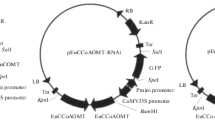
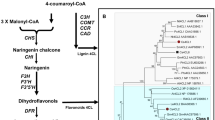
Abstract
The maize (Zea mays L.) caffeic acid O-methyl-transferase (COMT) is a key enzyme in the biosynthesis of lignin. In this work we have characterized the involvement of COMT in the lignification process through the study of the molecular mechanisms involved in its regulation. The examination of the maize COMT gene promoter revealed a putative ACIII box, typically recognized by R2R3-MYB transcription factors. We used the sequence of known R2R3-MYB factors to isolate five maize R2R3-MYB factors (ZmMYB2, ZmMYB8, ZmMYB31, ZmMYB39, and ZmMYB42) and study their possible roles as regulators of the maize COMT gene. The factors ZmMYB8, ZmMY31, and ZmMYB42 belong to the subgroup 4 of the R2R3-MYB family along with other factors associated with lignin biosynthesis repression. In addition, the induction pattern of ZmMYB31 and ZmMYB42 gene expression on wounding is that expected for repressors of the maize COMT gene. Arabidopsis thaliana plants over-expressing ZmMYB31 and ZmMYB42 down-regulate both the A. thaliana and the maize COMT genes. Furthermore, the over-expression of ZmMYB31 and ZmMYB42 also affect the expression of other genes of the lignin pathway and produces a decrease in lignin content of the transgenic plants.
Similar content being viewed by others
References
Anterola AM, Lewis NG (2002) Trends in lignin modifications: a comprehensive analysis of the effects of genetics manipulations/mutations on lignification and vascular integrity. Phytochemistry 61:221–294
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546
Boudet AM, Kajita S, Grima-Pettenati J, Goffner D (2003) Lignins and lignocellulosics: a better control of synthesis for new and improved uses. Trends Plant Sci 8:576–581
Bugos RC, Chiang VL, Campbell WH (1991) cDNA cloning, sequence analysis and seasonal expression of lignin-bispecific caffeic acid/5-hydroxyferulic acid O-methyltransferase of aspen. Plant Mol Biol 17:1203–1215
Caparrós-Ruiz D, Fornalé S, Civardi L, Puigdomènech P, Rigau J (2006) Isolation and characterisation of a family of laccases in maize. Plant Sci 171:217–225
Capellades M, Torres MA, Bastisch I, Stiefel V, Vignols F, Bruce WB, Peterson D, Puigdomenech P, Rigau J (1996) The maize caffeic acid O-methyltransferase gene promoter is active in transgenic tobacco and maize plant tissues. Plant Mol Biol 31:307–332
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6:325–330
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Collazo P, Montoliu L, Puigdomènech P, Rigau J (1992) Structure and expression of the lignin O-methyltransferase gene from Zea mays L. Plant Mol Biol 20:857–867
Cone KC, Cocciolone SM, Burr FA, Burr B (1993) Maize anthocyanin regulatory gene pI is a duplicate of c1 that functions in the plant. Plant Cell 5:1795–1805
de Obeso M, Caparrós-Ruiz D, Vignols F, Puigdomènech P, Rigau J (2003) Characterisation of maize peroxidases having differential patterns of mRNA accumulation in relation to lignifying tissues. Gene 309:23–33
Douglas CJ (1996) Phenylpropanoid metabolism and lignin biosynthesis: from weeds to trees. Trends Plant Sci 1:171–178
Feuillet C, Lauvergeat V, Deswarte C, Pilate G, Boudet A, Grima-Pettenati J (1995) Tissue- and cell-specific expression of a cinnamyl alcohol dehydrogenase promoter in transgenic poplar plants. Plant Mol Biol 27:651–667
Franken P, Schrell S, Peterson PA, Saedler H, Wienand U (1989) Molecular analysis of protein domain function encoded by the myb-homologous maize genes C1, Zm 1 and Zm 38. Plant J 6:21–30
Goicoechea M, Lacombe E, Legay S, Mihaljevic S, Rech P, Jauneau A, Lapierre C, Pollet B, Verhaegen D, Chaubet-Gigot N, Grima-Pettenati G (2005) EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J 43:553–567
Grotewold E, Athma P, Peterson T (1991) Alternatively spliced products of the maize P gene encode proteins with homology to the DNA-binding domain of myb-like transcription factors. Proc Natl Acad Sci USA 88:4587–4591
Grotewold E, Drummond BJ, Bowen B, Peterson T (1994) The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76:543–553
Guindon S, Lethiec F, Duroux P, Gascuel O (2005) PHYML Online-a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33(Web Server issue):W557–W559
Halpin C, Holt K, Chojecki J, Olivier D, Chabbert B, Monties B, Edwards K, Barakate A, Foxon GA (1998) Brown-midrib maize (bm1): a mutation affecting cinnamyl alcohol dehydrogenase gene. Plant J 14:545–553
Hauffe KD, Lee SP, Subramaniam R, Douglas CJ (1993) Combinatorial interactions between positive and negative cis-acting elements control spatial patterns of 4CL-1 expression in transgenic tobacco. Plant J 4:235–253
Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000a) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42:819–832
Hellens R, Mullineaux P, Klee H (2000b) A guide to Agrobacterium binary Ti vectors. Trends Plant Sci 5:446–451
Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensibility of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res 22:4673–4680
Hofgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16:9877
Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. Plant J 26:205–216
Karpinska B, Karlsson M, Srivastava M, Stenberg A, Schrader J, Sterky F, Bhalerao R, Wingsle G (2004) MYB transcription factors are differentially expressed and regulated during secondary vascular tissue development in hybrid aspen. Plant Mol Bio l56:255–270
Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, Smeekens S, Tonelli C, Paz-Ares J, Weisshaar B (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 16:263–76
Lewis NG, Yamamoto E (1990) Lignin: occurrence, biogenesis and biodegradation. Annu Rev Plant Physiol Plant Mol Biol 41:455–496
Marocco A, Wissenbach M, Becker D, Paz-Ares J, Saedler H, Salamini F, Rohde W (1989) Multiple genes are transcribed in Hordeum vulgare and Zea mays that carry the DNA binding domain of the myb oncoproteins. Mol Gen Genet 216:183–187
Mellerowicz EJ, Baucher M, Sundberg B, Boerjan W (2001) Unravelling cell wall formation in the woody dicot stem. Plant Mol Biol 47:239–274
Miyake K, Ito T, Senda M, Ishikawa R, Harada T, Niizeki M, Akada S (2003) Isolation of a subfamily of genes for R2R3-MYB transcription factors showing up-regulated expression under nitrogen nutrient-limited conditions. Plant Mol Biol 53:237–245
Morrow MP, Mascia P, Self KP, Altschuler M (1997) Molecular characterization of a brown midrib deletion mutation in maize. Mol Breed 3:351–357
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Patzlaff A, Newman LJ, Dubos C, Whetten RW, Smith C, McInnis S, Bevan MW, Sederoff RR, Campbell MM (2003a) Characterisation of Pt MYB1, an R2R3-MYB from pine xylem. Plant Mol Biol 53:597–608
Patzlaff A, McInnis S, Courtenay A, Surman C, Newman LJ, Smith C, Bevan MW, Mansfield S, Whetten RW, Sederoff RR, Campbell MM (2003b) Characterisation of a pine MYB that regulates lignification. Plant J 36:743–754
Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H (1987) The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J 6:3553–3558
Piquemal J, Chamayou S, Nadaud I, Beckert M, Barriere Y, Mila I, Lapierre C, Rigau J, Puigdomènech P, Jauneau A, Digonnet C, Boudet AM, Goffner D, Pichon M (2002) Down-regulation of caffeic acid o-methyltransferase in maize revisited using a transgenic approach. Plant Physiol 130:1675–1685
Pilu R, Piazza P, Petroni K, Ronchi A, Martin C, Tonelli C (2003) pl-bol3, a complex allele of the anthocyanin regulatory pl1 locus that arose in a naturally occurring maize population. Plant J 36:510–521
Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW (2004) AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J 40:979–995
Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133:1051–1071
Reichel C, Mathur J, Eckes P, Langenkemper K, Koncz C, Schell J, Reiss B, Maas C (1996) Enhanced green fluorescence by the expression of an Aequorea Victoria green fluorescent protein mutant in mono- and dicotyledonous plant cells. Proc Natl Acad Sci USA 93:5888–5893
Romero I, Fuertes A, Benito MJ, Malpica JM, Leyva A, Paz-Ares J (1998) More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J 14:273–284
Sablowski RW, Moyano E, Culianez-Macia FA, Schuch W, Martin C, Bevan M (1994) A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J 13:128–37
Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C (1998a) The AmMYB308 and AmMYB330 transcription factors from antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10:135–54
Tamagnone L, Merida A, Stacey N, Plaskitt K, Parr A, Chang CF, Lynn D, Dow JM, Roberts K, Martin C (1998b) Inhibition of phenolic acid metabolism results in precocious cell death and altered cell morphology in leaves of transgenic tobacco plants. Plant Cell 10:1801–16
Vignols F, Rigau J, Torres MA, Capellades M, Puigdomenech P (1995) The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7:407–416
Vignols F, José-Estanyol M, Caparrós-Ruiz D, Rigau J, Puigdomènech P (1999) Involvement of a maize proline-rich protein in secondary cell wall formation as deduced from its specific mRNA localisation. Plant Mol Biol 39:945–952
Whetten RW, MacKay JJ, Sederoff RR (1998) Recent advances in understanding lignin biosynthesis. Annu Rev Plant Physiol Plant Mol Biol 49:585–609
Whitmore FW (1978) Lignin-carbohydrate complex formed in isolated cell walls of callus. Phytochemistry 17:421–425
Ye ZH, Kneusel RE, Matern U, Varner JE (1994) An alternative methylation pathway in lignin biosynthesis in Zinnia. Plant Cell 6:1427–1439
Acknowledgments
We are indebted to Dr. Sheen (MGH, USA) for kindly providing us with the synthetic GFP (sGFP) as well as the John Innes Center (Norwich, UK) for providing us with the pGREENII-0029 vector. We want to thank Dr. Piulachs (IBMB-CSIC) for her advice and comments on the phylogenetic analyses performed in this work and Dr. Pons for the confocal microscopy analyses. We are also indebted to the sequencing and greenhouse teams of IBMB-CSIC, as well as Dr. Burgess for the English correction of this manuscript. This work was funded by the Spanish “Ministerio de Ciencia y Tecnología” (BIO2001-1140). S-F was financed by a post-doctoral grant from the Autonomous Government of Catalonia (2003PIV-A-00033). F.M-S was financed by the European Project INCO-II (ICA4-CT-2000-30017) and by the “Consorci CSIC-IRTA” laboratory. T-M was financed by the European Project MapMaize (PL 962312). D-C.R was financed by the Spanish “Ministerio de Ciencia y Tecnología” (“Ramón y Cajal” Program). This work was carried out within the framework of the “Centre de Referència de Biotecnologia” (CERBA) from the Autonomous Government of Catalonia.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fornalé, S., Sonbol, FM., Maes, T. et al. Down-regulation of the maize and Arabidopsis thaliana caffeic acid O-methyl-transferase genes by two new maize R2R3-MYB transcription factors. Plant Mol Biol 62, 809–823 (2006). https://doi.org/10.1007/s11103-006-9058-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-006-9058-2