Abstract
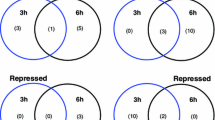
Salinity is one of the major constraints adversely influencing crop productivity. Saltol QTL is a major QTL associated with Na+-K+ ratio and seedling stage salinity tolerance in rice. With an aim to understand the contribution of individual genes localized within saltol towards salinity tolerance, we analysed the transcript abundance of a set of these genes in seedlings of contrasting genotypes of rice. We hypothesize that this approach may be helpful in identifying new ‘candidate genes’ for improving salinity tolerance in crops. For this purpose, seedlings of Oryza sativa cv. IR64 (sensitive) and the landrace Pokkali (tolerant) were subjected to short/long durations of salinity. qRT-PCR analysis clearly exhibited differential regulation of genes encoding signaling related protein (SRPs), where higher transcript abundance for most of them was observed in Pokkali than IR64 under non-stress conditions, thereby indicating towards well preparedness of the former to handle stress, in anticipation. Genes encoding proteins of unknown function (PUFs), though, constitute a considerable portion of plant genome, have so far been neglected in most studies. Time course analysis of these genes showed a continuous increase in their abundance in Pokkali, while in IR64, their abundance increased till 24 h followed by a clear decrease, thereby justifying their nomenclature as ‘salinity induced factors’ (SIFs). This is the first report showing possible involvement of SIFs localized within salinity related QTL towards salinity stress response. Based on the phenotypes of insertional mutants, it is proposed that these SIFs may have a putative function in vegetative growth (SIFVG), fertility (SIFF), viability (SIFV) or early flowering (SIFEF).






Similar content being viewed by others
References
Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris, Life Sciences 316:1194–1199
Bohnert HJ, Gong Q, Li P, Ma S (2006) Unravelling abiotic stress tolerance mechanisms-getting genomic going. Curr Opin Plant Biol 9:180–188
Bonilla P, Dvorak J, Mackill D, Deal K, Gregorio G (2002) RFLP and SSLP mapping of salinity tolerance genes in chromosome 1 of rice (Oryza sativa L.) using recombinant inbred lines. Philippine J Agric Sci 85:68–76
Claes B, Dekeyser R, Villarroel R, Van den Bulcke M, Bauw G, Van Montagu M, Caplan A (1990) Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. Plant Cell 2:19–27
Cui F, Liu L, Zhao Q, Zhang Z, Lia Q, Lin B, Wu Y, Tang S, Xie Q (2012) Arabidopsis ubiquitin conjugase UBC32 Is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 24(1):233–244
Diédhiou CJ, Popova OV, Dietz KJ, Golldack D (2008) The SNF1-type serine-threonine protein kinase SAPK4regulates stress-responsive gene expression in rice. BMC Plant Biol 8:49
Elge S, Brearley C, Xia HJ, Kehr J, Xue HW, Müller-Röber B (2001) An Arabidopsis inositol phospholipid kinase strongly expressed in procambial cells: synthesis of PtdIns(4,5) P2 and PtdIns(3,4,5) P3 in insect cells by 5-phosphorylation of precursors. Plant J 26:561–571
Flowers TJ, Colmer TD (2008) Salinity tolerance in halophytes. New Phytol 179:945–963
Flowers TJ, Yeo AR (1981) Variability in the resistance of sodium chloride salinity within rice (Oryza sativa L.) varieties. New Phytol 88:363–373
Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, OhmeTakagi M, Tran LSP, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39:863–876
Gregorio GB (1997) Tagging salinity tolerance genes in rice using amplified fragment length polymorphism (AFLP). Ph.D. Thesis, University of the Philippines, Los Banos, Laguna, Philippines
Gregorio GB, Senadhira D, Mendoza RD, Manigbas NL, Roxas JP, Guerta CQ (2002) Progress in breeding for salinity tolerance and associated abiotic stresses in rice. Field Crops Res 76:91–101
Guo G, Ge P, Ma C, Li X, Lv D, Wang S, Ma W, Yan Y (2012) Comparative proteomic analysis of salt response proteins in seedling roots of two wheat varieties. J Proteomics 75(6):1867–1885
He ZH, Cheeseman I, He D, Kohorn BD (1999) A cluster of five cell wall-associated receptor kinase genes, Wak1-5, are expressed in specific organs of Arabidopsis. Plant Mol Biol 39:1189–1196
Karan R, Singla-Pareek SL, Pareek A (2009) Histidine kinase and response regulator genes as they relate to salinity tolerance in rice. Funct Integr Genomics 9(3):411–417
Koyama ML, Levesley A, Koebner RM, Flowers TJ, Yeo AR (2001) Quantitative trait loci for component physiological traits determining salt tolerance in rice. Plant Physiol 125:406–422
Kulik A, Wawer I, Krzywińska E, Bucholc M, Dobrowolska G (2011) SnRK2 protein kinases—key regulators of plant response to abiotic Stresses. OMICS 15(12):859–872
Kumar G, Purty RS, Singla-Pareek SL, Pareek A (2009) Maintenance of stress related transcripts in tolerant cultivar at a level higher than sensitive one appears to be a conserved salinity response among plants. Plant Signal Behav 4(5):431–434
Kumar G, Kushwaha HR, Panjabi-Sabharwal V, Kumari S, Joshi R, Karan R, Mittal S, Singla-Pareek SL, Pareek A (2012a) Clustered metallothionein genes are co-regulated in rice and ectopic expression of OsMT1e-P confers multiple abiotic stress tolerance in tobacco via ROS scavenging. BMC Plant Biol 12:107
Kumar R, Mustafiz A, Sahoo K, Sharma V, Samanta S, Sopory SK, Pareek A, Singla-Pareek SL (2012b) Functional screening of cDNA library from a salt tolerant rice genotype Pokkali identifies mannose-1-phosphate guanyl transferase gene (OsMPG1) as a key member of salinity stress response. Plant Mol Biol 79(6):555–568
Kumari S, Panjabi Nee Sabharwal V, Kushwaha HR, Sopory SK, Singla-Pareek SL, Pareek A (2009) Transcriptome map for seedling stage specific salinity stress response indicates a specific set of genes as candidate for saline tolerance in Oryza sativa L. Funct Integr Genomics 9:109–123
Kushwaha HR, Singh AK, Sopory SK, Singla-Pareek SL, Pareek A (2009) Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genomics 10:200
Liang C, Zhang X, Chi X, Guan X, Li Y, Qin S, Shao HB (2011) Serine/threonine protein kinase SpkG Is a candidate for high salt resistance in the unicellular cyanobacterium Synechocystis sp. PCC 6803. PLoS One 6(5):e18718
Lin HX, Zhu MZ, Yano M, Gao JP, Liang ZW, Su WA, Hu XH, Ren ZH, Chao DY (2004) QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor Appl Genet 108:253–260
Liu H, Zhang H, Yang Y, Li G, Yang Y, Wang X, Basnayake BMVS, Li D, Song F (2008) Functional analysis reveals pleiotropic effects of rice RING-H2 finger protein gene OsBIRF1 on regulation of growth and defense responses against abiotic and biotic stresses. Plant Mol Biol 68:17–30
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆C(T) method. Methods 25(4):402–408
Luhua S, Ciftci-Yilmaz S, Harper J, Cushman J, Mittler R (2008) Enhanced tolerance to oxidative stress in transgenic arabidopsis plants expressing proteins of unknown function. Plant Physiol 148:280–292
Lutts S, Kinet JM, Bouharmont J (1995) Changes in plant response to NaCl during development of rice (Oryza sativus L.) varieties differing in salinity resistance. J Exp Bot 46:1843–1852
Maas EV, Hoffman GJ (1977) Crop salt tolerance, current assessment. J Irrig Drain Div ASCE 103:115–134
Martin RC, Cutter KG, Baldwin JC, Dombrowski JE (2012) Identification and characterization of a salt stress-inducible zinc finger protein from Festuca arundinacea. BMC Research Notes 5:66
Martinoia E, Maeshima M, Neuhaus HE (2007) Vacuolar transporters and their essential role in plant metabolism. J Exp Bot 58:83–102
Mikami K, Katagiri T, Luchi S, Yamaguchi-Shinozaki K, Shinozaki K (1998) A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J 15:563–568
Moons A, Prinsen E, Bauw G, Van Montagu M (1997) Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transcripts in rice roots. Plant Cell 9:2243–2259
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Nakamura A, Fujioka S, Sunohara H, Kamiya N, Hong Z, Inukai Y, Miura K, Takatsuto S, Yoshida S, Ueguchi-Tanaka M, Hasegawa Y, Kitano H, Makoto M (2006) The Role of OsBRI1 and its homologous Genes, OsBRL1 and OsBRL3, in rice. Plant Physiol 140(2):580–590
Nongpiur R, Soni P, Singla-Pareek SL, Pareek A (2012) Histidine kinases in plants: cross talk between hormone and stress responses. Plant Signal Behav 7(10):1230–1237
Pandit A, Rai V, Bal S, Sinha S, Kumar V et al (2010) Combining QTL mapping and transcriptome profiling of bulked RILs for identification of functional polymorphism for salt tolerance genes in rice (Oryzasativa L.). Mol Genet Genomics 284(2):121–136
Pawlowski K (2008) Uncharacterized/hypothetical proteins in biomedical ‘omics’ experiments: Is novelty being swept under the carpet? Brief Funct Genomic Proteomic 7(4):283–290
Plett D, Cotsaftis O, Johnson AA, Walia H, Wilson C, Ismail AM, Close TJ, Tester M, Baumann U (2011) Root-specific transcript profiling of contrasting rice genotypes in response to salinity stress. Mol Plant 4(1):25–41
Rabbani MS, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133:1755–1767
Rikke BA, Johnson TE (1998) Towards the cloning of genes underlying murine QTLs. Mamm Genome 9:963–968
Ron M, Weller JI (2007) From QTL to QTN identification in livestock “Winning by points rather than knock-out”. Animal Genetics: a review 38(5):429–439
Sahi C, Singh A, Kumar K, Blumwald E, Grover A (2006) Salt stress response in rice: genetics, molecular biology, and comparative genomics. Funct Integr Genomics 6:263–284
Salvi S, Tuberosa R (2005) To clone or not to clone plant QTLs: present and future challenges. Trends Plant Sci 10:297–304
Singh RK, Flowers TJ (2010) The physiology and molecular biology of the effects of salinity on rice. In: Pessarakli M (ed) Handbook of plant and crop stress, 3rd edn. Taylor and Francis, Florida, pp 901–942
Singh AK, Kumar R, Pareek A, Sopory SK, Singla-Pareek SL (2012) Overexpression of rice CBS domain containing protein improves salinity, oxidative and heavy metal tolerance in transgenic tobacco. Mol Biotechnol 52(3):205–216
Souer E, van Houwelingen A, Kloos D, Mol J, Koes R (1996) The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at merstem and primordia boundaries. Cell 85:159–170
Sreenivasulu N, Sopory SK, Kavi Kishor PB (2007) Deciphering the regulatory mechanisms of abiotic stress tolerance in plants by genomic approaches. Gene 388:1–13
Takehisa H, Shimodate T, Fukuta Y, Ueda T, Yano M, Yamaya T, Kameya T, Sato T (2004) Identification of quantitative trait loci for plant growth of rice in paddy field flooded with salt water. Field Crops Res 89:85–95
Thomson MJ, Ocampo MD, Egdane J, Rahman MA, Sajise AG, Adorada DL, Tumimbang-Raiz E, Blumwald E, Seraj ZI, Singh RK, Gregorio GB, Ismail AM (2010) Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice 3:148–160
Toivola DM, Strnad P, Habtezion A, Omary MB (2010) Intermediate filaments take the heat as stress proteins. Trends Cell Biol 20(2):79–91
Tóth G, Montanarella L, Rusco E (2008) Updated map of salt affected soils in the European union threats to soil quality in Europe. European Communities 61–74
Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC (2003) The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15:1563–1577
Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Zeng L, Wanamaker SI, Mandal J, Xu J, Cui X, Close TJ (2005) Close comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol 139:822–835
Walia H, Wilson C, Liu X, Ismail A, Close TJ (2007) Transcriptional profiling of rice under salinity stress during panicle initiation stage. Plant Mol Biol 63(5):609–623
Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410:380–383
Welters P, Takegawa K, Emr SD, Chrispeels MJ (1994) ATVPS34, a PtdIns 3-kinase of Arabidopsis thaliana is an essential protein with homology to a calcium-dependent lipid-binding domain. Proc Natl Acad Sci USA 91:11398–11402
Williams ME, Torabinejad J, Cohick E, Parker K, Drake EJ, Thompson JE, Hortter M, DeWald DB (2005) Mutations in the Arabidopsis phosphoinositide phosphatase gene SAC9 lead to overaccumulation of PtdIns(4,5)P2 and constitutive expression of the stress-response pathway. Plant Physiol 138(2):686–700
Zhang T, Zhao X, Wang W, Pan Y, Huang L, Liu X, Zong Y, Zhu L, Yang D, Fu B (2012) Comparative transcriptome profiling of chilling stress responsiveness in two contrasting rice genotypes. PLoS One 7(8):e43274
Acknowledgments
Authors would like to thank financial and Infrastructure support received from JNU through PURSE & UGC-RNW and ICGEB. Department of Science and Technology (DST) and Department of Biotechnology (DBT), Govt. of India is also acknowledged for financial support. N. S. and P.S. would like to thank University Grants Commission and Council of Scientific and Industrial Research, New Delhi, India respectively, for the award of their Research Fellowships.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Soda, N., Kushwaha, H.R., Soni, P. et al. A suite of new genes defining salinity stress tolerance in seedlings of contrasting rice genotypes. Funct Integr Genomics 13, 351–365 (2013). https://doi.org/10.1007/s10142-013-0328-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-013-0328-1