Abstract
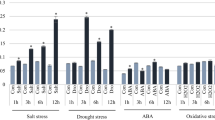
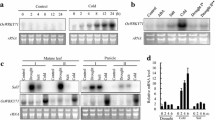
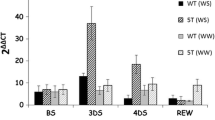
The rice OsMyb4 gene, which encodes a Myb transcription factor (TF), improves the stress tolerance/resistance when expressed in both monocotyledonous and dicotyledonous transgenic plants. In this study, a phylogenetic analysis showed the existence of putative OsMyb4 homologues in monocot and dicot species. In particular, the analysis revealed that OsMyb4 belongs to a small rice gene subfamily conserved among monocots. The expression analyses of the OsMyb4-like genes in rice, wheat, and Arabidopsis indicated that these genes are involved in the response to dehydration, cold, and wounding. Moreover, the in silico analysis of the 5′ upstream regions of the Osmyb4-like genes highlighted that the positions of some cis-elements involved in the stress response were conserved among the putative promoters, especially between OsMyb4 and its putative paralog Os02g41510. Finally, our transient expression assays in tobacco protoplasts demonstrated that OsMyb4 is able to repress the activity of both its own promoter and the Os02g41510 promoter by acting on the same binding site. A compensatory mechanism of auto-regulation is consistent with the well-known complexity of the OsMyb4-activated pathway, and this mechanism could regulate the transcription of other genes belonging to the family.
Similar content being viewed by others
Abbreviations
- ABA:
-
abscisic acid
- BS:
-
bluescript
- CaMV:
-
cauliflower mosaic virus
- GUS:
-
β-glucuronidase
- MBS:
-
Myb binding site
- qRT-PCR:
-
quantitative reverse transcription-polymerase chain reaction
- RH:
-
relative humidity
- TF:
-
transcription factor
References
Agarwal, M., Hao, Y., Kapoor, A., Dong, C.H., Fujii, H., Zheng, X., Zhu, J.K.: A R2R23 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. — J. biol. Chem. 281: 37636–37645, 2006.
Agarwal, P.K., Jha, B.: Transcription factors in plants and ABA dependent and independent abiotic stress signaling. — Biol. Plant. 54: 201–212, 2010.
Caruso, P., Baldoni, E., Mattana, M., Pietro Paolo, D., Genga, A., Coraggio, I., Russo, G., Picchi, V., Reforgiato Recupero, G., Locatelli, F.: Ectopic expression of a rice transcription factor, Mybleu, enhances tolerance of transgenic plants of Carrizo citrange to low oxygen stress. — Plant Cell Tissue Organ Cult. 109: 327–339, 2012.
Chang, W.C., Lee, T.Y., Huang, H.D., Huang, H.Y., Pan, R.L.: Plant PAN: plant promoter analysis navigator, for identifying combinatorial cis-regulatory elements with distance constraint in plant gene groups. — BMC Genom. 9: 561, 2008.
Chen, Y., Chen, Z., Kang, J., Kang, D., Gu, H., Qin, G.: AtMYB14 regulates cold tolerance in Arabidopsis. — Plant mol. Biol. Rep. 31: 87–97, 2013.
Chen, Y., Zhang, X., Wu, W., Chen, Z., Gu, H., Qu, L.J.: Overexpression of the wounding-responsive gene AtMYB15 activates the shikimate pathway in Arabidopsis. — J. Integr. Plant Biol. 48: 1084–1095, 2006.
Ding, Z., Li, S., An, X., Liu, X., Qin, H., Wang, D.: Transgenic expression of Myb15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. — J. Genet. Genom. 36: 17–29, 2009.
Docimo, T., Mattana, M., Fasano, R., Consonni, R., De Tommasi, N., Coraggio, I., Leone, A.: Ectopic expression of the Osmyb4 rice gene enhances synthesis of hydroxycinnamic acid derivatives in tobacco and clary sage. — Biol. Plant. 57: 179–183, 2013.
Dubouzet, J.G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E.G., Miura, S., Seki, M., Shinozaki, K., Yamaguchi-Shinozaki, K.: OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. — Plant J. 33: 751–763, 2003.
Espley, R.V., Brendolise, C., Chagné, D., Kutty-Amma, S., Green, S., Volz, R., Putterill, J., Schouten, H.J., Gardiner, S.E., Hellens, R.P., Allan, A.C.: Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. — Plant Cell 21: 168–183, 2009.
Felsenstein, J.: PHYLIP: phylogeny inference package. — Cladistics 5: 164–166, 1989.
Fornalé, S., Sonbol, F.M., Maes, T., Capellades, M., Puigdomènech, P., Rigau, J., Caparrós-Ruiz, D.: Downregulation of the maize and Arabidopsis thaliana caffeic acid O-methyl-transferase genes by two new maize R2R3-MYB transcription factors. — Plant. mol. Biol. 62: 809–823, 2006.
Fujita, M., Fujita, Y., Noutoshi, Y., Takahashi, F., Narusaka, Y., Yamaguchi-Shinozaki, K., Shinozaki, K.: Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. — Curr. Opin. Plant Biol. 9: 436–442, 2006.
Gális, I., Šimek, P., Narisawa, T., Sasaki, M., Horiguchi, T., Fukuda, H., Matsuoka, K.: A novel R2R3 MYB transcription factor NtMYBJS1 is a methyl jasmonatedependent regulator of phenylpropanoid-conjugate biosynthesis in tobacco. — Plant J. 46: 573–592, 2006.
Giovinazzo, G., Manzocchi, L.A., Bianchi, M.W., Coraggio, I., Viotti, A.: Functional analysis of the regulatory region of a zein gene in transiently transformed protoplasts. — Plant mol. Biol. 19: 257–263, 1992.
Guerra, J., Withers, D.A., Boxer, L.M.: MYB binding sites mediate negative regulation of c-MYB expression in T-cell lines. — Blood 86: 1873–1880, 1995.
He, Y., Li, W., Lv, J., Jia, Y., Wang, M., Xia, G.: Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. — J. exp. Bot. 63: 1511–1522, 2012.
Jain, M., Nijhawan, A., Tyagi, A.K., Khurana, J.P.: Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. — Biochem. biophys. Res. Commun. 345: 646–651, 2006.
James, P.L., Le Strat, L., Ellervik, U., Bratwall, C., Nordén, B., Brown, T., Fox, K.R.: Effects of a hairpin polyamide on DNA melting: comparison with distamycin and Hoechst 33258. — Biophys. Chem. 111: 205–212, 2004.
Katiyar, A., Smita, S., Lenka, S.K., Rajwanshi, R., Chinnusamy, V., Bansal, K.C.: Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. — BMC Genom. 13: 544, 2012.
Kops, G.J.P.L., Dansen, T.B., Polderman, P.E., Saarloos, I., Wirtz, K.W.A., Coffer, P.J., Huang, T.T., Bos, J.L., Medema, R.H., Burgering, B.M.T.: Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. — Nature 419: 316–321, 2002.
Larkin, M.A., Blackshields, G., Brown, N.P., Chenna, R., McGettigan, P.A., McWilliam, H., Valentin, F., Wallace, I.M., Wilm, A., Lopez, R., Thompson, J.D., Gibson, T.J., Higgins, D.G.: Clustal W and Clustal X version 2.0. — Bioinformatics 23: 2947–2948, 2007.
Laura, M., Consonni, R., Locatelli, F., Fumagalli, E., Allavena, A., Coraggio, I., Mattana, M.: Metabolic response to cold and freezing of Osteospermum ecklonis overexpressing Osmyb4. — Plant Physiol. Biochem. 48: 764–771, 2010.
Liu, R., Lü, B., Wang, X., Zhang, C., Zhang, S., Qian, J., Chen, L., Shi, H., Dong, H.: Thirty-seven transcription factor genes differentially respond to a harpin protein and affect resistance to the green peach aphid in Arabidopsis. — J. Biosci. 35: 435–450, 2010.
Livak, K.J., Schmittgen, T.D.: Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. — Methods 25: 402–408, 2001.
Locatelli, F., Vannini, C., Magnani, E., Coraggio, I., Bracale, M.: Efficiency of transient transformation in tobacco protoplasts is independent of plasmid amount. — Plant Cell Rep. 21: 865–871, 2003.
Lolas, I.B., Himanen, K., Grønlund, J.T., Lynggaard, C., Houben, A., Melzer, M., Van Lijsebettens, M., Grasser, K.D.: The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. — Plant J. 61: 686–697, 2010.
Lourenço, T., Saibo, N., Batista, R., Pinto Ricardo, C., Oliveira, M.M.: Inducible and constitutive expression of HvCBF4 in rice leads to differential gene expression and drought tolerance. — Biol. Plant. 55: 653–663, 2011.
Mao, X., Jia, D., Li, A., Zhang, H., Tian, S., Zhang, X., Jia, J., Jing, R.: Transgenic expression of TaMYB2A confers enhanced tolerance to multiple abiotic stresses in Arabidopsis. — Funct. Integr. Genom. 11: 445–465, 2011.
Mattana, M., Biazzi, E., Consonni, R., Locatelli, F., Vannini, C., Provera, S., Coraggio, I.: Overexpression of Osmyb4 enhances compatible solute accumulation and increases stress tolerance of Arabidopsis thaliana. — Physiol. Plant. 125: 212–223, 2005.
Mejia-Guerra, M.K., Pomeranz, M., Morohashi, K., Grotewold, E.: From plant gene regulatory grids to network dynamics. — Biochim. biophys. Acta 1819: 454–465, 2012.
Miura, K., Jin, J.B., Lee, J., Yoo, C.Y., Stirm, V., Miura, T., Ashworth, E.N., Bressan, R.A., Yun, D.J., Hasegawa, P.M.: SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. — Plant Cell 19: 1403–1414, 2007.
Movahedi, S., Sayed Tabatabaei, B.E., Alizade, H., Ghobadi, C., Yamchi, A., Khaksar, G.: Constitutive expression of Arabidopsis DREB1B in transgenic potato enhances drought and freezing tolerance. — Biol. Plant. 56: 37–42, 2012.
Murashige, T., Skoog, F.: A revised medium for rapid growth and bioassays with tobacco tissue cultures. — Physiol. Plant. 15: 473–497, 1962.
Nicolaides, N.C., Gualdi, R., Casadevall, C., Manzella, L., Calabretta, B.: Positive autoregulation of c-myb expression via Myb binding sites in the 5′ flanking region of the human c-myb gene. — Mol. cell. Biol. 11: 6166–6176, 1991.
Page, R.D.M.: TREEVIEW: an application to display phylogenetic trees on personal computers. — Computer Appl. Biosci. 12: 357–358, 1996.
Pandolfi, D., Solinas, G., Valle, G., Coraggio, I.: Cloning of a cDNA encoding a novel myb gene (acc. No. Y11414) highly expressed in cold stressed rice coleoptiles (PGR 97-079). — Plant Physiol. 114: 747, 1997.
Paolacci, A.R., Tanzarella, O.A., Porceddu, E., Ciaffi, M.: Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. — BMC mol. Biol. 10: 11–38, 2009.
Park, M.R., Yun, K.Y., Mohanty, B., Herath, V., Xu, F., Wijaya, E., Bajic, V.B., Yun, S.J., De Los Reyes, B.G.: Supra-optimal expression of the cold-regulated OsMyb4 transcription factor in transgenic rice changes the complexity of transcriptional network with major effects on stress tolerance and panicle development. — Plant Cell Environ. 33: 2209–2230, 2010.
Pasquali, G., Biricolti, S., Locatelli, F., Baldoni, E., Mattana, M.: Osmyb4 expression improves adaptive responses to drought and cold stress in transgenic apples. — Plant Cell Rep. 27: 1677–1686, 2008.
Qin, Y., Wang, M., Tian, Y., He, W., Han, L., Xia, G.: Overexpression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. — Mol. Biol. Rep. 39: 7183–7192, 2012.
Rosenfeld, N., Elowitz, M.B., Alon, U.: Negative autoregulation speeds the response times of transcription networks. — J. mol. Biol. 323: 785–793, 2002.
Rushton, P.J., Reinstädler, A., Lipka, V., Lippok, B., Somssich, I.E.: Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. — Plant Cell 14: 749–762, 2002.
Soltész, A., Vágújfalvi, A., Rizza, F., Kerepesi, I., Galiba, G., Cattivelli, L., Coraggio, I., Crosatti, C.: The rice Osmyb4 gene enhances tolerance to frost and improves germination under unfavourable conditions in transgenic barley plants. — J. appl. Genet. 53: 133–143, 2012.
Sugimoto, K., Takeda, S., Hirochika, H.: Myb-related transcription factor NtMyb2 induced by wounding and elicitors is a regulator of the tobacco retrotrasposon Tto1 and defense-related genes. — Plant Cell 12: 2511–2527, 2000.
Sugimoto, K., Takeda, S., Hirochika, H.: Transcriptional activation mediated by binding of a plant GATA-type zinc finger protein AGP1 to the AG-motif (AGATCCAA) of the wound-inducible Myb gene NtMyb2. — Plant J. 36: 550–564, 2003.
Todaka, D., Nakashima, K., Shinozaki, K., Yamaguchi-Shinozaki, K.: Toward understanding transcriptional regulatory networks in abiotic stress responses and tolerance in rice. — Rice 5: 6, 2012.
Vannini, C., Campa, M., Iriti, M., Genga, A., Faoro, F., Carravieri, S., Rotino, G.L., Rossoni, M., Spinardi, A., Bracale, M.: Evaluation of transgenic tomato plants ectopically expressing the rice Osmyb4 gene. — Plant Sci. 173: 231–239, 2007.
Vannini, C., Iriti, M., Bracale, M., Locatelli, F., Faoro, F., Croce, P., Pirona, R., Di Maro, A., Coraggio, I., Genga, A.: The ectopic expression of the rice Osmyb4 gene in Arabidopsis increases tolerance to abiotic, environmental and biotic stresses. — Physiol. mol. Plant Pathol. 69: 26–42, 2006.
Vannini, C., Locatelli, F., Bracale, M., Magnani, E., Marsoni, M., Osnato, M., Mattana, M., Baldoni, E., Coraggio, I.: Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. — Plant J. 37: 115–127, 2004.
Yang, Y., Klessing, D.F.: Isolation and characterization of a tobacco mosaic virus-inducible myb oncogene homolog from tobacco. — Proc. nat. Acad. Sci. USA 93: 14972–14977, 1996.
Yang, A., Dai, X., Zhang, W.H.: A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. — J. exp. Bot. 63: 2541–2556, 2012.
Zhang, L., Zhao, G., Jia, J., Liu, X., Kong, X.: Molecular characterization of 60 isolated Myb genes and analysis of their expression during abiotic stress. — J. exp. Bot. 63: 203–214, 2012.
Zhao, J., Zhang, W., Zhao, Y., Gong, X., Guo, L., Zhu, G., Wang, X., Gong, Z. Schumaker, K.S., Guo, Y.: SAD2, an importin β-like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. — Plant Cell 19: 3805–3818, 2007.
Zhou, M.Q., Shen, C., Wu, L.H., Tang, K.X, Lin, J.: CBFdependent signalling pathway: a key responder to low temperature stress in plants. — Crit. Rev. Biotechnol. 31: 186–192, 2011.
Author information
Authors and Affiliations
Corresponding author
Additional information
Acknowledgements: This work was partially supported by the FIRB-Strategic Project “Post-genome”, a programme of the Italian Ministry of University and Scientific Research [RBNE01LAC].
Rights and permissions
About this article
Cite this article
Baldoni, E., Genga, A., Medici, A. et al. The OsMyb4 gene family: stress response and transcriptional auto-regulation mechanisms. Biol Plant 57, 691–700 (2013). https://doi.org/10.1007/s10535-013-0331-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10535-013-0331-3