually highly condensed heterochromatin, we wanted to explore the correlation between 5-methylcytosine distribution and these unusual cytological features. Immunostaining was performed on meiotic pachytene chromosomes using an antibody against 5-methylcytosine (Fig. 8). Signals were distributed across all the chromosomes, although more intense signals were observed in all pericentromeric regions. In contrast, centromeric regions had no or very weak 5-methylcytosine signals (arrowheads on Fig. 8a). Heterochromatic knobs at chromosomal termini, composed of 18S rDNA, and the one in the long arm of chromosome 4 showed very strong 5-methylcytosine signals.
Discussion
Molecular cytogenetics has been useful to correlate genome sequences with chromosomal structures, which can lead to a more comprehensive understanding of the functional and structural properties of genomes. We used genomic resources and cytogenetics to advance our knowledge of cowpea chromosome and genome structure and to integrate the genetic and chromosomal maps.
Chromosome identification is the basis of much molecular cytogenetics work. Especially for species with small chromosomes (2–5 μm of mitotic metaphase chromosomes), it is nearly impossible to distinguish individual chromosomes using classical cytogenetics approaches, e.g., Giemsa and acetocarmine staining and length, arm ratios, and banding patterns. Pachytene chromosomes are much longer than mitotic metaphase chromosomes and provide higher resolution for analysis of cytological features (reviewed in de Jong et al. 1999; Jiang and Gill 2006). For example, cowpea pachytene chromosomes are >30 times longer than mitotic metaphase chromosomes. Therefore, we used pachytene chromosomes to provide more detailed description of chromosome structures of cowpea.
For several plants, chromosome-specific BAC clones have been used to identify individual chromosomes and integrate genetic and cytological maps (Dong et al. 2000; Cheng et al. 2001b; Chang et al. 2007; Koo et al. 2008; Tang et al. 2009; Fonseca et al. 2010; Xiong and Pires 2011). The 22 BAC clones used as a FISH probes in this study enabled us to integrate the genetic and chromosome maps and will accelerate cytogenetics studies in cowpea and other Vigna species that require chromosome identification. Vasconcelos et al. (2015) recently showed overall macrosynteny as well as several chromosomal rearrangements between common bean and cowpea based on cytogenetics mapping using common bean BAC clones. The tools developed here can be also used to investigate the conservation and evolution of chromosomal structures within Vigna species as well as to the closely related genus Phaseolus.
Genetic maps are mathematical representations of recombination and may not reflect actual physical distribution of recombination along a chromosome that can be biased due to nonrandom distribution of crossovers. Thus, FISH mapping on pachytene chromosomes using BAC clones containing genetic markers can reveal the distribution of recombination along a chromosome (Walling et al. 2006) which can provide insight into recombination hot/cold spots and the coverage of genetic map along individual chromosomes. In this study, we mapped genetically anchored BAC clones located near the ends of linkage groups to subtelomeric regions of chromosomes, which indicates that those linkage groups span the entire chromosomes.
Variation of recombination frequency can be associated with local chromosomal structures. Typically, recombination frequencies decrease at heterochromatic pericentromeres and centromeres, while increasing at distal ends of chromosomes in species such as maize, wheat, barley, sorghum, tomato, and rice (Gill et al. 1996; Wang et al. 2006a; Kim et al. 2005; Künzel et al. 2000; Szinay et al. 2008; Zhao et al. 2002). In addition to pericentromeres, recombination frequencies might be attenuated in two other chromosomal regions of cowpea accession used in this study such as the following: (1) unpaired termini of chromosomes 4, 6, and 10 and (2) the heterochromatic knob on the long arm of chromosome 4. The short arms of chromosome 4, 6, and 10 were often unpaired at pachytene, the stage where homologous chromosome synapse and recombination occur. We do not think that this is a cytological artifact, as it was consistently observed and only for a few chromosomes. It would be interesting to see if the unpaired termini are found in other cowpea strains or are specific to IT97K-499-35. Since IT97K-499-35 is a homozygous strain, based on SNP analysis (personal communication with Dr. Timothy Close, University of California, Riverside), we do not expect that this is due to structural heterozygosity. Further experiment, such as immunofluorescence with elements of synaptonemal complex such as meiotic asynaptic mutant 1 protein (ASY1) and zipper 1-like protein (ZYP1) (Armstrong et al. 2002; Higgins et al. 2005), may help in our understanding of this phenomena.
We found at least two different Ty3 gypsy LTR retrotransposons occupying the pericentromeres of cowpea that have very different distribution patterns—one highly condensed into knob-like structures and the other dispersed throughout the pericentromeres. Repetitive DNA sequences constitute a large fraction of plant genomes and contribute to variation in genome size and content among related species. Pericentromeric heterochromatic regions in plant genomes are composed primarily of highly repetitive sequences, including retrotransposons and tandem repeats (Jiang et al. 2002; Feng et al. 2002; Lin et al. 2005; Wang et al. 2006b; Chang et al. 2008). While the structure of pachytene chromosomes in cowpea resembles other plants with similar genome sizes, what was unique was the presence of very conspicuous heterochromatic knob-like structures flanking all centromeres. Based on cytogenetics and sequence analysis of BAC clone VUH2_70J18 that specifically hybridized to the knob-like structures, we confirmed that retrotransposons are major constituents of these structures. Given that pachytene chromosomes of common bean do not have this highly condensed knob-like structures in pericentromeres (unpublished data), we hypothesize that this retrotransposon inserted and then rapidly amplified to form these highly distinct heterochromatic regions in cowpea pericentromeres after divergence from common bean (∼5 MYA). What the functional significance of these structures might be is still unclear.
Heterochromatic knobs at chromosomal termini and/or in euchromatic regions have been observed in maize, Arabidopsis, rice, Antirrhinum majus, common bean, tomato, and several crucifer species plant genomes (Fransz et al. 2000; Cheng et al. 2001b; Lamb et al. 2007; Zhang et al. 2008; Chang et al. 2008; Mandakova and Lysak 2008; David et al. 2009). In these species, the heterochromatic knobs are composed of either rDNA or tandem repeats. In cowpea, there were heterochromatic knobs at chromosomal termini composed of rDNA and an interstitial knob at the short arm of chromosome 4. The genomic component of the interstitial knob is still unknown, but the knob likely arose after the divergence from common bean based on the observation of common bean pachytene chromosomes (David et al. 2009; Fonseca et al. 2014; our unpublished data). In common bean, there are small heterochromatic knobs at most chromosomal termini composed of the khipu tandem repeat (David et al. 2009). Consistent with Southern analysis and FISH experiment (David et al. 2009; Vasconcelos et al. 2015), we did not observe any khipu-related heterochromatic knobs at chromosomal termini of cowpea. Given that common bean and cowpea have high levels of synteny (http://harvest.ucr.edu/; Vasconcelos et al. 2015), repetitive sequences must have evolved rapidly to form species-specific sequences and chromosome structures after the divergence of common bean and cowpea ∼5 MYA.
In plants, centromeres typically consist of megabase-sized arrays of satellite repeats and centromere-specific retrotransposons (reviewed in Jiang et al. 2003). On pachytene chromosomes, centromeres can be visualized after DAPI staining as condensed or decondensed structures depending on the species and even individual chromosomes (Cheng et al. 2001b; Chang et al. 2008). Observation of DAPI-stained pachytene chromosomes of cowpea showed that all 11 centromeric regions were highly decondensed, but not appearing like “typical” euchromatin or heterochromatin, and much larger than typical plant centromeres. It will be interesting to investigate if the centromeric histone H3, which is a mark of functional centromeres, uniformly occupies the entire centromeric regions or partially resides in the regions in “beads on a string” pattern such as seen in Pisum sativum (Neumann et al. 2012; 2015).
From our repeat analysis, we found a 455-bp tandem repeat that was highly abundant in the genome and mainly localized at seven of the 11 pairs of centromeres. The other four centromeres showed very weak FISH signals, indicating that the 455-bp tandem repeat is not a major component of these centromeres. We do not know what sequences underlie the other four centromere pairs as we did not uncover any other obvious high-copy tandem repeats except a 176-bp tandem repeat related to the rDNA intergenic spacer subrepeat. Major component of these four pairs of centromeres might consist of nonrepetitive sequences or nontandem repeats such as transposons. It is also possible that the pVuKB1 sequence, which is present at centromeres of other cowpea accessions, might be present at these centromeres. One hypothesis for centromere evolution is that centromeres evolved from “neocentromeres” that originally formed from single- or low-copy sequences to satellite repeat-based centromeres via invasion and fixation of the repeats at centromeres. This hypothesis is strongly supported in potato where there are six satellite repeat-based centromeres and five repeat-free centromeres (Gong et al. 2012). The presence of the 455-bp tandem repeat at a subset of centromeres indicates that this tandem repeat may still be in the process of fixation by accumulating at nonrepetitive centromeres or by competing with other repetitive sequences to dominate all centromeres.
High levels of cytosine DNA methylation (5-methylcytosine) have been found at repeat-rich heterochromatic pericentromeres in plants (Zhang et al. 2006; Fransz et al. 2006; Yan et al. 2010). Hypermethylation was seen in cowpea pericentromeres and the heterochromatic knob-like structures at chromosomal termini and chromosome 4. In contrast, hypomethylation was observed in cowpea centromeres. Hypomethylation of the centromeric DNA was also found in maize and Arabidopsis, despite the fact that centromeres are composed primarily of mega-sized arrays of tandem repeats and retrotransposons which are typically methylated (Zhang et al. 2008; Koo and Jiang 2009). In rice, hypermethylation and hypomethylation of centromeric DNA were observed and appeared to depend on the DNA composition at individual functional centromeres (Yan et al. 2010). Nucleosomes by virtue of their position determine accessibility to DNA methyltransferases and play a prominent role in determining the methylation pattern of a genome (Chodavarapu et al. 2010). The differences in nucleosome positioning/structure can either facilitate or prevent methylation of the associated DNA and thus may contribute to differences in the methylation patterns of centromeric and pericentromeric chromatin of cowpea. In addition to DNA methylation, histone modification patterns in centromeric chromatin are different from both euchromatin and flanking heterochromatin in human and Drosophila melanogaster (Sullivan and Karpen 2004), which may explain the distinct appearance of DAPI-stained centromeric chromatin.
This study provides foundational knowledge of cowpea chromosome structure and molecular cytogenetics tools for further chromosome studies in Vigna species. Cowpea has interesting chromosomal structures that merit further investigation, including the dense heterochromatic knobs flanking the highly de-condensed and large centromeric regions. The information provided here should aid in the ongoing genome sequencing project by understanding chromosome structure and the distribution of a few of the major repeat families.
Abbreviations
- ASY1:
-
Meiotic asynaptic mutant 1 protein
- BAC:
-
Bacterial artificial chromosome
- BESs:
-
Bacterial artificial chromosome end sequences
- CDS:
-
Coding segments
- CL:
-
Cluster
- DAPI:
-
4′,6-Diamidino-2-phenylindole
- FISH:
-
Fluorescence in situ hybridization
- MYA:
-
Million years ago
- rDNA:
-
Ribosomal DNA
- TRF:
-
Tandem repeat finder
- ZYP1:
-
Zipper 1-like protein
References
Ananiev EV, Phillips RL, Rines HW (1998) Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc Natl Acad Sci U S A 95:13073–13078
Armstrong SJ, Caryl AP, Jones GH, Franklin FCH (2002) Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci 115:3645–3655
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Report 9:208–218
Bao Z, Eddy SR (2002) Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res 12:1269–1276
Barone A, Saccardo F (1990) Pachytene morphology of cowpea chromosomes. In: Ng NQ, Monti LM (eds) Cowpea Genet Resour. ITTA, Ibadna, pp 137–143
Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580
Chang SB, Anderson LK, Sherman JD et al (2007) Predicting and testing physical locations of genetically mapped loci on tomato pachytene chromosome 1. Genetics 176:2131–2138
Chang SB, Yang TJ, Datema E et al (2008) FISH mapping and molecular organization of the major repetitive sequences of tomato. Chromosom Res 16:919–933
Cheng Z, Buell CR, Wing RA et al (2001a) Toward a cytological characterization of the rice genome. Genome Res 11:2133–2141
Cheng ZK, Presting GG, Buell CR et al (2001b) High-resolution pachytene chromosome mapping of bacterial artificial chromosomes anchored by genetic markers reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice. Genetics 157:1749–1757
Cheng Z, Dong F, Langdon T et al (2002) Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14:1691–1704
Chodavarapu RK, Feng S, Bernatavichute YV et al (2010) Relationship between nucleosome positioning and DNA methylation. Nature 466:388–392
David P, Chen NWG, Pedrosa-Harand A et al (2009) A nomadic subtelomeric disease resistance gene cluster in common bean. Plant Physiol 151:1048–1065
de Jong H, Fransz P, Zabel P (1999) High resolution FISH in plants—techniques and applications. Trends Plant Sci 4:258–263
Dong F, Song J, Naess SK et al (2000) Development and applications of a set of chromosome-specific cytogenetic DNA markers in potato. Theor Appl Genet 101:1001–1007
Du J, Grant D, Tian Z et al (2010) SoyTEdb: a comprehensive database of transposable elements in the soybean genome. BMC Genomics 11:113
Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185
Feng Q, Zhang YJ, Hao P et al (2002) Sequence and analysis of rice chromosome 4. Nature 420:316–320
Findley SD, Cannon S, Varala K et al (2010) A fluorescence in situ hybridization system for karyotyping soybean. Genetics 185:727–744
Fonseca A, Ferreira J, dos Santos TR et al (2010) Cytogenetic map of common bean (Phaseolus vulgaris L.). Chromosom Res 18:487–502
Fonseca A, Richard MMS, Geffroy V, Pedrosa-Harand A (2014) Epigenetic analyses and the distribution of repetitive DNA and resistance genes reveal the complexity of common bean (Phaseolus vulgaris L., Fabaceae) heterochromatin. Cytogenet Genome Res 143:168–178
Fransz PF, Armstrong S, de Jong JH et al (2000) Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere region. Cell 100:367–376
Fransz P, ten Hoopen R, Tessadori F (2006) Composition and formation of heterochromatin in Arabidopsis thaliana. Chromosom Res 14:71–82
Galasso I, Schmidt T, Pignone D, Heslopharrison JS (1995) The molecular cytogenetics of Vigna-unguiculata (L) Walp—the physical organization and characterization of 18s-5.8s-25s ribosomal-RNA genes, 5s ribosomal-RNA genes, telomere-like sequences, and a family of centromeric repetitive DNA sequences. Theor Appl Genet 91:928–935
Galasso I, Harrison GE, Pignone D et al (1997) The distribution and organization of Ty1-copia-like retrotransposable elements in the genome of Vigna unguiculata (L) Walp (cowpea) and its relatives. Ann Bot 80:327–333
Genome sequencing—cowpea retrieved from https://www.integratedbreeding.net/126/communities/genomics-crop-info/agricultural-genomics/genome-sequencing/cowpea. Accessed 01 Nov 2015
Gill KS, Gill BS, Endo TR, Boyko EV (1996) Identification and high-density mapping of gene-rich regions in chromosome group 5 of wheat. Genetics 143:1001–1012
Gill N, Findley S, Walling JG et al (2009) Molecular and chromosomal evidence for allopolyploidy in soybean. Plant Physiol 151:1167–1174
Gong Z, Wu Y, Koblízková A et al (2012) Repeatless and repeat-based centromeres in potato: implications for centromere evolution. Plant Cell 24:3559–3574
Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202
Guerra M (2001) Fluorescent in situ hybridization in plant polytene chromosomes. Methods Cell Sci 23:133–138
Guerra M, Kenton A, Bennett MD (1996) rDNA sites in mitotic and polytene chromosomes of Vigna unguiculata (L) Walp and Phaseolus coccineus L revealed by in situ hybridization. Ann Bot 78:157–161
Hall AE (2004) Breeding for adaptation to drought and heat in cowpea. Eur J Agron 21:447–454
Higgins JD, Sanchez-Moran E, Armstrong SJ et al (2005) The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev 19:2488–2500. doi:10.1101/gad.354705
Iovene M, Cavagnaro PF, Senalik D et al (2011) Comparative FISH mapping of Daucus species (Apiaceae family). Chromosom Res 19:493–506
Iwata A, Tek AL, Richard MM et al (2013) Identification and characterization of functional centromeres of the common bean. Plant J 76:47–60
Jiang J, Gill BS (2006) Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49:1057–1068
Jiang N, Bao Z, Temnykh S et al (2002) Dasheng: a recently amplified nonautonomous long terminal repeat element that is a major component of pericentromeric regions in rice. Genetics 161:1293–1305
Jiang J, Birchler J, Parrott W, Kelly D (2003) A molecular view of plant centromeres. Trends Plant Sci 8:570–575
Kang YJ, Kim SK, Kim MY et al (2014) Genome sequence of mungbean and insights into evolution within Vigna species. Nat Commun 5:5443
Kang YJ, Satyawan D, Shim S et al (2015) Draft genome sequence of adzuki bean, Vigna angularis. Sci Rep 5:8069
Kim JS, Klein PE, Klein RR et al (2005) Molecular cytogenetic maps of sorghum linkage groups 2 and 8. Genetics 169:955–965
Kocsis E, Trus BL, Steer CJ et al (1991) Image averaging of flexible fibrous macromolecules: the clathrin triskelion has an elastic proximal segment. J Struct Biol 107:6–14
Koo DH, Jiang J (2009) Super-stretched pachytene chromosomes for fluorescence in situ hybridization mapping and immunodetection of DNA methylation. Plant J 59:509–516
Koo DH, Jo SH, Bang JW et al (2008) Integration of cytogenetic and genetic linkage maps unveils the physical architecture of tomato chromosome 2. Genetics 179:1211–1220
Kulikova O, Gualtieri G, Geurts R et al (2001) Integration of the FISH pachytene and genetic maps of Medicago truncatula. Plant J 27:49–58
Künzel G, Korzun L, Meister A (2000) Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics 154:397–412
Lamb JC, Meyer JM, Corcoran B et al (2007) Distinct chromosomal distributions of highly repetitive sequences in maize. Chromosom Res 15:33–49
Lin JY, Jacobus BH, SanMiguel P et al (2005) Pericentromeric regions of soybean (Glycine max L. Merr.) chromosomes consist of retroelements and tandemly repeated DNA and are structurally and evolutionarily labile. Genetics 170:1221–1230
Lin JY, Stupar RM, Hans C et al (2010) Structural and functional divergence of a 1-Mb duplicated region in the soybean (Glycine max) genome and comparison to an orthologous region from Phaseolus vulgaris. Plant Cell 22:2545–2561
Lucas MR, Diop N-N, Wanamaker S et al (2011) Cowpea–soybean synteny clarified through an improved genetic map. Plant Genome J 4:218–225
Lysak M, Fransz P, Schubert I (2006) Cytogenetic analyses of Arabidopsis. Methods Mol Biol 323:173–186
Mandakova T, Lysak MA (2008) Chromosomal phylogeny and karyotype evolution in x = 7 crucifer species (Brassicaceae). Plant Cell 20:2559–2570
Martinez-Zapater JM, Estelle MA, Somerville CR (1986) A highly repeated DNA sequence in Arabidopsis thaliana. Mol Gen Genet 204:417–423
McCarthy EM, McDonald JF (2003) LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics 19:362–367
Melters DP, Bradnam KR, Young HA et al (2013) Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol 14:R10
Muchero W, Diop NN, Bhat PR et al (2009) A consensus genetic map of cowpea [Vigna unguiculata (L) Walp.] and synteny based on EST-derived SNPs. Proc Natl Acad Sci U S A 106:18159–18164
Murata M, Ogura Y, Motoyoshi F (1994) Centromeric repetitive sequences in Arabidopsis thaliana. Jpn J Genet 69:361–370
Naylor RL, Falcon WP, Goodman RM et al (2004) Biotechnology in the developing world: a case for increased investments in orphan crops. Food Policy 29:15–44
Neumann P, Navrátilová A, Schroeder-Reiter E et al (2012) Stretching the rules: monocentric chromosomes with multiple centromere domains. PLoS Genet 8:e1002777
Neumann P, Pavlíková Z, Koblížková A et al (2015) Centromeres off the hook: massive changes in centromere size and structure following duplication of CenH3 gene in Fabeae species. Mol Biol Evol 32:1862–1879
Novak P, Neumann P, Macas J (2010) Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 11:378
Novak P, Neumann P, Pech J et al (2013) RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29:792–793
Ohmido N, Ishimaru A, Kato S et al (2010) Integration of cytogenetic and genetic linkage maps of Lotus japonicus, a model plant for legumes. Chromosom Res 18:287–299
Pignone D, Cifarelli S, Perrino P (1990) Chromosome identification in Vigna unguiculata (L.) Walp. In: N.Q. Ng and L.M. Monti (eds) Cowpea genetic resources. IITA Ibadan, Nigeria. pp. 144–150
Ross KJ, Fransz P, Jones GH (1996) A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosom Res 4:507–516
Saccardo F, del Giudice A, Galasso I (1992) Cytogenetics of cowpea. In: Thottappilly G, Monti LM, Mohan Raj DR, Moore AW (eds) Biotechnol enhancing Res Trop Crop Africa. CTS¥A/IITA, Ibadan, pp 89–98
SanMiguel PJ, Ramakrishna W, Bennetzen JL et al (2002) Transposable elements, genes and recombination in a 215-kb contig from wheat chromosome 5A(m). Funct Integr Genomics 2:70–80
Schmutz J, Cannon SB, Schlueter J et al (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Sullivan BA, Karpen GH (2004) Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11:1076–1083
Szinay D, Chang S-B, Khrustaleva L et al (2008) High-resolution chromosome mapping of BACs using multi-colour FISH and pooled-BAC FISH as a backbone for sequencing tomato chromosome 6. Plant J 56:627–637
Tang XM, de Boer JM, van Eck HJ et al (2009) Assignment of genetic linkage maps to diploid Solanum tuberosum pachytene chromosomes by BAC-FISH technology. Chromosom Res 17:899–915
Unfried K, Schiebel K, Hemleben V (1991) Subrepeats of rDNA intergenic spacer present as prominent independent satellite DNA in Vigna radiata but not in Vigna angularis. Gene 99:63–68
Vasconcelos EV, de Andrade Fonsêca AF, Pedrosa-Harand A et al (2015) Intra- and interchromosomal rearrangements between cowpea [Vigna unguiculata (L.) Walp.] and common bean (Phaseolus vulgaris L.) revealed by BAC-FISH. Chromosom Res 23:253–266
Venora G, Padulosi S (1997) Karyotypic analysis of wild taxa of V. unguiculata (L.) Walpers. Caryologia 50(2):125–138
Walling JG, Pires JC, Jackson SA (2005) Preparation of samples for comparative studies of plant chromosomes using in situ hybridization methods. Mol Evol Prod Biochem Data B 395:443–460
Walling JG, Shoemaker R, Young N et al (2006) Chromosome-level homeology in paleopolyploid soybean (Glycine max) revealed through integration of genetic and chromosome maps. Genetics 172:1893–1900
Wang C, Harper L, Cande W (2006a) High-resolution single-copy gene fluorescence in situ hybridization and its use in the construction of a cytogenetic map of maize chromosome 9. Plant Cell 18:529–544
Wang Y, Tang X, Cheng Z et al (2006b) Euchromatin and pericentromeric heterochromatin: comparative composition in the tomato genome. Genetics 172:2529–2540
Xiong Z, Pires JC (2011) Karyotype and identification of all homoeologous chromosomes of allopolyploid Brassica napus and its diploid progenitors. Genetics 187:37–49
Xu Z, Wang H (2007) LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res 35:W265–W268
Yan H, Kikuchi S, Neumann P et al (2010) Genome-wide mapping of cytosine methylation revealed dynamic DNA methylation patterns associated with genes and centromeres in rice. Plant J 63:353–365
Zhang D, Yang Q, Bao W et al (2005) Molecular cytogenetic characterization of the Antirrhinum majus genome. Genetics 169:325–335
Zhang X, Yazaki J, Sundaresan A et al (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126:1189–1201
Zhang W, Lee HR, Koo DH, Jiang J (2008) Epigenetic modification of centromeric chromatin: hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize. Plant Cell 20:25–34
Zhao Q, Zhang Y, Cheng Z et al (2002) A fine physical map of the rice chromosome 4. Genome Res 12:817–823
Acknowledgments
We would like to acknowledge the generous support of the USDA-NIFA grant 2009-01860 to S.A.J. and the National Science Foundation (MCB 1229956). We thank Timothy Close (University of California, Riverside) for providing chromosome-specific BAC clones, an updated version of genetic map, and for comments on this project. We thank Brian Abernathy (University of Georgia) for assistance in bioinformatics and Douglas R. Cook and Varma Penmetsa (University of California, Davis) for providing the pericentromeric VUH2 BAC clone.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Jiming Jiang
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Figure 1
FISH images of 18S (green) and 5S (red) rDNAs on meiotic pachytene chromosomes. Bar = 10 μm. In addition to four major 18S rDNA loci that overlapped with large heterochromatic knobs at chromosomal termini, one strong signal was detected in pericentromeric heterochromatin (arrow) and one weak signal was detected at a small heterochromatic knob on chromosome 9S (arrowhead). (GIF 24 kb) (GIF 71 kb)
Supplemental Figure 2
FISH image showing the position of BAC clone, M051D16 on chromosome 7. The BAC clone is anchored to short arm of linkage group 7 (3.3 cM), but physically mapped onto the long arm of chromosome 7. (A) Merged image of chromosome (blue) and the BAC signal (red). Arrowhead indicates centromere of chromosome 7. (B) DAPI image. Bar = 10 μm. (GIF 24 kb)
Supplemental Figure 3
Sequence analysis of BAC VUH2_81M23. Top panel, a 500-bp non-overlapping sliding window (x-axis) was used to map the number of BLASTN hits (y-axis) from a search against the cowpea repeat database (generated by RECON analysis of VUH2 BES database) along the length of VUH2_81M23. Annotation of VUH2_81M23 is shown at bottom. Yellow pentagons represent genes 1- 7, and genes 1, 4, and 6 are duplicates shown with asterisks. Two light green boxes are intact LTR retrotransposons A and B (Gypsy). Dark green boxes are four truncated LTR retrotransposons a-d. Pink box shows the position of a repeat sequence, fam26-16498, with multiple hits against the customized repeat database. (PPTX 80 kb)
Supplemental Figure 4
FISH analysis of repetitive fraction of pericentromeric BAC clones on pachytene chromosomes counterstained with DAPI (blue). (A and B) FISH image of LTR region in VUH2_81M23 (red) (C and D). FISH image of gag-pol region in VUH2_70J18 (red). Bar = 10 μm. (GIF 112 kb)
Supplemental Figure 5
FISH analysis of 176-bp tandem repeat (green) and 18S rDNA (red) on mitotic metaphase chromosomes. (C) Signals of 176-bp tandem repeat. Arrows indicate independent amplification of 176-bp tandem repeat from ribosomal genes. (D) Signals of 18S rDNA. Bar = 5 μm. (GIF 39 kb)
Supplemental Figure 6
FISH image of 285-bp tandem repeat (green) on pachytene chromosomes (Arrow). 285-bp tandem repeat signal overlaps with pericentromeric heterochromatin. Bar = 10 μm. (GIF 45 kb)
ESM 1
(PDF 13 kb)
ESM 2
(PDF 10 kb)
ESM 3
(PDF 9 kb)
ESM 4
(PPTX 241 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Iwata-Otsubo, A., Lin, JY., Gill, N. et al. Highly distinct chromosomal structures in cowpea (Vigna unguiculata), as revealed by molecular cytogenetic analysis. Chromosome Res 24, 197–216 (2016). https://doi.org/10.1007/s10577-015-9515-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-015-9515-3