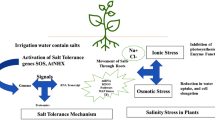
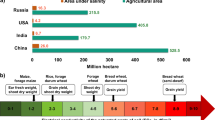
Abstract
Significant progress has been made in unraveling the molecular biology of rice in the past two decades. Today, rice stands as a forerunner amongst the cereals in terms of details known on its genetics. Evidence show that salt tolerance in plants is a quantitative trait. Several traditional cultivars, landraces, and wild types of rice like Pokkali, CSR types, and Porteresia coarctata appear as promising materials for donation of requisite salt tolerance genes. A large number of quantitative trait loci (QTL) have been identified for salt tolerance in rice through generation of recombinant inbred lines and are being mapped using different types of DNA markers. Salt-tolerant transgenic rice plants have been produced using a host of different genes and transcript profiling by micro- and macroarray-based methods has opened the gates for the discovery of novel salt stress mechanisms in rice, and comparative genomics is turning out to be a critical input in this respect. In this paper, we present a comprehensive review of the genetic, molecular biology, and comparative genomics effort towards the generation of salt-tolerant rice. From the data on comprehensive transcript expression profiling of clones representing salt-stress-associated genes of rice, it is shown that transcriptional and translational machineries are important determinants in controlling salt stress response, and gene expression response in tolerant and susceptible rice plants differs mainly in quantitative terms.




Similar content being viewed by others
References
Abbasi FM, Komatsu S (2004) A proteomic approach to analyze salt-responsive proteins in rice leaf sheath. Proteomics 4:2072–2081
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Agarwal S, Grover A (2005) Isolation and transcription profiling of anaerobic stress associated cDNA clones from flooding stress tolerant FR13A rice genotype. Ann Bot 96:831–844
Akbar M, Gunawardena IE, Ponnamperuma FN (1986a) Breeding for soil stresses. In: International Rice Research Institute (ed) Progress in rainfed lowland rice. International Rice Research Institute, Manila, Philippines, pp 263–272
Akbar M, Khush GS, Hillerislambers D (1986b) Genetics of salt tolerance in rice. In: International Rice Research Institute (ed) Progress in rainfed lowland rice. International Rice Research Institute, Manila, Philippines, pp 399–409
Akbar M, Jena KK, Seshu DV (1987) Salt tolerance in wild rices. Int Rice Res Newsl 12:15
Alia, KondoY, Sakamoto A, Nonaka H, Hayashi H, Saradhi PP, Chen THH, Murata N (1999) Enhanced tolerance to light stress of transgenic Arabidopsis plants that express the codA gene for a bacterial choline oxidase. Plant Mol Biol 40:279–288
Altschul SF, Madden TL, Scheffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Amaya I, Botella MA, De La Calle M, Medina MI, Heredia A, Bressan RA, Hasegawa PM, Quesada MA, Valpuesta V (1999) Improved germination under osmotic stress of tobacco plants overexpressing a cell wall peroxidase. FEBS Lett 457:80–84
Anderson P, Kedersha N (2002) Stressful initiations. J Cell Sci 115:3227–3234
Anil VS, Krishnamurthy P, Kuruvilla S, Sucharitha K, Thomas G, Mathew MK (2005) Regulation of the uptake and distribution of Na+ in shoots of rice (Oryza sativa) variety Pokkali: role of Ca2+ in salt tolerance response. Physiol Plant 124:451–464
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285:1256–1258
Apse MP, Sottosanto JB, Blumwald E (2003) Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J 36:229–239
Asadulghani, Nitta K, Kaneko Y, Kojima K, Fukuzawa H, Kosaka H, Nakamoto H (2004) Comparative analysis of the hspA mutant and wild-type Synechocystissp. Strain PCC 6803 under salt stress: evaluation of the role of hspA in salt-stress management. Arch Microbiol 182:487–497
Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24:23–58
Bhumbla D, Abrol I (1978) Saline and sodic soils. In: Soils and rice. International Rice Research Institute, Manila, Philippines, pp 719–738
Blumwald E, Grover A (2006) Salt tolerance. In: Halford NG (ed) Plant biotechnology: current and future uses of genetically modified crops. John Wiley and Sons Ltd., UK, pp 206–224
Bohnert HJ, Ayoubi P, Borchert C, Bressan RA, Burnap RL, Cushman JC, Cushman, MA, Deyholos M, Fischer R, Galbraith DW, Hasegawa PM, Jenks M, Kawasaki S, Koiwa H, Kore-Eda S, Lee B-H, Michalowski CB, Misawa E, Nomura M, Ozturk N, Postier B, Prade R, Song CP, Tanaka Y, Wang H, Zhu JK (2001) A genomics approach toward salt stress tolerance. Plant Physiol Biochem 39:295–311
Bonilla P, Dvorak J, Mackill D, Deal K, Gregorio G (2002) RFLP and SSLP mapping of salinity tolerance genes in chromosome 1 of rice (Oryza sativa L.) using recombinant inbred lines. Philipp Agric Sci 85:68–76
Bressan RA, Zhang C, Zhang H, Hasegawa PM, Bohnert HJ, Zhu JK (2001) Learning from the Arabidopsis experience. The next gene search paradigm. Plant Physiol 127:1354–1360
Causse M, Fulton T, Cho Y, Ahn S, Chunwongse J, Wu K, Xiao J, Yu Z, Ronald P, Harrington S, Second G, McCouch S, Tanksley S (1994) Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138:1251–1274
Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA, Budworth PR, Tao Y, Xie Z, Chen X, Lam S, Kreps JA, Harper JF, Si-Ammour A, Mauch-Mani B, Heinlein M, Kobayashi K, Hohn T, Dangl JL, Wang X, Zhu T (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 3:559–574
Chinnusamy V, Jagendorf A, Zhu JK (2005) Understanding and improving salt tolerance in plants. Crop Sci 45:437–448
Claes B, Dekeyser R, Villarroel R, van den Bulcke M, Bauw G, Van Montagu M (1990) Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. Plant Cell 2:19–27
Colmer TD, Munns R, Flowers TJ (2005) Improving salt tolerance of wheat and barley: future prospects. Aust J Exp Agric 45:1425–1443
Cooper B, Clarke J D, Budworth P, Kreps J, Hutchison D, Park S, Guimil S, Dunn M, Luginbühl P, Ellero C, Goff SA, Glazebrook J (2003) A network of rice genes associated with stress response and seed development. Proc Natl Acad Sci USA 100:4945–4950
Denekamp M, Smeekens SC (2003) Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiol 132:1415–1423
Dombrowski JE (2003) Salt stress activation of wound-related genes in tomato plants. Plant Physiol 132:2098–2107
Dubey H, Grover A (2001) Current initiatives in proteomics research: the plant perspective. Curr Sci 80:262–269
Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33:751–763
Estruch F (2000) Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev 25:469–486
Flowers TJ (2004) Improving crop salt tolerance. J Exp Bot 55:307–319
Flowers TJ, Yeo AR (1981) Variability in the resistance of sodium chloride salinity within rice (Oryza sativa L.) varieties. New Phytol 88:363–373
Flowers TJ, Yeo AR (1995) Breeding for salinity tolerance in crop plants. Aust J Plant Physiol 22:875–884
Flowers TJ, Flowers SA, Hajibagheri MA, Yeo AR (1990) Salt tolerance in the halophytic wild rice Porteresia coarctata Tateoka. New Phytol 114:675–684
Flowers TJ, Koyama ML, Flowers SA, Sudhakar C, Singh KP, Yeo AR (2000) QTL: their place in engineering tolerance of rice to salinity. J Exp Bot 51:99–106
Forster BP, Ellis RP, Thomas WTb, Newton AC, Tuberosa R, This D, El-Enein RA, Bahri MH, Salem MB (2000) The development and application of molecular markers for abiotic stress tolerance in barley. J Exp Bot 51:19–27
Francki M, Appels R (2002) Wheat functional genomics and engineering crop improvement. Genome Biol 3:1013.1–1013.5
Freemont PS (2000) Ubiquitination: RING for destruction? Curr Biol 10:84–87
Fukuda A, Nakamura A, Tagiri A, Tanaka H, Miyao A, Hirochika H, Tanaka Y (2004) Function, intracellular localization and the importance in salt tolerance of a vacuolar Na(+)/H(+) antiporter from rice. Plant Cell Physiol 45:146–159
Gao X, Ren Z, Zhao Y, Zhang H (2003) Overexpression of SOD2 increases salt tolerance of Arabidopsis. Plant Physiol133:1873–1881
Garcia AB, de Almeida JE, Iyer S, Gerats T, Van Montagu M, Caplan AB (1997) Effects of osmoprotectants upon NaCl stress in rice. Plant Physiol 115:159–169
Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, Wu RJ (2002) Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA 99:15898–15903
Gaxiola RA, Li J, Undurraga S, Dang LM, Allen GJ, Alper SL, Fink GR (2001) Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc Natl Acad Sci USA 98:11444–11449
Gepstein S, Grover A, Blumwald E (2006) Producing biopharmaceuticals in the desert: building an abiotic stress tolerance in plants for salt, heat and drought. In: Knablein J, Muller RH (eds) Modern biopharmaceuticals. Wiley-VCH Verlag GmbH, Weinhaum, pp 967–994
Glenn EP, Brown JJ, Blumwald E (1999) Salt tolerance and crop potential of halophytes. Crit Rev Plant Sci 18:227–255
Gong Z, Koiwa H, Cushman MA, Ray A, Bufford D, Kore-eda S, Matsumota TK, Zhu J, Cushman JC, Bressan RA, Hasegawa PM (2001) Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants. Plant Physiol 126:363–375
Goossens A, Dever TE, Pascual-Ahuir A, Serrano R (2001) The protein kinase Gcn2p mediates sodium toxicity in yeast. J Biol Chem 276:30753–30760
Greenway H, Munns R (1980) Mechanisms of salt tolerance in nonhalophytes. Annu Rev Plant Physiol 31:149–190
Gregorio GB, Senadhira D, Mendoza RD, Manigbas NL, Roxas JP, Guerta CQ (2002) Progress in breeding for salinity tolerance and associated abiotic stresses in rice. Field Crops Res 76:91–101
Grover A (2002) Molecular biology of stress responses. Cell Stress Chaperones 7:1–15
Grover A, Pental D (2003) Breeding objectives and requirements for producing transgenic for the major field crops of India. Curr Sci 84:310–320
Grover A, Pareek A, Maheshwari SC (1993) Molecular approaches for genetically engineering plants tolerant to salt stress. Proc Indian Natl Sci Acad B 59:113–127
Harding HP, Calfon M, Urano F, Novoa I, Ron D (2002) Transcriptional and translational control in the mammalian unfolded protein response. Annu Rev Cell Dev Biol 18:575–599
Harushima Y, Yano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin SY, Antonio BA, Parco A, Kajiya H, Huang N, Yamamoto K, Nagamura Y, Kurata N, Khush GS, Sasaki T (1998) A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148:479–494
Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51:463–499
Hayashi H, Alia Mustardy L, Deshnium PG, Ida M, Murata N (1997) Transformation of Arabidopsis thaliana with the codA gene for choline oxidase; accumulation of glycinebetaine and enhanced tolerance to salt and cold stress.Plant J 12:133–142
He C, Yan J, Shen G, Fu L, Holaday AS, Auld D, Blumwald E, Zhang H (2005) Expression of an Arabidopsis vacuolar sodium/proton antiporter gene in cotton improves photosynthetic performance under salt conditions and increases fibre yield in the field. Plant Cell Physiol 46:1848–1854
Hirochika H, Guiderdoni E, AnG, Hsing Y, Eun MY, Han C, Upadhyaya N, Ramachandran R, Zhang Q, Pereira A, Sundaresan V, Leung H (2004) Rice mutant resources for gene discovery. Plant Mol Biol 54:325–334
Holmstrom KO, Somersalo S, Manda A, Palva TE, Welin B (2000) Improved tolerance to salinity and low temperature in transgenic tobacco producing glycine betaine. J Exp Bot 51:177–185
Hoshida H, Tanaka Y, Hibino T, Hayashi Y, Tanaka A, Takabe T, Takabe T (2000) Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase. Plant Mol Biol 43:103–111
Huang J, Hirji R, Adam L, Rozwadowski KL, Hammerlindl JK, Keller WA, Selvaraj G (2000) Genetic engineering of glycinebetaine production toward enhancing stress tolerance in plants: metabolic limitations. Plant Physiol 122:747–756
International Rice Genome Sequencing Project (2005) The map based sequence of the rice genome. Nature 436:793–800
Jaiswal P, Ni J, Yap I, Ware D, Spooner W, Youens-Clark K, Ren L, Liang C, Zhao W, Ratnapu K, Faga B, Canaran P, Fogleman M, Hebbard C, Avraham S, Schmidt S, Casstevens TM, Buckler ES, Stein L, McCouch S (2006) Gramene: a bird’s eye view of cereal genomes. Nucleic Acids Res 34:D717–D723
Jang IC, Oh SJ, Seo JS, Choi WB, Song SI, Kim CH, Kim YS, Seo HS, Choi YD, Nahm BH, Kim JK (2003) Expression of a bifunctional fusion of the Escherichia coli genes for trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase in transgenic rice plants increases trehalose accumulation and abiotic stress tolerance without stunting growth. Plant Physiol 131:516–524
Jia GX, Zhu ZQ, Chang FQ, Li YX (2002) Transformation of tomato with the BADH gene from Atriplex improves salt tolerance. Plant Cell Rep 21:141–146
Kanesaki Y, Suzuki I, Allakhverdiev SI, Mikami K, Murata N (2002) Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem Biophys Res Commun 290:339–348
Kanno A, Hirai A (1993) A transcription map of the chloroplast genome from rice (Oryza sativa). Curr Genet 23:166–174
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcriptional factor. Nat Biotechnol 17:287–291
Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith D, Bohnert HJ (2001) Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13:889–905
Kawaura K, Mochida K, Yamazaki Y, Ogihara Y (2006) Transcriptome analysis of salinity stress responses in common wheat using a 22k oligo-DNA microarray. Funct Integr Genomics 6:132–142
Khan MSA, Hamid A, Karim MA (1997) Effect of sodium chloride on germination and seedling characters of different types of rice (Oryza sativa L.). J Agron Crop Sci 179:163–169
Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, Hotta I, Kojima K, Namiki T, Ohneda E, Yahagi W, Suzuki K, Li CJ, Ohtsuki K, Shishiki T, Otomo Y, Murakami K, Iida Y, Sugano S, Fujimura T, Suzuki Y, Tsunoda Y, Kurosaki T, Kodama T, Masuda H, Kobayashi M, Xie Q, Lu M, Narikawa R, Sugiyama A, Mizuno K, Yokomizo S, Niikura J, Ikeda R, Ishibiki J, Kawamata M, Yoshimura A, Miura J, Kusumegi T, Oka M, Ryu R, Ueda M, Matsubara K, Kawai J, Carninci P, Adachi J, Aizawa K, Arakawa T, Fukuda S, Hara A, Hashizume W, Hayatsu N, Imotani K, Ishii Y, Itoh M, Kagawa I, Kondo S, Konno H, Miyazaki A, Osato N, Ota Y, Saito R, Sasaki D, Sato K, Shibata K, Shinagawa A, Shiraki T, Yoshino M, Hayashizaki Y, Yasunishi A (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301:376–379
Kim KY, Park SW, Chung YS, Chung CH, Kim JI, Lee JH (2004) Molecular cloning of low-temperature-inducible ribosomal proteins from soybean. J Exp Bot 55:1153–1155
Kishore PBK, Hong Z, Miao GH, Hu CAA, Verma DPS (1995) Overexpression of Δ-1-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol 108:1387–1394
Komatsu S, Konishi H, Shen S, Yang G (2003) Rice proteomics. Mol Cell Proteomics 2.1:2–10
Kong-ngern K, Daduang S, Wongkham C, Bunnag S, Kosittrakun M, Theerakulpisut (2005) Protein profiles in response to salt stress in leaf sheaths of rice seedlings. Sci Asia 31:403–408
Kore-Eda S, Cushman MA, Akselrod I, Bufford D, Fredrickson M, Clark E, Cushman JC (2004) Transcript profiling of salinity stress responses by large-scale expressed sequence tag analysis in Mesembryanthemum crystallinum. Gene 341:83–92
Koyama ML, Levesley A, Koebner RM, Flowers TJ, Yeo AR (2001) Quantitative trait loci for component physiological traits determining salt tolerance in rice. Plant Physiol 125:406–422
Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130:2129–2141
Kurata N, Nagamura Y, Yamamoto K, Harushima Y, Sue N, Wu J, Antonio BA, Shomura A, Shimizu T, Lin SY, Inoue T, Fukuda A, Shimano T, Kuboki Y, Toyama T, Miyamoto Y, Kirihara T, Hayasaka K, Miyao A, Monna L, Zhong HS, Tamura Y, Wang ZX, Momma T, Umehara Y, Yano M, Sasaki T, Minobe Y (1994) A 300 kilobase interval genetic map of rice including 883 expressed sequences. Nat Genet 8:365–372
Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK (2001) The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev 15:912–924
Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P (2004) SMART 4.0: towards genomic data integration. Nucleic Acids Res 32:D142–D144
Li L, Wang X, Stolc V, Li X, Zhang D, Su N, Tongprasit W, Li S, Cheng Z, Wang J, Deng XW (2006) Genome-wide transcription analyses in rice using tiling microarrays. Nat Genet 38:124–129
Lilius G, Holmberg N, Bulow L (1996) Enhanced NaCl stress tolerance in transgenic tobacco expressing bacterial choline dehydrogenase. Biotechnology 14:177–180
Lin HX, Zhu MZ, Yano M, Gao JP, Liang ZW, Su WA, Hu XH, Ren ZH, Chao DY (2004) QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor Appl Genet 108:253–260
Lorieux M, Petror M, Huang N, Guiderdoni E, Ghesquier A (1996) Aroma in rice: genetic analysis of a quantitative trait. Theor Appl Genet 93:1145–1151
Lu SY, Jing YX, Shen SH, Zhao HY, Ma LQ, Zhou XJ, Ren Q, Li YF (2005) Antiporter gene from Hordum brevisubulatum (Trin.) Link and its overexpression in transgenic tobaccos. J Integr Plant Biol 47:343–349
Lutts S, Kinet JM, Bouharmont J (1995) Changes in plant response to NaCl during development of rice (Oryza sativa L.) varieties differing in salinity resistance. J Exp Bot 46:1843–1852
Mahalingam R, Gomez-Buitrago A, Eckardt N, Shah N, Guevara-Garcia A, Day P, Raina R, Fedoroff NV (2003) Characterizing the stress/defense transcriptome of Arabidopsis. Genome Biol 4:R20
Majee M, Maitra S, Dastidar KG, Pattnaik S, Chatterjee A, Hait NC, Das KP, Majumder AL (2004) A novel salt-tolerant l-myo-inositol-1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka, a halophytic wild rice: molecular cloning, bacterial overexpression, characterization, and functional introgression into tobacco-conferring salt tolerance phenotype. J Biol Chem 279:28539–28552
McCouch, SR, Kochert G, Yu ZH, Wang ZY, Khush GS, Coffiman WR, Tanksley SD (1988) Molecular mapping of rice chromosomes. Theor Appl Genet 76:815–829
McCouch S, Teytelman L, Xu Y, Lobos K, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, Zhang Q, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L (2002) Development of 2,240 new SSR markers for rice (Oryza sativa L.) DNA Res 9:199–207
Marin K, Kanesaki Y, Los DA, Murata N, Suzuki I, Hagemann M (2004) Gene expression profiling reflects physiological processes in salt acclimation of Synechocystis sp. strain PCC 6803. Plant Physiol 136:3290–3300
Minhas D, Grover A (1999) Transcript levels of genes encoding various glycolytic and fermentation enzymes change in response to abiotic stress. Plant Sci 146:41–51
Mishra B, Singh RK (2000) CSR27, a fine grain salt tolerant variety of rice released. Salinity News 11:3
Mittler R (2002) Oxidative stress, antioxidants, and stress tolerance. Trends Plant Sci 7:405–410
Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H (2003) Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15:1771–1780
Mohanty A, Kathuria H, Ferjani A, Sakamoto A, Mohanty P, Murata N, Tyagi AK (2002) Transgenics of an elite indica rice variety Pusa Basmati 1 harbouring the codA gene are highly tolerant to salt stress. Theor Appl Genet 106:51–57
Montero-Lomeli M, Morais BLB, Figueiredo DL, Neto DCS, Martins JRP, Masuda CA (2002) The initiation factor eIF4A is involved in the response to lithium stress in Saccharomyces cerevisiae. J Biol Chem 277:21542–21548
Moons A, Bauw G, Prinsen E, Van-Montagu M, Straeten VDD (1995) Molecular and physiological responses to abscisic acid and salt in roots in salt-sensitive and salt tolerant indica rice varieties. Plant Physiol 107:177–186
Munns R (2005) Genes and salt tolerance: bringing them together. New Phytol 167:645–663
Nakayama H, Yoshida, K, Ono H, Murooka Y, Shinmyo A (2000) Ectoine, the compatible solute of Halomonas elongata, confers hyperosmotic tolerance in cultured tobacco cells. Plant Physiol 122:1239–1247
Nandi S, Subudhi PK, Senadhira D, Maigbas NL, Sen-Mandi S, Huang N (1997) Mapping QTLs for submergence tolerance in rice by AFLP analysis and selective genotyping. Mol Gen Genet 255:1–8
Nanjo T, Kobayashi M, Yoshiba Y, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (1999) Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana.FEBS Lett 461:205–210
Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, Nakazono M, Hirai A, Kadowaki K (2002) The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Gen Genet 268:434–445
O’ Mahony PJ, Oliver MJ (1999) The involvement of ubiquitin in vegetative desiccation tolerance. Plant Mol Biol 41:657–667
Ohta M, Hayashi Y, Nakashima A, Hamada A, Tanaka A, Nakamura T, Hayakawa T (2002) Introduction of a Na+/H+ antiporter gene from Atriplex gmelini confers salt tolerance to rice. FEBS Lett 532:279–282
Oono Y, Seki M, Nanjo T, Narusaka M, Fujita M, Satoh R, Satou M, Sakurai T, Ishida J, Akiyama K, Iida K, Maruyama K, Satoh S, Yamaguchi-Shinozaki K, Shinozaki K (2003) Monitoring expression profiles of Arabidopsis gene expression during rehydration process after dehydration using a 7000 full-length cDNA microarray. Plant J 34:868–887
Pardo JM, Reddy MP, Yang S, Maggio A, Huh GH, Matsumoto T, Coca MA, Paino-D’Urzo M, Koiwa H, Yun DJ, Watad AA, Bressan RA, Hasegawa PM (1998) Stress signaling through Ca2+/calmodulin-dependent protein phosphatase calcineurin mediates salt adaptation in plants. Proc Natl Acad Sci USA 95:9681–9686
Pareek A, Singla SL, Grover A (1995) Immunological evidence for accumulation of two high-molecular-weight (104 and 90 kDa) HSPs in response to different stresses in rice and in response to high temperature stress in different plant genera. Plant Mol Biol 29:293–301
Pareek A, Singla SL, Grover A (1997) Salt responsive proteins/genes in crop plants. In: Jaiwal PK, Singh RP, Gulati A (eds) Strategies for improving salt tolerance in higher plants. Science Publishers, USA, pp 365–382
Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH (2001) Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13:1035–1046
Park S, Li J, Pittman JK, Berkowitz GA, Yang H, Undurraga S, Morris J, Hirschi KD, Gaxiola RA (2005) Up-regulation of a H+-pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants. Proc Natl Acad Sci USA 102:18830–18835
Parker R, Flowers TJ, Moore AL, Harpham NVJ (2006) An accurate and reproducible method for proteome profiling of the effects of salt stress in the rice leaf lamina. J Exp Bot 57:1109–1118
Prasad SR, Bagali PG, Hittalmani S, Shashidhar HE (2000a) Molecular mapping of quantitative trait loci associated with seedling tolerance to salt (Oryza sativa L.). Curr Sci 78:162–164
Prasad KVSK, Sharmila P, Kumar PA, Saradhi PP (2000b) Transformation of Brassica juncea (L.) Czern with bacterial codA gene enhances its tolerance to salt stress. Mol Breed 6:489–499
Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Monitoring expression profiles of rice genes under cold, drought, and high- salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133:1755–1767
Rausell A, Kanhonou R, Yenush L, Serrano R, Ros R (2003) The translation initiation factor eIF1A is an important determinant in the tolerance to NaCl stress in yeast and plants. Plant J 34:257–267
Reddy AR, Ramakrishna W, Chandra Shekhar A, Ithal N, Babu PR, Bonaldo FM, Soares B, Bennetzen JL (2002) Novel genes are enriched in normalized cDNA libraries from drought stressed seedlings of indica rice (Oryza sativa L. subsp. indica cv. Nagina 22). Genome 45:204–211
Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37:1141–1146
Revenkova E, Masson J, Koncz C, Afsar K, Jakovleva L, Paszkowski J (1999) Involvement of Arabidopsis thaliana ribosomal protein S27 in mRNA degradation triggered by genotoxic stress. EMBO J 18:490–499
Roxas VP, Smith RK, Allen ER, Allen RD (1997) Overexpression of glutathione S-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nat Biotechnol 15:988–991
Roy M, Jain RK, Rohila JS, Wu R (2000) Production of agronomically superior transgenic rice plants using Agrobacterium transformation methods: present status and future perspectives. Curr Sci 79:954–960
Sahi C, Agarwal M, Reddy MK, Sopory SK, Grover A (2003) Isolation and expression analysis of salt stress-associated ESTs from contrasting rice cultivars using a PCR-based subtraction method. Theor Appl Genet 106:620–628
Sahi C, Singh A, Blumwald E, Grover A (2006) Beyond osmolytes and transporters: novel plant salt stress tolerance-related genes from transcriptional profiling data. Physiol Plant 127:1–9
Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Over-expression of a single Ca2+dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23:319–327
Sakamoto A, Alia, Murata N, Murata A (1998) Metabolic engineering of rice leading to biosynthesis of glycinebetaine and tolerance to salt and cold. Plant Mol Biol 38:1011–1019
Sallaud C, Gay C, Larmande P, Bes M, Piffanelli P, Piegu B, Droc G, Regad F, Bourgeois E, Meynard D, Perin C, Sabau X, Ghesquiere A, Glaszmann JC, Delseny M, Guiderdoni E (2004) High throughput T-DNA insertion mutagenesis in rice: a first step towards in silico reverse genetics. Plant J 39:450–464
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, New York
Sanan-Mishra N, Pham XH, Sopory SK, Tuteja N (2005) Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc Natl Acad Sci USA 102:509–514
Santerre A, Britt AB (1994) Cloning of a 3-methyladenine-DNA glycosylase from Arabidopsis thaliana. Proc Natl Acad Sci USA 91:2240–2244
Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninici P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray.Plant Cell 13:61–72
Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nokajima M, Enju A, Sakurai T, Satou M, Akiyama K, Taji T, Yamaguchi-Shinozaki K, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31:279–292
Shi H, Lee BH, Wu SJ, Zhu JK (2003) Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol 21:81–85
Shimamoto K (1999) Molecular biology of rice, Springer-Verlag, Tokyo
Shiozaki N, Yamada M, Yoshiba Y (2005) Analysis of salt-stress-inducible ESTs isolated by PCR-subtraction in salt-tolerant rice. Theor Appl Genet 110:1177–1186
Singla-Pareek SL, Reddy MK, Sopory SK (2003) Genetic engineering of the glyoxylase pathway in tobacco leads to enhanced salinity tolerance. Proc Natl Acad Sci USA 100:14672–14677
Sivakumar P, Sharmila P, Saradhi PP (1998) Proline suppresses Rubisco activity in higher plants. Biochem Biophys Res Commun 252:428–432
Sottosanto JB, Gelli A, Blumwald E (2004) DNA array analyses of Arabidopsis thaliana lacking a vacuolar Na/H antiporter: impact of AtNHX1 on gene expression. Plant J 40:752–771
Sun W, Bernard C, van de Cotte B, Van Montagu M, Verbruggen N (2001) At-HSP17.6A, encoding a small heat-shock protein in Arabidopsis can enhance osmotolerance upon overexpression. Plant J 27:407–415
Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, Narusaka Y, Narusaka M, Zhu JK, Shinozaki K (2004) Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol 135:1697–1709
Takeda S, Sugimoto K, Kakutani T, Hirochika H (2001) Linear DNA intermediates of the Tto1 retrotransposon in Gag particles accumulated in stressed tobacco and Arabidopsis thaliana. Plant J 28:307–317
Takehisa H, Shimodate T, Fukuta Y, Ueda T, Yano M, Yamayad T, Kameya T, Sato T (2004) Identification of quantitative trait loci for plant growth of rice in paddy field flooded with salt water. Field Crops Res 89:85–95
Tanaka Y, Hibin T, Hayashi Y, Tanaka A, Kishitani S. Takabe T, Yokota S, Takabe T (1999) Salt tolerance of transgenic rice overexpressing yeast mitochondrial Mn-SOD in chloroplasts. Plant Sci 148:131–138
Tarczynski MC, Jensen RG, Bohnert H (1993) Stress protection of transgenic tobacco by production of the osmolyte mannitol. Science 259:508–510
Tsunematsu H, Yoshimura A, Horushima Y, Nagamura Y, Kurata N, Yano M, Sasaki T, Iwata N (1996) RFLP framework map using recombinant inbred lines in rice. Breed Sci 46:279–284
Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki B, Hirayama T, Shinozaki K (1999) A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11:1743–1754
Veena J, Reddy VS, Sopory SK (1999) Glyoxalase I from Brassica juncea: molecular cloning, regulation and its over-expression confer tolerance in transgenic tobacco under stress. Plant J 17:385–395
Vernon DM, Tarczynski, MC, Jensen RG, Bohnert HJ (1993) Cyclitol production in transgenic tobacco. Plant J 4:199–205
Vierstra RD, Callis J (1999) Polypeptide tags, ubiquitous modifiers for plant protein regulation. Plant Mol Biol 41:435–442
Vinocur B, Altman A (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol 16:123–132
Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Zeng L, Wanamaker SI, Mandal J, Xu J, Cui X, Close TJ (2005) Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol 139:822–835
Walia H, Wilson C, Wahi A, Condamine P, Cui X, Close TJ (2006) Expression analysis of barley (Hordeum vulgare L.) during salinity stress. Funct Integr Genomics 6:143–156
Wang GL, Mackill DJ, Bonman JM, McCouch SR, Champoux MC, Nelson RJ (1994) RFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivar. Genetics 136:1421–1434
Wang J, Zuo K, Wu W, Song J, Sun X, Lin J, Li X, Tang K (2004) Expression of a novel antiporter gene from Brassica napus resulted in enhanced salt tolerance in transgenic tobacco plants. Biol Plant 48:509–515
Wendler WM, Kremmer E, Forster R, Winnacker EL (1997) Identification of pirin, a novel highly conserved nuclear protein. J Biol Chem 272:8482–8489
Widawsky DA, O’Toole JC (1990) Prioritizing rice biotechnology research agenda for Eastern India. The Rockefeller Foundation, New York, USA
Wood AJ, Oliver MJ (1999) Translational control in plant stress: formation of messenger ribonucleoprotein complexes (mRNPs) in Tortula ruralis in response to desiccation.Plant J 18:359–370
Wood AJ, Duff RJ, Oliver MJ (2000) The translational apparatus of Tortula ruralis: polysomal retention of transcripts encoding the ribosomal proteins RPS14, RPS16 and RPL23 in desiccated and rehydrated gametophytes. J Exp Bot 51:1655–1662
Wool IG (1996) Extraribosomal functions of ribosomal proteins. Trends Biochem Sci 21:164–165
Wu CA, Yang GD, Meng QW, Zheng CC (2004) The cotton GhNHX1 gene encoding a novel putative tonoplast Na+/K+ antiporter plays an important role in salt stress. Plant Cell Physiol 45:600–607
Wu L, Fan Z, Guo L, Li Y, Chen ZL, Qu LJ (2005) Overexpression of the bacterial nhaA gene in rice enhances salt and drought tolerance. Plant Sci. 168:297–302
Xiao J, Li J, Yuan L, Tanksley SD (1996) Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor Appl Genet 92:230–244
Xu D, Duan X, Wang B, Hong B, Ho THD, Wu R (1996) Expression of a late embryogenesis abundant (LEA) protein gene, HVA1, from barley confers tolerance to drought and salinity in transgenic rice. Plant Physiol 110:249–257
Xue ZY, Zhi DY, Xue GP, Zhang H, Zhao YX, Xia GM (2004) Enhanced salt tolerance of transgenic wheat (Tritivum aestivum L.) expressing a vacuolar Na+/H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na+. Plant Sci 167:849–859
Yadav R, Courtois B, Huang N, McLaren G (1997) Mapping genes controlling root morphology and root distribution in a doubled-haploid population of rice. Theor Appl Genet 94:619–632
Yeo AR, Caporn SJM, Flowers TJ (1985) The effect of salinity upon photosynthesis in rice (Oryza sativa L.): gas exchange by individual leaves in relation to their salt content. J Exp Bot 36:1240–1248
Yilmaz JL, Bulow L (2002) Enhanced stress tolerance in Escherichia coli and Nicotiana tabacum expressing a betaine aldehyde dehydrogenase/choline dehydrogenase fusion protein. Biotechnol Prog 18:1176–1182
Yin XY, Yang AF, Zhang KW, Zhang JR (2004) Production and analysis of transgenic maize with improved salt tolerance by the introduction of AtNHX1gene. Acta Bot Sin 46:854–861
Zhang HX, Blumwald E (2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19:765–768
Zhang G-Y, Guo Y, Chen S-L, Chen S-Y (1995) RFLP tagging of a salt tolerant gene in rice. Plant Sci 110:227–224
Zhang HX, Hodson JN, Williams JP, Blumwald E (2001) Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc Natl Acad Sci USA 98:6896–6901
Zhao F, Guo S, Zhang H, Zhao Y (2006a) Expression of yeast SOD2 in transgenic rice results in increased salt tolerance. Plant Sci 170:216–224
Zhao F, Wang Z, Zhang Q, Zhao Y, Zhang H (2006b) Analysis of the physiological mechanism of salt-tolerant transgenic rice carrying a vacuolar Na+/H+ antiporter gene from Suaeda salsa. J Plant Res 119:95–104
Zhu B, Su J, Chang MC, Verma DPS, Fan YL, Wu R (1998) Overexpression of a pyrroline-5-carboxylate synthetase gene and analysis of tolerance to water and salt stress in transgenic rice. Plant Sci 139:41–48
Acknowledgements
AG is thankful to the Department of Biotechnology (DBT), Government of India and National Agriculture Technology Project (NATP), Indian Council of Agricultural Research (ICAR), Government of India, for the financial support. CS and AS acknowledge the Council of Scientific and Industrial Research (CSIR), New Delhi for the fellowship grants.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sahi, C., Singh, A., Kumar, K. et al. Salt stress response in rice: genetics, molecular biology, and comparative genomics. Funct Integr Genomics 6, 263–284 (2006). https://doi.org/10.1007/s10142-006-0032-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-006-0032-5