Abstract
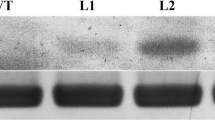
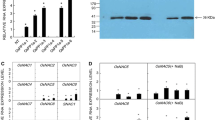
We have recently identified and classified a cystathionine β-synthase domain containing protein family in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa L.). Based on the microarray and MPSS data, we have suggested their involvement in stress tolerance. In this study, we have characterized a rice protein of unknown function, OsCBSX4. This gene was found to be upregulated under high salinity, heavy metal, and oxidative stresses at seedling stage. Transgenic tobacco plants overexpressing OsCBSX4 exhibited improved tolerance toward salinity, heavy metal, and oxidative stress. This enhanced stress tolerance in transgenic plants could directly be correlated with higher accumulation of OsCBSX4 protein. Transgenic plants could grow and set seeds under continuous presence of 150 mM NaCl. The total seed yield in WT plants was reduced by 80%, while in transgenic plants, it was reduced only by 15–17%. The transgenic plants accumulated less Na+, especially in seeds and maintained higher net photosynthesis rate and Fv/Fm than WT plants under NaCl stress. Transgenic seedlings also accumulated significantly less H2O2 as compared to WT under salinity, heavy metal, and oxidative stress. OsCBSX4 overexpressing transgenic plants exhibit higher abiotic stress tolerance than WT plants suggesting its role in abiotic stress tolerance in plants.







Similar content being viewed by others
References
Gollery, M., Harper, J., Cushman, J., Mittler, T., Girke, T., Zhu, J. K., et al. (2006). What makes species unique? The contribution of proteins with obscure features. Genome Biology, 7, R57.
Luhua, S., Ciftci-Yilmaz, S., Harper, J., Cushman, J., & Mittler, R. (2008). Enhanced tolerance to oxidative stress in transgenic Arabidopsis plants expressing proteins of unknown function. Plant Physiology, 148, 280–292.
Pawlowski, K. (2008). Uncharacterized/hypothetical proteins in biomedical ‘omics’ experiments: Is novelty being swept under the carpet? Briefings in Functional Genomics and Proteomics, 7, 283–290.
Kushwaha, H. R., Singh, A. K., Sopory, S. K., Singla-Pareek, S. L., & Pareek, A. (2009). Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genomics, 10, 200.
Bateman, A. (1997). The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends in Biochemical Sciences, 22, 12–13.
Ignoul, S., & Eggermont, J. (2005). CBS domains: Structure, function, and pathology in human proteins. American Journal of Physiology and Cell Physiology, 289, C1369–C1378.
Wang, X., Ren, X., Zhu, L., & He, G. (2004). OsBi1, a rice gene, encodes a novel protein with a CBS-like domain and its expression is induced in responses to herbivore feeding. Plant Science, 166, 1581–1588.
Gissot, L., Polge, C., Jossier, M., Girin, T., Bouly, J. P., Kreis, M., et al. (2006). AKINβγ contributes to SnRK1 heterotrimeric complexes and interacts with two proteins implicated in plant pathogen resistance through its KIS/GBD sequence. Plant Physiology, 142, 931–944.
Lv, Q., Tang, R., Liu, H., Gao, X., Li, Y., Zheng, H., et al. (2009). Cloning and molecular analyses of the Arabidopsis thaliana chloride channel gene family. Plant Science, 176, 650–661.
Espinoza, C., Medina, C., Somerville, S., & Arce-Johnson, P. (2007). Senescence-associated genes induced during compatible viral interactions with grapevine and Arabidopsis. Journal of Experimental Botany, 58, 3197–3212.
Fabro, G., Di Rienzo, J. A., Voigt, C. A., Savchenko, T., Dehesh, K., Somerville, S., et al. (2008). Genome-wide expression profiling Arabidopsis at the stage of Golovinomyces cichoracearum haustorium formation. Plant Physiology, 146, 1421–1439.
Zolla, L., Rinalducci, S., Antonioli, P., & Righetti, P. G. (2008). Proteomics as a complementary tool for identifying unintended side effects occurring in transgenic maize seeds as a result of genetic modifications. Journal of Proteome Research, 7, 1850–1861.
Liu, L., Zhou, Y., Zhou, G., Ye, R., Zhao, L., Li, X., et al. (2008). Identification of early senescence-associated genes in rice flag leaves. Plant Molecular Biology, 67, 37–55.
Kumari, S., Sabharwal, V. P., Kushwaha, H. R., Sopory, S. K., Singla-Pareek, S. L., & Pareek, A. (2009). Transcriptome map for seedling stage specific salinity stress response indicates a specific set of genes as candidate for saline tolerance in Oryza sativa L. Functional & Integrative Genomics, 9, 109–123.
Sahu, B. B., & Shaw, B. P. (2009). Isolation, identification and expression analysis of salt-induced genes in Suaeda maritima, a natural halophyte, using PCR-based suppression subtractive hybridization. BMC Plant Biology, 9, 69.
Wang, Q., Lai, T., Qin, G., & Tian, S. (2009). Response of jujube fruits to exogenous oxalic acid treatment based on proteomic analysis. Plant and Cell Physiology, 50, 230–242.
Sambrook, J., & Russell, D. W. (2001). Molecular cloning: A laboratory manual (3rd ed.). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Singla-Pareek, S. L., Reddy, M. K., & Sopory, S. K. (2003). Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proceedings of the National Academy of Sciences of United States of America, 100, 14672–14677.
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiology, 24, 1–15.
Deunff, E. L., Davoine, C., Dantec, C. L., Billard, J. P., & Huault, C. (2004). Oxidative burst and expression of germin/oxo genes during wounding of ryegrass leaf blades: Comparison with senescence of leaf sheaths. The Plant Journal, 38, 421–431.
Kumar, G., Purty, R. S., Sharma, M. P., Singla-Pareek, S. L., & Pareek, A. (2009). Physiological responses among Brassica species under salinity stress show strong correlation with transcript abundance for SOS pathway-related genes. Journal of Plant Physiology, 166, 507–520.
Yoshida, S., Forno, D. A., Cock, J. H., & Gomez, K. A. (1972). Laboratory manual for physiological studies of rice. Manila, Philippines: International Rice Research Institute.
Jain, M., Nijhawan, A., Tyagi, A. K., & Khurana, J. P. (2006). Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochemical and Biophysical Research Communications, 345, 646–651.
Pfaffl, M. W., Horgan, G. W., & Dempfle, L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Research, 30, e36.
Finn, R. D., Mistry, J., Tate, J., Coggill, P., Heger, A., Pollington, J. E., et al. (2010). The Pfam protein families database. Nucleic Acids Research, 38, D211–D222.
Collart, F. R., Osipiuk, J., Trent, J., Olsen, G. J., & Huberman, E. (1996). Cloning and characterization of the gene encoding IMP dehydrogenase from Arabidopsis thaliana. Gene, 174, 217–220.
Hechenberger, M., Schwappach, B., Fischer, W. N., Frommer, W. B., Jentsch, T. J., & Steinmeyer, K. (1996). A family of putative chloride channels from Arabidopsis and functional complementation of a yeast strain with a CLC gene disruption. The Journal of Biological Chemistry, 271, 33632–33638.
Diédhiou, C. J., & Golldack, D. (2006). Salt-dependent regulation of chloride channel transcripts in rice. Plant Science, 170, 793–800.
Majee, M., Maitra, S., Dastidar, K. G., Pattnaik, S., Chatterjee, A., et al. (2004). A novel salttolerant l-myo-inositol-1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka, a halophytic wild rice: Molecular cloning, bacterial overexpression, characterization, and functional introgression into tobacco-conferring salt tolerance phenotype. The Journal of Biological Chemistry, 279, 28539–28552.
Das-Chatterjee, A., Goswami, L., Maitra, S., Dastidar, K. G., Ray, S., et al. (2006). Introgression of a novel salt tolerant l-myo-inositol 1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka (PcINO1) confers salt tolerance to evolutionary diverse organisms. FEBS Letters, 580, 3980–3988.
Sahi, C., Agarwal, M., Reddy, M. K., Sopory, S. K., & Grover, A. (2003). Isolation and expression analysis of salt stress-associated ESTs from contrasting rice cultivars using a PCR-based subtraction method. Theoretical and Applied Genetics, 106, 620–628.
Walia, H., Wilson, C., Condamine, P., Liu, X., Ismail, A. M., Zeng, L., et al. (2005). Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiology, 139, 822–835.
Zhang, H. X., Hodson, J. N., Williams, J. P., & Blumwald, E. (2001). Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proceedings of the National Academy of Sciences of United States of America, 98, 12832–12836.
Santos, C. V. (2004). Regulation of chlorophyll biosynthesis and degradation by salt stress in sunflower leaves. Scientia Horticulturae, 103, 93–99.
Singla-Pareek, S. L., Yadav, S. K., Pareek, A., Reddy, M. K., & Sopory, S. K. (2008). Enhancing salt tolerance in a crop plant by overexpression of glyoxalase II. Transgenic Research, 17, 171–180.
Singla-Pareek, S. L., Yadav, S. K., Pareek, A., Reddy, M. K., & Sopory, S. K. (2006). Transgenic tobacco overexpressing glyoxalase pathway enzymes grow and set viable seeds in zinc-spiked soils. Plant Physiology, 140, 613–623.
Shen, B., Jensen, R. G., & Bohnert, H. J. (1997). Increased resistance to oxidative stress in transgenic plants by targeting mannitol biosynthesis to chloroplasts. Plant Physiology, 113, 1177–1183.
Lee, S.-H., Ahsan, N., Lee, K.-W., Kim, D.-H., Lee, D.-G., Kwak, S.-S., et al. (2007). Simultaneous overexpression of both CuZn superoxide dismutase and ascorbate peroxidase in transgenic tall fescue plants confers increased tolerance to a wide range of abiotic stresses. Journal of Plant Physiology, 164, 1626–1638.
Shi, H., Lee, B. H., Wu, S. J., & Zhu, J. K. (2003). Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nature Biotechnology, 21, 81–85.
Yang, L., Tang, R., Zhu, J., Liu, H., Mueller-Roeber, B., Xia, H., et al. (2008). Enhancement of stress tolerance in transgenic tobacco plants constitutively expressing AtIpk2beta, an inositol polyphosphate 6-/3-kinase from Arabidopsis thaliana. Plant Molecular Biology, 66, 329–343.
Srinivasan, T., Kumar, K. R. R., Meur, G., & Kirti, P. B. (2009). Heterologous expression of Arabidopsis NPR1 (AtNPR1) enhances oxidative stress tolerance in transgenic tobacco plants. Biotechnology Letters, 31, 1343–1351.
Pitzschke, A., & Hirt, H. (2006). Mitogen-activated protein kinases and reactive oxygen species signaling in plants. Plant Physiology, 141, 351–356.
Maathuis, F. J. M., & Amtmann, A. (1999). K+ nutrition and Na+ toxicity: The basis of cellular K+/Na+ ratios. Annals of Botany, 84, 123–133.
He, C., Yan, J., Shen, G., Fu, L., Holaday, A. S., Auld, D., et al. (2005). Expression of an Arabidopsis vacuolar sodium/proton antiporter gene in cotton improves photosynthetic performance under salt conditions and increases fiber yield in the field. Plant and Cell Physiology, 46, 1848–1854.
Rajagopal, D., Agarwal, P., Tyagi, W., Singla-Pareek, S. L., Reddy, M. K., & Sopory, S. K. (2007). Pennisetum glaucum Na+/H+ antiporter confers high level of salinity tolerance in transgenic Brassica juncea. Molecular Breeding, 19, 15–137.
Verma, D., Singla-Pareek, S. L., Rajagopal, D., Reddy, M. K., & Sopory, S. K. (2007). Functional validation of a novel isoform of Na+/H+ antiporter from Pennisetum glaucum for enhancing salinity tolerance in rice. Journal of Biosciences, 32, 621–628.
Obata, T., Kitamoto, H. K., Nakamura, A., Fukuda, A., & Tanaka, Y. (2007). Rice shaker potassium channel OsKAT1 confers tolerance to salinity stress on yeast and rice cells. Plant Physiology, 144, 1978–1985.
Brini, F., Hanin, M., Mezghani, I., Berkowitz, G. A., & Masmoudi, K. (2007). Overexpression of wheat Na+/H+ antiporter TNHX1 and H+-pyrophosphatase TVP1 improve salt- and drought-stress tolerance in Arabidopsis thaliana plants. Journal of Experimental Botany, 58, 301–308.
Garg, A. K., Kim, J. K., Owens, T. G., Ranwala, A. P., Choi, Y. D., Kochian, L. V., et al. (2002). Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proceedings of the National Academy of Sciences of United States of America, 99, 15898–15903.
Sanan-Mishra, N., Pham, X. H., Sopory, S. K., & Tuteja, N. (2005). Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proceedings of the National Academy of Sciences of United States of America, 102, 509–514.
Chaves, M. M., Flexas, J., & Pinheiro, C. (2009). Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Annals of Botany, 103, 551–560.
Uddin, M. I., Qi, Y., Yamada, S., Shibuya, I., Deng, X. P., Kwak, S. S., et al. (2008). Overexpression of a new rice vacuolar antiporter regulating protein OsARP improves salt tolerance in tobacco. Plant and Cell Physiology, 49, 880–890.
Bowler, C., Montagu, M. V., & Inze, D. (1992). Superoxide dismutase and stress tolerance. Annual Review of Plant Physiology and Plant Molecular Biology, 43, 83–116.
Thordal-Christensen, H., Zhang, Z., Wei, Y., & Collinge, D. B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. The Plant Journal, 11, 1187–1194.
Rivero, R. M., Kojima, M., Gepstein, A., Sakakibara, H., Mittler, R., Gepstein, S., et al. (2007). Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proceedings of the National Academy of Sciences of United States of America, 104, 19631–19636.
Tarczynski, M. C., Jensen, R. G., & Bohnert, H. J. (1993). Stress protection of transgenic tobacco by production of the osmolyte mannitol. Science, 259, 508–510.
Apse, M. P., Aharon, G. S., Snedden, W. A., & Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science, 285, 1256–1258.
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., & Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nature Biotechnology, 17, 287–291.
Fukuoka, S., Saka, N., Koga, H., Ono, K., Shimizu, T., Ebana, K., et al. (2009). Loss of function of a proline-containing protein confers durable disease resistance in rice. Science, 325, 998–1001.
Dobrota, C. (2006). Energy dependent plant stress acclimation. Reviews in Environmental Science and Biotechnology, 5, 243–251.
Scott, J. W., Hawley, S. A., Green, K. A., Anis, M., Stewart, G., Scullion, G. A., et al. (2004). CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. Journal of Clinical Investigations, 113, 274–284.
Biemans-Oldehinkel, E., Mahmood, N. A. B. N., & Poolman, B. (2006). A sensor for intracellular ionic strength. Proceedings of the National Academy of Sciences of United States of America, 103, 10624–10629.
Acknowledgments
Authors thank Mr. Ramesh Singh for assistance in the transgenic plant analysis. Financial support received from Department of Biotechnology and Department of Science and Technology, Government of India is also acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singh, A.K., Kumar, R., Pareek, A. et al. Overexpression of Rice CBS Domain Containing Protein Improves Salinity, Oxidative, and Heavy Metal Tolerance in Transgenic Tobacco. Mol Biotechnol 52, 205–216 (2012). https://doi.org/10.1007/s12033-011-9487-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-011-9487-2