Abstract
Main conclusion
Using late blight resistance genes targeting conservative effectors of Phytophthora infestans and the constructing gene pyramids may lead to durable, broad-spectrum resistance, which could be accelerated through genetic engineering.
Abstract
Potato (Solanum tuberosum L.) is one of the most important food crops worldwide. In 2020, potato production was estimated to be more than 359 million tons according to the Food and Agriculture Organization (FAO). Potato is affected by many pathogens, among which Phytophthora infestans, causing late blight, is of the most economic importance. Crop protection against late blight requires intensive use of fungicides, which has an impact on the environment and humans. Therefore, new potato cultivars have been bred using resistance genes against P. infestans (Rpi genes) that originate from wild relatives of potato. Such programmes were initiated 100 years ago, but the process is complex and long. The development of genetic engineering techniques has enabled the direct transfer of resistance genes from potato wild species to cultivars and easier pyramiding of multiple Rpi genes, which potentially increases the durability and spectrum of potato resistance to rapidly evolving P. infestans strains. In this review, we summarize the current knowledge concerning Rpi genes. We also discuss the use of Rpi genes in breeding as well as their detection in existing potato cultivars. Last, we review new sources of Rpi genes and new methods used to identify them and discuss interactions between P. infestans and host.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Potato (Solanum tuberosum L.) plants are cultivated worldwide; the largest areas can be found in Asia and Europe and potato production is systematically increasing in Africa (Haverkort and Struik 2015). Late blight is the most economically important potato disease. Costs associated with crop loss and chemical control of late blight were estimated to be more than € 9 billion per year (Haverkort et al. 2016). Late blight is caused by Phytophthora infestans (Mont.) de Bary, an oomycete within the kingdom Stramenopiles, which also infects tomato (Solanum lycopersicum L.) plants. This pathogen can infect stems, berries, leaves and tubers, which leads to complete crop loss. In the nineteenth century, P. infestans caused severe destruction of potato crops in Europe, especially in Ireland, where potatoes were the staple food (Kamoun et al. 2015). Intensive research on potato late blight has led to the discovery of dominant resistance genes against P. infestans (Rpi genes) in potato wild species. Research was initiated to introduce the Rpi genes from Solanum demissum into potato cultivars (Black et al. 1953; Malcolmson and Black 1966). Potato cultivars carrying resistance genes derived from S. demissum, including Pentland Ace (R3), Pentland Dell (R1, R2 and R3) and Epoka (R4), have been registered and cultivated on a large scale in Europe (Malcolmson 1969; Rudkiewicz 1985). However, Rpi genes introduced from S. demissum were quickly overcome by new virulent P. infestans strains (Jo et al. 2014). Rpi gene introgression from wild relatives of the potato into commercial cultivars through crossing is time-consuming, especially in the case of species separated from potato with crossing barriers such as different endosperm balance numbers (EBNs). For example, the introgression of a single Rpi gene (Rpi-blb2) from the wild species Solanum bulbocastanum to potato cultivars Bionica and Toluca necessitated more than 45 years (Haverkort et al. 2016). Compared with conventional breeding, genetic engineering techniques such as cisgenesis facilitate a faster introduction of Rpi genes into commercial cultivars (Ghislain et al. 2019). However, to avoid a rapid overcoming of resistance in newly engineered cultivars, introducing not one but several Rpi genes at a time has been proposed (Haverkort et al. 2016).
In this review, we summarize the knowledge concerning Rpi genes. We discuss the use of Rpi genes in traditional and genetic modification-based breeding as well as their detection in existing potato cultivars. We present new sources of Rpi genes and new methods used to identify them and discuss the interactions between P. infestans and the host.
Sources of Rpi genes
Hawkes’s taxonomy originally distinguished 232 potato wild species (Hawkes 1990). However, recent morphological and molecular studies have reduced the number of potato wild species to 107 (Spooner et al. 2016). These wild species grow in America from the southwestern United States to central Argentina and Chile (Hijmans and Spooner 2001). The highest number of species (93) occurs in Peru, 43 of which can be described as rare. Another country where species richness is particularly high is Mexico, which has 36 potato wild species (Hijmans and Spooner 2001).
Wild relatives of the potato are unique sources of genetic variation. They are characterized as being highly resistant to various diseases, including late blight, and they have been used in breeding programmes for more than 100 years (Machida-Hirano 2015). To date, more than 70 Rpi genes have been identified and mapped in 32 Solanum species (Table 1). Most of the Rpi genes have been derived from tuber-bearing species (25): Mexican (9 species), Bolivian (6), Peruvian (4), Argentine (3), Paraguayan (1), USA (1) and one species found generally in the Andes. Novel Rpi genes were found also in S. tuberosum subspecies andigena and in Hungarian cultivar Sárpo Mira. Six Rpi genes were identified in four non-tuber-bearing species and five from the tomato wild species S. pimpinellifolium. Single resistance genes were identified in 15 potato wild species. Frequently, multiple functional Rpi genes have been found within a single species, e.g., S. demissum (14 Rpi genes), S. bulbocastanum (5), S. berthaultii (5), S. stoloniferum (4), S. edinense (4), S. venturii (4), S. hjertingii (3), S. chacoense (3), S. huancabambense (2), S. pinnatisectum (2) S. schenckii (2) and S. tarijense (2). The Rpi genes were mapped in clusters onto potato chromosomes I, IV, V, VI, VII, VIII, IX, X, and XI. For example, on chromosome IV, a total of 13 Rpi genes from seven potato wild species were found. Several Rpi genes have not yet been mapped, including the following: Rpi-pta2 from S. stoloniferum; R4BI and R4MA from S. demissum; Rpi-ber1.2, Rpi-ber1.3, and Rpi-ber1.4 from S. berthaultii; Rpi-tar1.3 from S. tarijense, Rpi-nrs1 from S. neorossii and putative novel Rpi genes from S. jamesii and S. tuberosum subsp. andigena (Table 1).
Recently, using advanced techniques, new Rpi genes have been identified. Through genetic linkage analysis and collinearity analysis, a new dominant resistance gene, Rpi2, from the Mexican diploid wild species S. pinnatisectum was mapped onto potato chromosome VII (Yang et al. 2017). The Rpi2 locus is different from the previously reported resistance locus Rpi1, which is on the same chromosome. Rpi2 provides broad-spectrum resistance against various P. infestans isolates, including those that overcome resistance conferred by R9. Resistance gene enrichment sequencing (RenSeq) was used to finely map onto chromosome X, the Rpi-rzc1 gene from S. ruiz-ceballosii, which confers high and broad-spectrum resistance to 500 diverse Polish P. infestans isolates, (Jupe et al. 2013; Brylińska et al. 2015). Two complementary enrichment strategies that target resistance genes (RenSeq) and single/low-copy number genes (GenSeq, generic-mapping enrichment sequencing) independently positioned the broad-spectrum resistance gene Rpi-ver1 from the Mexican wild species S. verrucosum on potato chromosome IX (Chen et al. 2018). Diploid wild potato, S. jamesii (JAMI-4) is completely resistant to the super virulent P. infestans isolate 2013–18-306, which can overcome the resistance conferred by the genes R1, R2, R3a, R3b, R4, R5, R6, R7, R8, R9, R10, and R11 (Zheng et al. 2020). Diagnostic RenSeq (dRenSeq) analysis demonstrated JAM1-4 harbors R3a. However, transgenic Désirée plants containing R3a are susceptible to the isolate 2013–18-306. The authors speculated that resistance in JAM1-4 was provided by uncharacterized novel resistance gene(s). Rpi-amr3 from S. americanum was identified and cloned via RenSeq and single-molecule real-time (SMRT) sequencing (SMRT RenSeq) (Witek et al. 2016). Bulked segregant analysis coupled with RenSeq mapped Rpi-amr3 on chromosome IV of the potato reference genome of a doubled monoploid clone of S. tuberosum group Phureja DM1–3 516 R44 (DM). Transgenic diploid potato carrying Rpi-amr3 showed resistance against three P. infestans isolates. Another new Rpi gene from S. americanum Rpi-amr1, was positionally cloned and mapped onto the short arm of chromosome XI (Witek et al. 2021). Using association genomics and long-read RenSeq, the authors identified three allele-specific proteins, which showed 100% identity to Rpi-amr1 protein, from two S. americanum assessions and one S. nodiflorum assession. Eight additional Rpi-amr1 allele-specific proteins, sharing 90% homology to Rpi-amr1 proteins, were identified from six accessions of S. americanum and two accessions of S. nigrescens and they all conferred late blight resistance to P. infestans isolate 88069 in transient assays. One homologue, Rpi-amr1-3409 from S. nigrescens, was mapped onto chromosome I based on the potato DM reference genome, suggesting that a fragment of DNA from the end of the short arm of chromosome XI in other resistant accessions was translocated to the end of the long arm of chromosome I in S. nigrescens. Moreover, the authors identified Rpi-amr1 homologues in hexaploid S. nigrum accessions, providing resistance to the P. infestans isolate 88069. Previous studies indicated that S. nigrum is a non-host to P. infestans and S. americanum may be the diploid ancestor of hexaploid S. nigrum (Colon et al. 1992; Poczai and Hyvönen 2010). The Rpi-amr1 homologues, which confer late blight resistance in S. nigrum, were most likely inherited from S. americanum (Witek et al. 2021). Rpi-amr1 confers broad-spectrum late blight resistance in cultivated potato. Stably transformed transgenic potato cultivar Maris Piper plants carrying Rpi-amr1, resist 19 P. infestans isolates tested, including those overcoming Rpi-vnt1, Rpi-blb1 and Rpi-blb2. In potato wild species S. chacoense, two resistance genes, Rpi-chc1.1 and Rpi-chc1.2 have been identified (Monino-Lopez et al. 2021). An allele-mining strategy allowed the identification of Rpi-chc1.1 orthologue in S. chacoense, S. berthaultii and S. tarijense accessions resistant to late blight. For many years, researchers have continued to search for new Rpi genes among wild potato. The largest collections of potato germplasm are available in International Potato Center (CIP) in Peru, the USDA Potato Genebank in Wisconsin, USA, and IPK Gatersleben Genebank in Germany (Karki et al. 2021b). An analysis of resistance to P. infestans carried out over a period of more than 20 years has shown that among 34 potato wild relatives there are accessions characterized by a high level of resistance, but the genes underlying this resistance are still unknown (Pérez et al. 1999; Zoteyeva et al. 2012; Khiutti et al. 2015; Bachmann-Pfabe et al. 2019; Zoteyeva 2020; Karki et al. 2021b). A list of such potato wild relatives is shown in Table 2. These species are native to Mexico, Argentina, Bolivia, Peru, Ecuador and Chile. Research using aggressive P. infestans isolates, showed that these species can contribute to development of new durable resistant cultivars. It is worth noting that species having 2EBN and 4EBN (Table 2) can cross with cultivated potato and can be used in potato breeding programs (Karki et al. 2021b). On the other hand, species with 1EBN cannot be crossed with cultivated potato and require application of other methods for the Rpi gene introgression. Recently, 189 potato genotypes, from 20 wild species and cultivated Solanum tuberosum from Andigenum and Chilotanum groups, were screened for their resistance against P. infestans (Duan et al. 2021). Ten genotypes from five wild species originating in Mexico showed a broad-spectrum resistance to all four P. infestans used, suggesting that each of these genotypes contains Rpi gene(s) other than R1-R11. They belong to S. bulbocastanum (3 genotypes), S. cardiophyllum (4), S. jamesii (1), S. brachycarpum (1) and S. trifidum (1). The other 127 genotypes displayed isolate-specific resistance.
Structure of Rpi genes and their distribution in the potato genome
Most of the plant resistance (R) genes are members of a large gene family that encodes nucleotide-binding site and leucine-rich repeat (NB-LRR; NLR) domain-containing proteins (Lozano et al. 2015). On the basis of the structure of NLR proteins, two main groups can be distinguished. The first is the so-called TIR-NB-LRRs (TNLs) with N-terminal domain homologous to the Drosophila Toll domain and human interleukin-1 receptor. The second group is non-TIR-NB-LRRs known as CNLs, which contains coiled coil (CC) structure or leucine zipper (LZ) motif in N-terminal region (Ballvora et al. 2002; Sekhwal et al. 2015).
Genome sequencing revealed that the diploid potato clone RH89-039-15 (S. tuberosum ssp. tuberosum) contains 738 partial or full-length NLR sequences (Bakker et al. 2011). In the potato reference genome DM, 438 out of 40,000 identified genes contain the characteristic NB-LRR domain (Jupe et al. 2012). The use of RenSeq led to an increase in the number of identified NLR in the DM reference genome from 438 to 755 (Jupe et al. 2013). All twelve potato chromosomes contain genes belonging to the CNL and TNL groups, except for chromosomes III and X, on which genes from the TNL group are not found. The majority of NLR genes were found on chromosomes IV (57) and XI (54). The fewest number of NLR genes (3) was found on chromosome III. Moreover, the greatest number of NLR gene clusters is on chromosome IV. There are 4.7 times more CNL genes than TNL genes in the analyzed potato genome (Jupe et al. 2012). Recently, using Illumina HiSeq 2000 technology, 585 NBS domains, including 11 not previously described, were analyzed in 96 potato genomes (Prakash et al. 2020).
To date, nearly 50 Rpi genes that are from Solanum species have been cloned. Most of the cloned genes belong to the CNL family, but several NLR genes remain unclassified (Table 3). The size and structure of different Rpi genes, as well as of different alleles of the same Rpi gene, are diverse. Examples of the longest Rpi genes include Rpi-amr1-2307 (7277 bp) from S. americanum and Rpi-blb2 from S. bulbocastanum (4858 bp) belonging to the CC-NB-LRR class, and R1 from S. demissum (4102 bp) which is part of LZ-NB-LRR group. The shortest genes include R2 family members, e.g., R2 from S. demissum (2538 bp), Rpi-edn1.1 from S. edinense (2544 bp), Rpi-hjt1.1, Rpi-hjt1.2 and Rpi-hjt1.3 from S. hjertingii (2544 bp). These genes are located on chromosome IV and are members of the LZ-NB-LRR group. Most of the Rpi genes are intron-free. Variation in the size and structure of Rpi gene alleles is well described for Rpi-amr1. The size of the identified alleles of this gene ranged from 2768 to 7277 bp and the number of introns range from one to four (Table 3). The listed homologues of the Rpi-amr1 gene have been identified in different species and in different accessions. Molecular cloning of the Rpi genes facilitates studies at the molecular level of the control of resistance to potato late blight (Ballvora et al. 2002). The cloned genes can be used in genetic engineering to develop late blight resistant cultivars.
Plant R genes encode proteins that directly or indirectly detect effector proteins introduced by pathogens (Sekhwal et al. 2015). This leads to the activation of effector-triggered immunity (ETI) and results in reactive oxygen species (ROS) production, callose deposition, and programmed cell death through the hypersensitive response (HR) (Turnbull et al. 2019). The mechanism of potato Rpi gene activation by P. infestans effectors is poorly understood. Studies conducted on Arabidopsis thaliana show that the conformation of NLR proteins may influence their function. The A. thaliana NLR protein Peronospora parasitica 1 protein (RPP1), which recognizes the ATR1 effector from Peronospora parasitica, remains in an inactive form in the absence of an effector in the cell environment. Binding of the effector to the LRR domain leads to oligomerization and activation of the RPP1 protein (Schreiber et al. 2016). Another A. thaliana NLR protein, ZAR1, in an inactive form, forms a multicomponent complex with resistance-related kinase 1 (RKS1) (Wang et al. 2019). Inactive ZAR1-RKS1 complex is activated by the effector AvrAC from Xanthomonas campestris. AvrAC uridylates the PBL2 kinase to produce PBL2UMP. Interactions between PBL2UMP and the inactive ZAR1-RKS1 complex then lead to conformational changes and the formation of the active pentameric ZAR1 resistosome (ZAR1-RKS1-PBL2UMP). The active resistosome has a funnel-shaped structure required for AvrAC-induced ZAR1 plasma membrane association, cell death, and resistance to X. campestris. Nonetheless, how widespread these models are and whether the mechanism of action is the same in potato are unknown (Wang et al. 2019). Activation of Rpi genes may also depend on external factors. Rpi-vnt1.1 requires light to confer resistance against P. infestans. In the dark, plants produce shortened chloroplast protein glycerate 3-kinase (GLYK), which does not bind Avrvnt1 and that results in a lack of activation of the Rpi-vnt1.1 protein (Gao et al. 2020).
Arms race
The host immune response against pathogen invasion can be summarized by the zig-zag model, which involves two steps. The first step relates to the detection of the conserved pathogen-associated molecular pattern (PAMP) which triggers PAMP-triggered immunity (PTI). To avoid PTI, the pathogen secretes effector proteins into the host cells to disrupt the immune response. Effectors are detected by the host's NLRs, leading to activate more robust and faster response termed ETI, which represents the second level of activation the host immune response. Interactions between R genes and effectors represent host–pathogen molecular co-evolution when effectors evolve to evade detection and R proteins evolve to establish or retain detection (Hein et al. 2009; Naveed et al. 2020).
During the pathogenesis of P. infestans, a key step is the formation of haustorium in potato tissue through which the pathogen secretes effectors. These proteins manipulate and alter the host's immune response to promote infection. Genes encoding pathogen effectors that induce R gene response are defined as avirulence (Avr) genes (Qutob et al. 2006). Cytoplasmic effectors secreted by P. infestans can be divided into two classes, CRN (crinkling, necrosis) and RxLR effectors. The effectors of RxLR type possess arginine-any amino acid residue-leucine–arginine motifs in N-terminal region. All known P. infestans effectors, which are recognized by the products of corresponding potato Rpi genes, belong to the RxLR class (Martynov and Chizhik 2020). The RxLR effectors contain the highly conserved N-terminal RxLR motif involved in the translocation of P. infestans effector proteins into plant cells, and the heterogeneous C-terminal region that can be recognized by plant R gene products (Dou et al. 2008).
The function of the P. infestans effector in the infection process has been defined for only a few examples. Avr2, by interacting with members of the BRI1-suppressor 1-like family proteins (BSL1, BSL2 and BSL3) from potato, inhibits the activity of the oomycete infestin 1 (INF1); as a result, programmed cell death does not occur (Turnbull et al. 2019). Avr3 inactivates the host ubiquitin E3 ligase CMPG1, leading to programmed cell death inhibition (Bos et al. 2010). The effector Avr3a-like, by stabilizing host cinnamyl alcohol dehydrogenase 7 (CAD7), limits the activation of defense mechanisms, such as callose deposition, the ROS burst and WRKY33 expression (Li et al. 2019).
In the genome of P. infestans, 563 effector genes with the RxLR motif have been identified (Haas et al. 2009). For 15 of them, the respective potato Rpi genes have been identified (Table 4). According to the gene-for-gene concept, specific pathogen effectors activate corresponding host plant R proteins (Flor 1971). However, some effectors can be recognized by products of multiple Rpi genes, or vice versa, a specific Rpi gene product can detect multiple effectors from the same or different Phytophthora pathogens. The Avr2 effector can be recognized by not only R2 protein from S. demissum but also by Rpi-blb3 from S. bulbocastanum, Rpi-mcq1 from S. mochiquense, Rpi-hcb1.1 and Rpi-hcb1.2 from S. huancabambense (Aguilera-Galvez et al. 2018, 2020). The Rpi genes encoding these proteins are located on different chromosomes, R2 and Rpi-blb3 on chromosome IV, Rpi-mcq1, Rpi-hcb1.1 and Rpi-hcb1.2 on chromosome IX (Table 1). Rpi-amr1 from S. americanum recognizes the effector Avramr1 from P. infestans but also the Avramr1 homologues from Phytophthora parasitica and Phytophthora cactorum (Witek et al. 2021). Likewise, recognition of Avramr3 by Rpi-amr3 from S. americanum activates resistance against not only P. infestans but also against other economically important Phytophthora pathogens, including P. parasitica and Phytophthora palmivora (Lin et al. 2021). The authors suggest that Rpi-amr1 and Rpi-amr3 provide non-host type resistance to multiple Phytophthora pathogens in S. americanum (Lin et al. 2021; Witek et al. 2021). In some cases, different alleles from the same Rpi gene can recognize different effectors which belong to the same RxLR family (Monino-Lopez et al. 2021). Protein products of Rpi-chc1.1 and Rpi-chc1.2 which are allelic variants recognize two different effectors within the same effector class Avrchc1.1 and Avrchc1.2, respectively. The LRR domain is involved in the recognition of cognate effector and changes in its structure may lead to the loss of the ability to recognize the effector or to shift the recognition ability from one effector to another. The exchange of the LRR domain in the chimeric receptors changed the recognition spectrum of the Avrchc1.1 to Avrchc1.2 (Monino-Lopez et al. 2021).
To date, several mechanisms have been identified that allow effectors to avoid recognition by corresponding host R proteins. The products of such unrecognized alleles act as virulence factors. P. infestans virulence can arise in multiple ways (Huang et al. 2019). The simplest one involves point mutations, e.g., the P. infestans effector Avr3a. In tested P. infestans populations, two alleles of Avr3a differing at the protein level by only two amino acids can be distinguished. Avr3aKI activates the potato resistance gene R3a, leading to a HR, while Avr3aEM is not recognized by the product of the R3a gene, leading to infection (Armstrong et al. 2005). Isolates with truncated Avr4 protein resulting from a frameshift generated by two single deletions are not detected by the product of the R4 gene in potato. Functional analysis shows that two single deletions do not affect the elicitor activity of the Avr4 protein (Van Poppel et al. 2008). P. infestans isolates that possess the Avr1 homologue Avr1-like (A-L) do not induce a resistance response in R1 potato. The sequence of the Avr1-like (A-L) effector is in 82% identical to that of the Avr1 protein but is truncated by the T region at the C-terminal end of Avr1 (Du et al. 2018). Due to changes in expression regulation, the Avrvnt1 effector is not detected by Rpi-vnt1 plants (Stefańczyk et al. 2017; Pais et al. 2018). Expression of Avrchc1.2 effector is rapidly downregulated in the first hours after inoculation with P. infestans isolates, which explains why the presence of Rpi-chc1.2 in the potato plants does not provide resistance to late blight (Monino-Lopez et al. 2021). The Rpi-blb1 protein recognizes ipiO (Avrblb1) effectors from classes I and II, resulting in a HR. Effectors from class III, i.e., ipiO4, are not recognized by the Rpi-blb1 protein and inhibit HR caused by classes I and II of the effector (Champouret 2010). Potentially, the effectors that are essential for infection could not be mutated without a fitness cost and loss of pathogenicity. R proteins recognizing such essential effectors would likely provide broad-spectrum and durable resistance. P. infestans Avr3a, especially virulent allele Avr3aEM, may be an example of an essential effector since this gene is conserved among diverse P. infestans strains and is highly expressed at the early stage of infection (Yin et al. 2017). Further searching and functional characterization of conserved effectors and corresponding Rpi genes may inform strategies for obtaining durable late blight resistance.
The adaptation of potato to the continuous evolution of the pathogen is through the diversification of R genes by recombination, gene conversion, duplication and/or selection (Jupe et al. 2012). While some of the S. demissum Rpi genes were found to be race-specific and rapidly became ineffective, the following genes have been described as providing a broad-spectrum of resistance against P. infestans: Rpi-blb1, Rpi-blb2 and Rpi-blb3 from S. bulbocastanum; R8 and R9 from S. demissum and Rpi-vnt1.1 from S. venturii (Vleeshouwers et al. 2011; Vossen et al. 2016). However, these genes have not yet been widely introduced into potato cultivars, in part because of crossing barriers. This continuous co-evolution of pathogen effectors and plant R genes represents a so-called arms race between plants and pathogens (Khavkin 2015).
Rpi genes in potato cultivars and breeding lines
Breeding potato cultivars with resistance genes against P. infestans gives opportunities to limit the use of fungicides (Haverkort et al. 2016). However, breeders often do not know what R genes are present in existing potato cultivars and which of them are effective against local P. infestans populations. Some potato cultivars show moderate or high levels of resistance to late blight, but the basis of their resistance remains unknown. Various methods, including PCR, effectoromics, transcriptomics, single nucleotide polymorphism (SNP) array genotyping or dRenSeq, have been used to determine which Rpi genes are present in potato cultivars (Table 5). Frequently, a combination of more than one method was used.
Analysis of 600 potato cultivars from Europe, Asia, and South America by PCR using gene-specific primers, allowed to detect R1 in 135 potato genotypes (Gebhardt et al. 2004). Using gene-specific markers, it was possible to confirm the presence of the R1 and R2-like genes in the Polish cultivar Bzura showing a high level of field resistance (Plich et al. 2015). The Mastenbroek potato late blight differential set is a group of 11 potato genotypes (MaR1-MaR11) expected to contain 11 individual S. demissum Rpi genes (Mastenbroek 1952). However, studies using Rpi gene-specific markers and agroinfiltration assay showed that differential plants harbor more than one Rpi gene. MaR8 and MaR9 plants, which have a broad-spectrum resistance in both the field and the greenhouse, contain four (R3a, R3b, R4, R8) and seven (R1, Rpi-abpt, R3a, R3b, R4, R8, R9) Rpi genes, respectively (Kim et al. 2012; Zhu et al. 2015). R1 was additionally found in MaR5 and MaR6 genotypes. These findings are consistent with those of Trognitz and Trognitz (2007), who found R1 in the R5, R6 and R9 plants also within the Scottish Black’s differential set. The Rpi-vnt1 gene from S. venturii and Rpi-phu1 from S. phureja were mapped to the same region on the potato chromosome IX and the nucleotide sequences of both genes are identical (Śliwka et al. 2006; Foster et al. 2009). The Rpi-vnt1.1 confers resistance to a wide range of P. infestans strains, except isolates EC1 and EC3626 from Ecuador (Foster et al. 2009; Witek et al. 2021). The presence of the Rpi-vnt1.1 gene has been confirmed in Dutch cultivar Alouette and in Polish cultivar Gardena using the PCR marker phu1_2069 (Stefańczyk et al. 2020). The Rpi-vnt1.1 have been also found in six other cultivars (Table 5). It is worth underlining that PCR markers designed on the basis of gene sequence may display low specificity. Detection of the Rpi-vnt1 gene in late blight susceptible cultivars, including Bintje and Early Rose, is most likely due to use of non-specific markers and the detection of non-functional homologues (Rogozina et al. 2021). Detection of the presence Rpi genes in potato cultivar with the use of PCR markers requires additional methods confirming resistance to P. infestans, as it is prone to false-positive results.
Another strategy is the effectoromics approach, where effectors are functionally tested in potato germplasms for their response to cognate R gene using agroinfiltration assay (Domazakis et al. 2017). More than 200 predicted RxLR effectors selected from P. infestans genome sequence were used in an agroinfiltration test, which allowed to detect five effective Rpi genes (R3a, R3b, R4, Rpi-Smira1, Rpi-Smira2) in the Sárpo Mira cultivar characterized by a high level of field resistance (Rietman et al. 2012). Four of them were pyramided qualitative Rpi genes. The remaining one, Rpi-Smira2, provides a quantitative field resistance, and its presence in potato cultivar can only be detected in field tests. R8 with nucleotide sequence identical to that of Rpi-Smira2 can be found also in the resistant potato cultivars Jacqueline Lee, Missaukee, PB-06 and S-60, by long-range PCR (Vossen et al. 2016). Agroinfiltration with ten P. infestans effectors revealed that Avr4 and Avr8 effector induce HR in potato cultivar Qingshu9, Avrvnt1.1 induces HR in Longshu7, Avr3aEM effector (i.e., virulent allele of Avr3aKI) induces HR in cultivars Qingshu9 and Longshu7 (Elnahal et al. 2020). Screening with over 50 different P. infestans RxLR effectors has shown a specific response to Avr2, which confirms that SW93-1015 contains a functional homologue of the R2 gene. Out of R2 gene homologues cloned from SW93-1015, one encoded a protein identical to Rpi-abpt. Transgenic potato cultivar Désirée with this gene was resistant to P. infestans (Lenman et al. 2016).
Analysis of the transcriptome of the Chinese cultivar Cooperation 88 (C88), which has been characterized as displaying durable late blight resistant for 20 years, revealed the presence of multiple Rpi genes (Hao et al. 2018). This cultivar is highly resistant to two super virulent P. infestans strains, IPO 428–2 and XA-4. Within 5 days of inoculation with XA-4, a change in the expression of Rpi genes was noted. These genes can be classified as R1, R2, R3a, Rpi-blb1, Rpi-blb2 and Rpi-vnt1 homologues (Hao et al. 2018).
SNP array genotyping can also be used to detect the R genes in the cultivated potato (Karki et al. 2021a). F1 population containing 79 progeny clones derived from crossing Payette Russet with A0012–5 was screened for resistance to the US-23 genotype of P. infestans in detached leaf assay. Linkage mapping using markers from the potato SNP array confirmed the presence of a single resistant gene on the short arm of chromosome IV of cultivar Payette Russet, in the same locus as that for R2, Rpi-abpt, and Rpi-blb3. Using the primers for Rpi-blib3, a PCR product of the expected size (~ 2500 bp) was obtained and sequenced. The Rpi gene allele from Payette Russet is identical to the Rpi-abpt sequence except for a synonymous C to T substitution at position 87 (Karki et al. 2021a).
A new tool used for genetic mapping, searching, and testing the functionality of resistance genes in cultivars and breeding lines is dRenSeq (Armstrong et al. 2019). dRenSeq has been used to identify and validate all currently known NLRs effective against potato virus X, the potato cyst nematode Globodera pallida and P. infestans. Screening by dRenSeq for the presence of 22 functional Rpi genes in 11 potato cultivars and one late blight differential line 2573 led to the identification of one to seven Rpi genes in each tested genotype (Table 5). Single Rpi genes were found in cultivars Craigs Snow White, Spunta and Toluca. Seven Rpi genes, i.e., R1, R1ΔT4109, R3a, R3bG1696/G3111, R8, R9a, and Rpi-abptT86, have been identified in the differential line 2573 (Armstrong et al. 2019).
Genetic improvement for durable P. infestans resistance
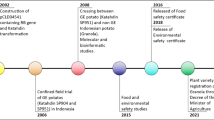
The introgression of Rpi genes into susceptible potato cultivars is limited by long breeding cycles and the high level of heterozygosity across the potato genome (Jo et al. 2014). An attempt to introgress the Rpi genes from S. bulbocastanum began in 1959. In the first step, a cross was made between S. bulbocastanum (B, 2x) bearing Rpi-blb2 and S. acaule (A, 4x) to obtain an AB (3x) plants, which after polyploidisation to the hexaploid level, was crossed with S. phureja (P, 2x) resulting in ABP (4x) material. Successive rounds of bridge crosses between ABP (4x) and S. tuberosum, led to produce ABPT plants which, after three backcrossing to S. tuberosum, eventually led to registration in 2006 and 2008 of two P. infestans resistant cultivars Toluca and Bionica (Haverkort et al. 2016). Another example of the introduction of Rpi genes into the potato gene pool is the 11-year-long project Bioimpuls, which resulted in the development of true seed population with single or multiple Rpi genes against late blight through classical breeding (Keijzer et al. 2021). In this project, three groups of sources of resistance to P. infestans were distinguished. The first group includes cultivars and advanced breeding clones containing the R8, Rpi-cap1, Rpi-chc1, Rpi-vnt1 and Rpi-blb2 genes that is ready for the commercial crossing. The second group includes breeding clones with R9 and Rpi-edn2 genes which require one or two rounds of backcrossing. The third group is potato wild species that have not been used so far, including S. brachycarpum, S. bukasovii, S. iopetalum, S. multiinterruptum, and S. sucrense. This group needs two or three additional rounds of backcrossing to be used for commercial crosses (Keijzer et al. 2021).
Different approaches have been developed to overcome crossing barriers and to shorten the time for introducing Rpi genes into susceptible cultivars, including the use of somatic hybrids, hybrid breeding and genetic engineering. To transfer late blight resistance from S. michoacanum to the gene pool of cultivated potato, somatic hybridization and backcrossing (BC) have been used (Smyda et al. 2013; Smyda-Dajmund et al. 2017). The genetic composition of the obtained somatic hybrids was analyzed using diversity array technology (DArT) (Smyda-Dajmund et al. 2016). Using somatic hybridization, crossing and backcrossing, four Rpi genes (Rpi-blb1, Rpi-blb3, R3a, R3b) were introduced into cultivated potato (Rakosy-Tican et al. 2020). The presence of these genes in the back-crossed progeny (BC1 and BC2) was confirmed via gene-specific markers. In addition, the functionality of Rpi-blb1, Rpi-blb3, R3a and R3b was confirmed via agroinfiltration with the corresponding Avr effectors (Avrblb1, Avr2, Avr3a, Avr3b).
Another method to facilitate transfer of resistance to late blight to potato cultivar is hybrid breeding. This method enables obtaining plants with single or pyramided Rpi genes without disrupting the genetic composition of the parental breeding lines that have good agronomic performance. Su et al. (2020) described the introgression of Rpi genes into homozygous diploid potato. First, four different Rpi genes, which were derived from S. avilesii, S. tarijense, S. venturii and S. chacoense, were introduced in three highly homozygous (e.g., with homozygosity scores as 88%, 88% and 79%) diploid potato breeding lines via marker-assisted introgression. After two backcrosses supported with marker selection and one selfing, parents with the homozygous resistance allele were produced and were used for crossing. The two backcrossing steps ensured the removal of most of the genome of the donor wild species parent, and the selfing step helped to remove any remaining unwanted introgressions. The hybrids were made by crossing two homologous parents, each having a different Rpi gene. Finally, hybrids with single Rpi gene (Rpi-avl1, Rpi-tar1, Rpi-vnt1.1, Rpi-chc1.1) and hybrids with combination of two Rpi genes (Rpi-avl1 and Rpi-chc1, Rpi-avl1 and Rpi-tar1, Rpi-avl1 and Rpi-vnt1.1, Rpi-tar1 and Rpi-vnt1.1, Rpi-vnt1.1 and Rpi-chc1) were obtained. The hybrids were tested for resistance to P. infestans in three separate field trials. The hybrids with two resistance genes were more resistant compared to the ones with the respective single Rpi gene. Hybrid breeding with the use of existing elite material and marker-assisted introgression allows obtaining resistant plants in a relatively short time (Su et al. 2020).
An alternative approach involves genetic engineering, which significantly shortens the long time to introgress resistance genes through breeding cycle for tetraploid potato plants (Van Esse et al. 2020). One such method is cisgenesis, i.e., the introduction of genetic material from the same species or from a crossable species (Hou et al. 2014). Transformed potato cultivars obtained by genetic engineering are shown in Table 6. Single Rpi genes have been introduced into several potato cultivars. Rpi-vnt1.1 or Rpi-sto1 have been introduced separately into the cultivars Atlantic, Bintje and Potae9 (Jo et al. 2014). The obtained transgenic plants were evaluated for late blight resistance in detached leaf assay and agroinfiltration assay using five P. infestans isolates. Transgenic Atlantic, Bintje and Potae9 with Rpi-sto1 were resistant to all tested isolate except pic99189. Transgenic Atlantic and Bintje with Rpi-vnt1.1 gene were resistant to all tested isolate except EC1 (Jo et al. 2014). Rpi-vnt1.1 and Rpi-mcq1 have been transformed separately into the cultivar Désirée and tested in field experiment (Jones et al. 2014). All transgenic plants with the Rpi-mcq1 gene were susceptible to late blight. Transformed Désirée plants with the Rpi-vnt1.1 gene remain fully resistant to P. infestans or have reduced disease severity compared to susceptible controls (Jones et al. 2014). In another study, whole-plant resistance assays were carried out in the confined biosafety greenhouse to evaluate the late blight resistance of the Désirée plants transformed with Rpi-vnt1 (Roman et al. 2017). Unexpectedly, 5 out of 52 transgenic events showed resistance to two Peruvian P. infestans isolates belonging to the EC-1 lineage. As reported previously, a different isolate of the EC-1 lineage (isolate EC1 from Ecuador, the only one tested from the lineage), in which the cognate effector gene Avrvnt1 was not expressed, was able to break the resistance conditioned by the Rpi-vnt1 gene in transformed Désirée plants based on a detached leaf assay (Foster et al. 2009; Pel et al. 2009. The authors inferred the EC-1 isolates used in Peru may differ in virulence within the EC-1 lineage (Roman et al. 2017).
The resistance provided by a single Rpi gene can be quickly overcome by an adapted P. infestans strain. A promising breeding approach involves pyramiding several different Rpi genes in one potato cultivar. In 2006, a Durable Resistance in potato against Phytophthora (DuRPh) project has been initiated at Wageningen University and aimed at developing durable resistance in existing potato cultivars by pyramiding Rpi genes via cisgenesis (Haverkort et al. 2009, 2016). Transgenic Désirée potato plants with two Rpi genes (Rpi-blb3:sto1, Rpi-vnt1:ch1 or Rpi-vnt1:sto1) and with three Rpi genes (Rpi-blb3:vnt1:sto1) have been produced. These plants have not been infected by P. infestans during the two-year field tests (Haverkort et al. 2016). Transgenic potato cultivar Désirée with three Rpi genes (Rpi-blb3, Rpi-vnt1.1 and Rpi-sto1) was obtained also by Haesaert et al. (2015). Plants with these genes showed complete resistance to late blight during two-year field trials in Belgium and the Netherlands (Haesaert et al. 2015). In another study, successful stacking of RB and Rpi-blb2 from S. bulbocastanum and Rpi-vnt1.1 from S. venturii in the susceptible potato cultivars Désirée and Victoria resulted in complete resistance to late blight (Ghislain et al. 2019). Compared with those with a single Rpi gene (RB, Rpi-blb2 or Rpi-vnt1.1), transgenic plants with three Rpi genes showed a significantly higher level of resistance. In three-year field trials involving transgenic plants in southwestern Uganda, no isolate of P. infestans that could overcome the resistance provided by the three Rpi genes was found (Ghislain et al. 2019). The same three Rpi genes, RB, Rpi-blb2 and Rpi-vnt1.1, were stacked in two popular Kenyan potato cultivars Tigoni and Shangi (Webi et al. 2019).
The level of resistance to late blight in transgenic plants not only is dependent on the recognition spectrum and activity of Rpi gene(s), but also can depend on the genetic background of the recipient genotype (Shandil et al. 2017). Potato F1 progenies, obtained from crossing transgenic cultivar Katahdin carrying an RB gene with non-transgenic susceptible cultivar Kufri Bahar or resistant cultivar Kufri Jyoti, were screened for resistance to late blight by whole plants assay. The cultivar Kufri Jyoti is late blight resistant, with resistance inherited from S. demissum containing three Rpi genes (R3, R4, R7). A high level of resistance was observed in the 85.2% of progeny plants from the cross with resistant cultivar Kufri Jyoti, while only 36.4% of progeny were resistant in a cross with the susceptible one. A few F1 genotypes with the RB transgene were highly resistance to late blight, while others were completely susceptible, despite having the RB transgene. The authors explain that by the effects of diversity in genetic background of parental cultivars including the genes involved in signal transduction cascade and encoding pathogenesis-related proteins (Shandil et al. 2017).
Gene editing techniques are an alternative approach to introducing Rpi genes into potato cultivar by conventional methods or by genetic engineering. Gene editing can be used to repair non-functional alleles of Rpi genes. In the study by Van Doorn (2020), a non-functional allele of the Rpi-chc1 was used from two, susceptible to late blight, cultivars Colomba and Altus. A chimeric receptor was created with exchanges in the LRR domain between the susceptible allele of the Rpi-chc1 gene and the functional Rpi-chc1.1 and Rpi-chc1.2 alleles from S. chacoense. This resulted in the restoration of the recognition of P. infestans effectors Avr-chc1.1 and Avr-chc1.2, which was associated with resistance to late blight (Van Doorn 2020).
Currently, transgenic potato is not available on the European market. In 2003, the transgenic potato cultivar Fortuna carrying Rpi-blb1 and Rpi-blb2 from S. bulbocastanum, which was produced by a German company BASF, was not approved for introduction in Europe (Storck et al. 2012). The American company Simplot developed Innate technology, which is used to improve known potato cultivars through genetic engineering. The second-generation Innate transgenic potato lines containing Rpi-vnt1.1 are resistant to P. infestans (Richael 2021).
In conclusion, searching for new Rpi genes among potato wild relatives and then applying these genes in potato cultivars represents an alternative to the use of fungicides for late blight control. Due to rapid evolution of new virulent isolates of P. infestans, potato breeding for durable late blight resistance is challenging. The use of Rpi genes recognizing conservative, essential effectors of P. infestans and the construction of Rpi gene pyramids may help to achieve durable, broad-spectrum late blight resistance, which could be accelerated through genetic engineering.
Author contribution statement
PP performed the literature analysis and wrote the manuscript. JŚ and ZY reviewed and corrected the manuscript. All authors contributed to the conception and design of the review.
Availability of data and material
Not applicable.
Code availability
Not applicable.
References
Aguilera-Galvez C, Champouret N, Rietman H, Lin X, Wouters D, Chu Z, Jones JDG, Vossen JH, Visser RGF, Wolters PJ, Vleeshouwers V (2018) Two different R gene loci co-evolved with Avr2 of Phytophthora infestans and confer distinct resistance specificities in potato. Stud Mycol 89:105–115. https://doi.org/10.1016/j.simyco.2018.01.002
Aguilera-Galvez C, Chu Z, Omy SH, Wouters D, Gilroy EM, Vossen JH, Visser RGF, Birch P, Jones JDG, Vleeshouwers VGAA (2020) The Rpi-mcq1 resistance gene family recognizes Avr2 of Phytophthora infestans but is distinct from R2. bioRxiv. https://doi.org/10.1101/2020.10.08.331181
Akino S, Takemoto D, Hosaka K (2014) Phytophthora infestans: a review of past and current studies on potato late blight. J Gen Plant Pathol 80:24–37. https://doi.org/10.1007/s10327-013-0495-x
Antonova O, Klimenko N, Evdokimova Z, Kostina L, Gavrilenko T (2018) Finding RB/Rpi-blb1/Rpi-sto1-like sequences in conventionally bred potato varieties. Vavilov J Genet Breed 22:693–702. https://doi.org/10.18699/VJ18.412
Armstrong MR, Whisson SC, Pritchard L, Bos JI, Venter E, Avrova AO, Rehmany AP, Böhme U, Brooks K, Cherevach I, Hamlin N, White B, Fraser A, Lord A, Quail MA, Churcher C, Hall N, Berriman M, Huang S, Kamoun S, Beynon JL, Birch PRJ (2005) An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc Natl Acad Sci 102:7766–7771. https://doi.org/10.1073/pnas.0500113102
Armstrong MR, Vossen J, Lim TY, Hutten RCB, Xu J, Strachan SM, Harrower B, Champouret N, Gilroy EM, Hein I (2019) Tracking disease resistance deployment in potato breeding by enrichment sequencing. Plant Biotechnol J 17:540–549. https://doi.org/10.1111/pbi.12997
Bachmann-Pfabe S, Hammann T, Kruse J, Dehmer KJ (2019) Screening of wild potato genetic resources for combined resistance to late blight on tubers and pale potato cyst nematodes. Euphytica 215:48. https://doi.org/10.1007/s10681-019-2364-y
Bakker E, Borm T, Prins P, van der Vossen E, Uenk G, Arens M, de Boer J, van Eck H, Muskens M, Vossen J, van der Linden G, van Ham R, Klein-Lankhorst R, Visser R, Smant G, Bakker J, Goverse A (2011) A genome-wide genetic map of NB-LRR disease resistance loci in potato. Theor Appl Genet 123:493–508. https://doi.org/10.1007/s00122-011-1602-z
Ballvora A, Ercolano MR, Weiss J, Meksem K, Bormann CA, Oberhagemann P, Salamini F, Gebhardt C (2002) The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J 30:361–371. https://doi.org/10.1046/j.1365-313x.2001.01292.x
Black W, Mastenbroek C, Mills WR, Peterson LC (1953) A proposal for an international nomenclature of races of Phytophthora infestans and of genes controlling immunity in Solanum demissum derivatives. Euphytica 2:173–179. https://doi.org/10.1007/BF00053724
Bonde R, Murphy EF (1952) Resistance of certain tomato varieties and crosses to late blight. Maine Agr Expl Sta Bull 497:5–15
Bos JI, Armstrong MR, Gilroy EM, Boevink PC, Hein I, Taylor RM, Zhendong T, Engelhardt S, Vetukuri RR, Harrower B, Dixelius C, Bryan G, Sadanandom A, Whisson SC, Kamoun S, Birch PRJ (2010) Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc Natl Acad Sci 107:9909–9914. https://doi.org/10.1073/pnas.0914408107
Bradshaw JE, Bryan GJ, Lees AK, McLean K, Solomon-Blackburn RM (2006) Mapping the R10 and R11 genes for resistance to late blight (Phytophthora infestans) present in the potato (Solanum tuberosum) R-gene differentials of black. Theor Appl Genet 112:744–751. https://doi.org/10.1007/s00122-005-0179-9
Brown-Donovan KM, Porter GA, Tan EH (2021) Late blight resistance profiles of elite potato germplasm in the United States. Am J Potato Res 98:232–245. https://doi.org/10.1007/s12230-021-09837-0
Brylińska M, Tomczyńska I, Jakuczun H, Wasilewicz-Flis I, Witek K, Jones JDG, Śliwka J (2015) Fine mapping of the Rpi-rzc1 gene conferring broad-spectrum resistance to potato late blight. Eur J Plant Pathol 143:193–198. https://doi.org/10.1007/s10658-015-0663-2
Champouret N (2010) Functional genomics of Phytophthora infestans effectors and Solanum resistance genes. Doctor thesis, Wageningen University
Chen X, Lewandowska D, Armstrong MR, Baker K, Lim TY, Bayer M, Harrower B, McLean K, Jupe F, Kamil Witek K, Lees AK, Jones JD, Bryan GJ, Hein I (2018) Identification and rapid mapping of a gene conferring broad-spectrum late blight resistance in the diploid potato species Solanum verrucosum through DNA capture technologies. Theor Appl Genet 131:1287–1297. https://doi.org/10.1007/s00122-018-3078-6
Chunwongse J, Chunwongse C, Black L, Hanson P (2002) Molecular mapping of the Ph-3 gene for late blight resistance in tomato. J Hortic Sci Biotechnol 77:281–286. https://doi.org/10.1080/14620316.2002.11511493
Colon IT, Eijlander R, Budding DJ, Van Ijzendoorn MT, Pieters MMJ, Hoogendoorn J (1992) Resistance to potato late blight (Phytophthora infestans (Mont.) de Bary) in Solanum nigrum, S. villosum and their sexual hybrids with S. tuberosum and S. demissum. Euphytica 66:55–64. https://doi.org/10.1007/BF00023508
Danan S, Veyrieras JB, Lefebvre V (2011) Construction of a potato consensus map and QTL meta-analysis offer new insights into the genetic architecture of late blight resistance and plant maturity traits. BMC Plant Biol 11:1–17. https://doi.org/10.1186/1471-2229-11-16
De Vetten N, Verzaux E, Vossen J, Hendrik Rietman H, Vleeshouwers VGAA, Evert Jacobsen E, Visser R (2014) Cloning and exploitation of functional R-gene from Solanum x edniense. US patent 2014/0041072A1.
Domazakis E, Lin X, Aguilera-Galvez C, Wouters D, Bijsterbosch G, Wolters PJ, Vleeshouwers VGAA (2017) Effectoromics-based identification of cell surface receptors in potato. Methods Mol Biol 1578:337–353. https://doi.org/10.1007/978-1-4939-6859-6_29
Dou D, Kale SD, Wang X, Chen Y, Wang Q, Wang X, Jiang RHY, Arredondo FD, Anderson RG, Thakur PB, McDowell JM, Wang Y, Tyler BM (2008) Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell 20:1118–1133. https://doi.org/10.1105/tpc.107.057067
Du Y, Weide R, Zhao Z, Msimuko P, Govers F, Bouwmeester K (2018) RXLR effector diversity in Phytophthora infestans isolates determines recognition by potato resistance proteins; the case study AVR1 and R1. Stud Mycol 89:85–93. https://doi.org/10.1016/j.simyco.2018.01.003
Duan Y, Duan S, Armstrong MR, Xu J, Zheng J, Hu J, Chen X, Hein I, Li G, Jin L (2020) Comparative transcriptome profiling reveals compatible and incompatible patterns of potato toward Phytophthora infestans. G3 (bethesda) 10:623–634. https://doi.org/10.1534/g3.119.400818
Duan Y, Duan S, Xu J, Zheng J, Hu J, Li X, Li B, Li G, Jin L (2021) Late blight resistance evaluation and genome-wide assessment of genetic diversity in wild and cultivated potato species. Front Plant Sci 12:710468. https://doi.org/10.3389/fpls.2021.710468
El-Kharbotly A, Palomino-Sánchez C, Salamini F, Jacobsen E, Gebhardt C (1996) R6 and R7 alleles of potato conferring race-specific resistance to Phytophthora infestans (Mont.) de Bary identified genetic loci clustering with the R3 locus on chromosome XI. Theor Appl Genet 92:880–884. https://doi.org/10.1007/bf00221901
Elnahal ASM, Li J, Wang X, Zhou C, Wen G, Wang J, Lindqvist-Kreuze H, Meng Y, Shan W (2020) Identification of natural resistance mediated by recognition of Phytophthora infestans effector gene Avr3aEM in potato. Front Plant Sci 11:919. https://doi.org/10.3389/fpls.2020.00919
Flor HH (1971) Current status of the gene-for-gene concept. Annu Rev Phytopathol 9:275–296. https://doi.org/10.1146/annurev.py.09.090171.001423
Foster SJ, Park TH, Pel M, Brigneti G, Sliwka J, Jagger L, Van der Vossen E, Jones JD (2009) Rpi-vnt1.1, a Tm-22 homolog from Solanum venturii, confers resistance to potato late blight. Mol Plant Microbe Interact 22:589–600. https://doi.org/10.1094/mpmi-22-5-0589
Gallegly M, Marvel M (1955) Inheritance of resistance to tomato race 0 of Phytophthora infestans. Phytopathology 45:103–109
Gao C, Xu H, Huang J, Sun B, Zhang F, Savage Z, Duggan C, Yan T, Wu C, Wang Y, Vleeshouwers VGAA, Kamoun S, Bozkurt TO, Dong S (2020) Pathogen manipulation of chloroplast function triggers a light-dependent immune recognition. Proc Natl Acad Sci USA 117:9613–9620. https://doi.org/10.1073/pnas.2002759117
Gebhardt C, Ballvora A, Walkemeier B, Oberhagemann P, Schüler K (2004) Assessing genetic potential in germplasm collections of crop plants by marker-trait association: a case study for potatoes with quantitative variation of resistance to late blight and maturity type. Mol Breed 13:93–102. https://doi.org/10.1023/B:MOLB.0000012878.89855.df
Ghislain M, Byarugaba AA, Magembe E, Njoroge A, Rivera C, Román ML, Tovar JC, Gamboa S, Forbes GA, Kreuze JF, Barekye A, Kiggundu A (2019) Stacking three late blight resistance genes from wild species directly into African highland potato varieties confers complete field resistance to local blight races. Plant Biotechnol J 17:1119–1129. https://doi.org/10.1111/pbi.13042
Golas TM, Sikkema A, Gros J, Feron RM, Van den Berg RG, Van der Weerden GM, Mariani C, Allefs JJ (2010) Identification of a resistance gene Rpi-dlc1 to Phytophthora infestans in European accessions of Solanum dulcamara. Theor Appl Genet 120:797–808. https://doi.org/10.1007/s00122-009-1202-3
Golas TM, Van de Geest H, Gros J, Sikkema A, D’Agostino N, Nap JP, Mariani C, Allefs JJ, Rieu I (2013) Comparative next-generation mapping of the Phytophthora infestans resistance gene Rpi-dlc2 in a European accession of Solanum dulcamara. Theor Appl Genet 126:59–68. https://doi.org/10.1007/s00122-012-1959-7
Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T, Bozkurt TO, Ah-Fong AM, Alvarado L, Anderson VL, Armstrong MR, Avrova A, Baxter L, Beynon J, Boevink PC, Bollmann SR, Bos JI, Bulone V, Cai G, Cakir C, Carrington JC, Chawner M, Conti L, Costanzo S, Ewan R, Fahlgren N, Fischbach MA, Fugelstad J, Gilroy EM, Gnerre S, Green PJ, Grenville-Briggs LJ, Griffith J, Grünwald NJ, Horn K, Horner NR, Hu CH, Huitema E, Jeong DH, Jones AM, Jones JD, Jones RW, Karlsson EK, Kunjeti SG, Lamour K, Liu Z, Ma L, Maclean D, Chibucos MC, McDonald H, McWalters J, Meijer HJ, Morgan W, Morris PF, Munro CA, O’Neill K, Ospina-Giraldo M, Pinzón A, Pritchard L, Ramsahoye B, Ren Q, Restrepo S, Roy S, Sadanandom A, Savidor A, Schornack S, Schwartz DC, Schumann UD, Schwessinger B, Seyer L, Sharpe T, Silvar C, Song J, Studholme DJ, Sykes S, Thines M, van de Vondervoort PJ, Phuntumart V, Wawra S, Weide R, Win J, Young C, Zhou S, Fry W, Meyers BC, van West P, Ristaino J, Govers F, Birch PR, Whisson SC, Judelson HS, Nusbaum C (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461:393–398. https://doi.org/10.1038/nature08358
Haesaert G, Vossen JH, Custers R, De Loose M, Haverkort A, Heremans B, Hutten R, Kessel G, Landschoot S, Van Droogenbroeck B, Visser RGF, Gheysen G (2015) Transformation of the potato variety Desiree with single or multiple resistance genes increases resistance to late blight under field conditions. Crop Prot 77:163–175. https://doi.org/10.1016/j.cropro.2015.07.018
Hao D, Yang J, Long W, Yi J, VanderZaag P, Li C (2018) Multiple R genes and phenolic compounds synthesis involved in the durable resistance to Phytophthora infestans in potato cv. Cooperation 88. Agri Gene 8:28–36. https://doi.org/10.1016/j.aggene.2018.04.001
Haverkort AJ, Struik PC (2015) Yield levels of potato crops: recent achievements and future prospects. Field Crop Res 182:76–85. https://doi.org/10.1016/j.fcr.2015.06.002
Haverkort AJ, Struik PC, Visser RGF, Jacobsen E (2009) Applied biotechnology to combat late blight in potato caused by Phytophthora infestans. Potato Res 52:249–264. https://doi.org/10.1007/s11540-009-9136-3
Haverkort AJ, Boonekamp PM, Hutten R, Jacobsen E, Lotz LAP, Kessel GJT, Vossen JH, Visser RGF (2016) Durable late blight resistance in potato through dynamic varieties obtained by cisgenesis: scientific and societal advances in the DuRPh project. Potato Res 59:35–66. https://doi.org/10.1007/s11540-015-9312-6
Hawkes J (1990) The potato: evolution, biodiversity and genetic resources. Belhaven Press, London
Hein I, Gilroy EM, Armstrong MR, Birch PR (2009) The zig-zag-zig in oomycete–plant interactions. Mol Plant Pathol 10:547–562. https://doi.org/10.1111/j.1364-3703.2009.00547.x
Hijmans RJ, Spooner DM (2001) Geographic distribution of wild potato species. Am J Bot 88:2101–2112. https://doi.org/10.2307/3558435
Hou H, Atlihan N, Lu ZX (2014) New biotechnology enhances the application of cisgenesis in plant breeding. Front Plant Sci 5:389. https://doi.org/10.3389/fpls.2014.00389
Huang S, Vleeshouwers VGAA, Werij JS, Hutten RC, Van Eck HJ, Visser RG, Jacobsen E (2004) The R3 resistance to Phytophthora infestans in potato is conferred by two closely linked R genes with distinct specificities. Mol Plant Microbe Interact 17:428–435. https://doi.org/10.1094/mpmi.2004.17.4.428
Huang S, Van der Vossen EA, Kuang H, Vleeshouwers VGAA, Zhang N, Borm TJ, Van Eck HJ, Baker B, Jacobsen E, Visser RG (2005) Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J 42:251–261. https://doi.org/10.1111/j.1365-313X.2005.02365.x
Huang G, Liu Z, Gu B, Zhao H, Jia J, Fan G, Meng Y, Du Y, Shan W (2019) An RXLR effector secreted by Phytophthora parasitica is a virulence factor and triggers cell death in various plants. Mol Plant Pathol 20:356–371. https://doi.org/10.1111/mpp.12760
Huang S (2005) Discovery and characterization of the major late blight resistance complex in potato: genomic structure, functional diversity, and implications. PhD thesis, Wageningen University
Jo KR, Arens M, Kim TY, Jongsma MA, Visser RG, Jacobsen E, Vossen JH (2011) Mapping of the S. demissum late blight resistance gene R8 to a new locus on chromosome IX. Theor Appl Genet 123:1331–1340. https://doi.org/10.1007/s00122-011-1670-0
Jo KR, Kim CJ, Kim SJ, Kim TY, Bergervoet M, Jongsma MA, Visser RG, Jacobsen E, Vossen JH (2014) Development of late blight resistant potatoes by cisgene stacking. BMC Biotechnol 14:50. https://doi.org/10.1186/1472-6750-14-50
Jo KR, Visser RG, Jacobsen E, Vossen JH (2015) Characterisation of the late blight resistance in potato differential MaR9 reveals a qualitative resistance gene, R9a, residing in a cluster of Tm-22 homologs on chromosome IX. Theor Appl Genet 128:931–941. https://doi.org/10.1007/s00122-015-2480-6
Jo KR (2013) Unveiling and deploying durability of late blight resistance in potato from natural stacking to cisgenic stacking. PhD thesis, Wageningen University
Jones JDG, Witek K, Verweij W, Jupe F, Cooke D, Dorling S, Tomlinson L, Smoker M, Perkins S, Foster S (2014) Elevating crop disease resistance with cloned genes. Philos Trans R Soc Lond B Biol Sci 369:20130087. https://doi.org/10.1098/rstb.2013.0087
Jones JDG, Foster S, Chu Z, Park T, Van der Vossen E, Pel M, Visser R (2009) Late blight resistance genes and methods. WO/2009/013468A2
Jupe F, Pritchard L, Etherington GJ, Mackenzie K, Cock PJ, Wright F, Sharma SK, Bolser D, Bryan GJ, Jones JD, Hein I (2012) Identification and localisation of the NB-LRR gene family within the potato genome. BMC Genom 13:75. https://doi.org/10.1186/1471-2164-13-75
Jupe F, Witek K, Verweij W, Śliwka J, Pritchard L, Etherington GJ, Maclean D, Cock PJ, Leggett RM, Bryan GJ, Cardle L, Hein I, Jones JDG (2013) Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB‐LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations.Plant J 76:530–544. https://doi.org/10.1111/tpj.12307
Kamoun S, Furzer O, Jones JD et al (2015) The top 10 oomycete pathogens in molecular plant pathology. Mol Plant Pathol 16:413–434. https://doi.org/10.1111/mpp.12190
Karki HS, Halterman DA, Endelman JB (2021a) Characterization of a late blight resistance gene homologous to R2 in potato variety Payette Russet. Am J Potato Res 98:78–84. https://doi.org/10.1007/s12230-020-09811-2
Karki HS, Jansky SH, Halterman DA (2021b) Screening of wild potatoes identifies new sources of late blight resistance. Plant Dis 105:368–376. https://doi.org/10.1094/pdis-06-20-1367-re
Keijzer P, van Bueren ET, Engelen CJM, Hutten RCB (2021) Breeding late blight resistant potatoes for organic farming—a collaborative model of participatory plant breeding: the Bioimpuls project. Potato Res. https://doi.org/10.1007/s11540-021-09519-8
Khavkin EE (2015) Potato late blight as a model of pathogen-host plant coevolution. Russ J Plant Physiol 62:408–419. https://doi.org/10.1134/S1021443715030103
Khiutti A, Spooner DM, Jansky SH, Halterman DA (2015) Testing taxonomic predictivity of foliar and tuber resistance to Phytophthora infestans in wild relatives of potato. Phytopathology 105:1198–1205. https://doi.org/10.1094/phyto-02-15-0046-r
Kim HJ, Lee HR, Jo KR, Mortazavian SM, Huigen DJ, Evenhuis B, Kessel G, Visser RG, Jacobsen E, Vossen JH (2012) Broad spectrum late blight resistance in potato differential set plants MaR8 and MaR9 is conferred by multiple stacked R genes. Theor Appl Genet 124:923–935. https://doi.org/10.1007/s00122-011-1757-7
Kuhl JC, Hanneman RE Jr, Havey MJ (2001) Characterization and mapping of Rpi1, a late-blight resistance locus from diploid (1EBN) Mexican Solanum pinnatisectum. Mol Genet Genom 265:977–985. https://doi.org/10.1007/s004380100490
Lenman M, Ali A, Mühlenbock P, Carlson-Nilsson U, Liljeroth E, Champouret N, Vleeshouwers VG, Andreasson E (2016) Effector-driven marker development and cloning of resistance genes against Phytophthora infestans in potato breeding clone SW93-1015. Theor Appl Genet 129:105–115. https://doi.org/10.1007/s00122-015-2613-y
Li G, Huang S, Guo X, Li Y, Yang Y, Guo Z, Kuang H, Rietman H, Bergervoet M, Vleeshouwers VGAA, van der Vossen EAG, Qu D, Visser RGF, Jacobsen E, Vossen JH (2011) Cloning and characterization of R3b; members of the R3 superfamily of late blight resistance genes show sequence and functional divergence. Mol Plant Microbe Interact 24:1132–1142. https://doi.org/10.1094/mpmi-11-10-0276
Li T, Wang Q, Feng R, Li L, Ding L, Fan G, Li W, Du Y, Zhang M, Huang G, Schäfer P, Meng Y, Tyler BM, Shan W (2019) Negative regulators of plant immunity derived from cinnamyl alcohol dehydrogenases are targeted by multiple Phytophthora Avr3a-like effectors. New Phytol. https://doi.org/10.1111/nph.16139
Lin X, Olave-Achury A, Heal R, Witek K, Karki H, Song T, Wu CH, Adachi H, Kamoun S, Vleeshouwers V, Jones J (2021) Rpi-amr3 confers resistance to multiple Phytophthora species by recognizing a conserved RXLR effector. bioRxiv 447899. https://doi.org/10.1101/2021.06.10.447899
Lokossou AA, Park TH, Van Arkel G, Arens M, Ruyter-Spira C, Morales J, Whisson SC, Birch PR, Visser RG, Jacobsen E, Van der Vossen EA (2009) Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. Mol Plant Microbe Interact 22:630–641. https://doi.org/10.1094/mpmi-22-6-0630
Lokossou A (2010) Dissection of the major late blight resistance cluster on potato linkage group IV. PhD thesis, Wageningen University
Lozano R, Hamblin MT, Prochnik S, Jannink JL (2015) Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genom 16:360. https://doi.org/10.1186/s12864-015-1554-9
Machida-Hirano R (2015) Diversity of potato genetic resources. Breed Sci 65:26–40. https://doi.org/10.1270/jsbbs.65.26
Malcolmson JF (1969) Races of Phytophthora infestans occurring in Great Britain. Trans Br Mycol Soc 53:417–423. https://doi.org/10.1016/S0007-1536(69)80099-9
Malcolmson JF, Black W (1966) New R genes in Solanum demissum lindl. And their complementary races of Phytophthora infestans (Mont.) de bary. Euphytica 15:199–203. https://doi.org/10.1007/BF00022324
Martynov V, Chizhik V (2020) Genetics of pathogen–host interaction by the example of potato late blight disease. Russ J Genet 56:261–268. https://doi.org/10.1134/S1022795420030102
Mastenbroek C (1952) Over de differentiatie van Phytophthora infestans (Mont.) en de vererving van de resistentie van Solanum demissum Lindl. PhD thesis, Fytopathologie, Landbouwhogeschool Wageningen, Wageningen, The Netherlands
Merk HL, Foolad MR (2012) Parent–offspring correlation estimate of heritability for late blight resistance conferred by an accession of the tomato wild species Solanum pimpinellifolium. Plant Breed 131:203–210. https://doi.org/10.1111/j.1439-0523.2011.01898.x
Monino-Lopez D, Nijenhuis M, Kodde L, Kamoun S, Salehian H, Schentsnyi K, Stam R, Lokossou A, Abd-El-Haliem A, Visser RGF, Vossen JH (2021) Allelic variants of the NLR protein Rpi-chc1 differentially recognize members of the Phytophthora infestans PexRD12/31 effector superfamily through the leucine-rich repeat domain. Plant J 107:182–197. https://doi.org/10.1111/tpj.15284
Naess SK, Bradeen JM, Wielgus SM, Haberlach GT, McGrath JM, Helgeson JP (2000) Resistance to late blight in Solanum bulbocastanum is mapped to chromosome 8. Theor Appl Genet 101:697–704. https://doi.org/10.1007/s001220051533
Nakitandwe J (2007) Molecular characterisation of Phytophthora infestans resistance genes in Solanum caripense and development of molecular markers for genetic mapping. Chapter 4: A major gene for resistance to Phytophthora infestans mapped on chromosome IX of Solanum caripense. PhD thesis, University of Natural Resources and Life Sciences
Naveed ZA, Wei X, Chen J, Mubeen H, Ali GS (2020) The PTI to ETI continuum in Phytophthora-plant interactions. Front Plant Sci 11:2030. https://doi.org/10.3389/fpls.2020.593905
Oosumi T, Rockhold DR, Maccree MM, Deahl KL, McCue KF, Belknap WR (2009) Gene Rpi-bt1 from Solanum bulbocastanum confers resistance to late blight in transgenic potatoes. Am J Potato Res 86:456. https://doi.org/10.1007/s12230-009-9100-4
Orbegozo J, Roman ML, Rivera C, Gamboa S, Tovar JC, Forbes GA, Lindqvist-Kreuze H, Kreuze JF, Ghislain M (2016) Rpi-blb2 gene from Solanum bulbocastanum confers extreme resistance to late blight disease in potato. Plant Cell Tiss Organ Cult 125:269–281. https://doi.org/10.1007/s11240-016-0947-z
Pais M, Yoshida K, Giannakopoulou A, Pel MA, Cano LM, Oliva RF, Witek K, Lindqvist-Kreuze H, Vleeshouwers V, Kamoun S (2018) Gene expression polymorphism underpins evasion of host immunity in an asexual lineage of the Irish potato famine pathogen. BMC Evol Biol 18:93. https://doi.org/10.1186/s12862-018-1201-6
Park TH, Vleeshouwers VGAA, Hutten RCB, Van Eck HJ, Van der Vossen EAG, Jacobsen E, Visser RGF (2005a) High-resolution mapping and analysis of the resistance locus Rpi-abpt against Phytophthora infestans in potato. Mol Breed 16:33–43. https://doi.org/10.1007/s11032-005-1925-z
Park TH, Gros J, Sikkema A, Vleeshouwers VG, Muskens M, Allefs S, Jacobsen E, Visser RG, Van der Vossen EAG (2005b) The late blight resistance locus Rpi-bib3 from Solanum bulbocastanum belongs to a major late blight R gene cluster on chromosome 4 of potato. Mol Plant Microbe Interact 18:722–729. https://doi.org/10.1094/mpmi-18-0722
Park TH, Foster S, Brigneti G, Jones JDG (2009) Two distinct potato late blight resistance genes from Solanum berthaultii are located on chromosome 10. Euphytica 165:269–278. https://doi.org/10.1007/s10681-008-9784-4
Pel MA, Foster SJ, Park TH, Rietman H, Van Arkel G, Jones JDG, Van Eck HJ, Jacobsen E, Visser RGF, Van Der Vossen EAG (2009) Mapping and cloning of late blight resistance genes from Solanum venturii using an interspecific candidate gene approach. Mol Plant Microbe Interact 22:601–615
Pérez W, Salas A, Raymundo R, Huaman Z, Nelson R, Bonierbale M (1999) Evaluation of wild potato species for resistance to late blight. CIP Program Rep 2000:49–62
Plich J, Tatarowska B, Lebecka R, Śliwka J, Zimnoch-Guzowska E, Flis B (2015) R2-like gene contributes to resistance to Phytophthora infestans in polish potato cultivar Bzura. Am J Potato Res 92:350–358. https://doi.org/10.1007/s12230-015-9437-9
Poczai P, Hyvönen J (2010) On the origin of Solanum nigrum: can networks help? Mol Biol Rep 38:1171–1185. https://doi.org/10.1007/s11033-010-0215-y
Van Poppel PM (2010) The Phytophthora infestans avirulence gene Avr4 and its potato counterpart R4. PhD thesis, Wageningen University
Prakash C, Trognitz FC, Venhuizen P, Von Haeseler A, Trognitz B (2020) A compendium of genome-wide sequence reads from NBS (nucleotide binding site) domains of resistance genes in the common potato. Sci Rep 10:11392. https://doi.org/10.1038/s41598-020-67848-z
Qutob D, Tedman-Jones J, Gijzen M (2006) Effector-triggered immunity by the plant pathogen Phytophthora. Trends Microbiol 14:470–473. https://doi.org/10.1016/j.tim.2006.09.004
Rakosy-Tican E, Thieme R, König J, Nachtigall M, Hammann T, Denes TE, Kruppa K, Molnár-Láng M (2020) Introgression of two broad-spectrum late blight resistance genes, Rpi-Blb1 and Rpi-Blb3, from Solanum bulbocastanum dun plus race-specific R genes into potato pre-breeding lines. Front Plant Sci 11:699. https://doi.org/10.3389/fpls.2020.00699
Richael CM (2021) Development of the genetically modified Innate® potato. Plant Breed Rev 44:57–78
Rietman H, Bijsterbosch G, Cano LM, Lee HR, Vossen JH, Jacobsen E, Visser RG, Kamoun S, Vleeshouwers VGAA (2012) Qualitative and quantitative late blight resistance in the potato cultivar Sárpo Mira is determined by the perception of five distinct RXLR effectors. Mol Plant Microbe Interact 25:910–919. https://doi.org/10.1094/mpmi-01-12-0010-r
Rietman H (2011) Putting the Phytophthora infestans genome sequence at work: multiple novel avirulence and potato resistance gene candidates revealed. PhD thesis, Wagenigen University
Rogozina EV, Beketova MP, Muratova OA, Kuznetsova MA, Khavkin EE (2021) Stacking resistance genes in multiparental interspecific potato hybrids to anticipate late blight outbreaks. Agronomy 11:115. https://doi.org/10.3390/agronomy11010115
Roman ML, Izarra M, Lindqvist-Kreuze H, Rivera C, Gamboa S, Tovar JC, Forbes GA, Krauze KF, Ghislain M (2017) R/Avr gene expression study of Rpi-vnt1. 1 transgenic potato resistant to the Phytophthora infestans clonal lineage EC-1. Plant Cell Tiss Organ Cul 131:259–268. https://doi.org/10.1007/s11240-017-1281-9
Rudkiewicz F (1985) Zaraza ziemniaka (Phytophthora infestans (Mont.) de Bary). Państwowe Wydawnictwo Naukowe, Warsaw
Sandbrink J, Colon L, Wolters P, Stiekema WJ (2000) Two related genotypes of Solanum microdontum carry different segregating alleles for field resistance to Phytophthora infestans. Mol Breed 6:215–225. https://doi.org/10.1023/A:1009697318518
Schreiber KJ, Bentham A, Williams SJ, Kobe B, Staskawicz BJ (2016) Multiple domain associations within the arabidopsis immune receptor RPP1 regulate the activation of programmed cell death. PLoS Pathog 12:e1005769. https://doi.org/10.1371/journal.ppat.1005769
Sekhwal MK, Li P, Lam I, Wang X, Cloutier S, You FM (2015) Disease resistance gene analogs (RGAs) in plants. Int J Mol Sci 16:19248–19290. https://doi.org/10.3390/ijms160819248
Shandil RK, Chakrabarti SK, Singh BP, Sharma S, Sundaresha S, Kaushik SK, Bhatt AK, Sharma NN (2017) Genotypic background of the recipient plant is crucial for conferring RB gene mediated late blight resistance in potato. BMC Genet 18:22. https://doi.org/10.1186/s12863-017-0490-x
Śliwka J, Jakuczun H, Lebecka R, Marczewski W, Gebhardt C, Zimnoch-Guzowska E (2006) The novel, major locus Rpi-phu1 for late blight resistance maps to potato chromosome IX and is not correlated with long vegetation period. Theor Appl Genet 113:685–695. https://doi.org/10.1007/s00122-006-0336-9
Śliwka J, Jakuczun H, Chmielarz M, Hara-Skrzypiec A, Tomczyńska I, Kilian A, Zimnoch-Guzowska E (2012a) Late blight resistance gene from Solanum ruiz-ceballosii is located on potato chromosome X and linked to violet flower colour. BMC Genet 13:11. https://doi.org/10.1186/1471-2156-13-11
Śliwka J, Jakuczun H, Chmielarz M, Hara-Skrzypiec A, Tomczyńska I, Kilian A, Zimnoch-Guzowska E (2012b) A resistance gene against potato late blight originating from Solanum × michoacanum maps to potato chromosome VII. Theor Appl Genet 124:397–406. https://doi.org/10.1007/s00122-011-1715-4
Smilde WD, Brigneti G, Jagger L, Perkins S, Jones JD (2005) Solanum mochiquense chromosome IX carries a novel late blight resistance gene Rpi-moc1. Theor Appl Genet 110:252–258. https://doi.org/10.1007/s00122-004-1820-8
Smyda P, Jakuczun H, Dębski K, Sliwka J, Thieme R, Nachtigall M, Wasilewicz-Flis I, Zimnoch-Guzowska E (2013) Development of somatic hybrids Solanum × michoacanum bitter. (Rydb.) (+) S. tuberosum L. and autofused 4x S. × michoacanum plants as potential sources of late blight resistance for potato breeding. Plant Cell Rep 32:1231–1241. https://doi.org/10.1007/s00299-013-1422-5
Smyda-Dajmund P, Śliwka J, Wasilewicz-Flis I, Jakuczun H, Zimnoch-Guzowska E (2016) Genetic composition of interspecific potato somatic hybrids and autofused 4x plants evaluated by DArT and cytoplasmic DNA markers. Plant Cell Rep 35:1345–1358. https://doi.org/10.1007/s00299-016-1966-2
Smyda-Dajmund P, Śliwka J, Wasilewicz-Flis I, Jakuczun H, Zimnoch-Guzowska E (2017) BC1 and F1 progeny from Solanum × michoacanum (+) S. tuberosum somatic hybrids, autofused 4× S. michoacanum and cultivated potato. Am J Potato Res 94:323–333. https://doi.org/10.1007/s12230-017-9568-2
Song J, Bradeen JM, Naess SK, Raasch JA, Wielgus SM, Haberlach GT, Liu J, Kuang H, Austin-Phillips S, Buell CR, Helgeson JP, Jiang J (2003) Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc Natl Acad Sci USA 100:9128–9133. https://doi.org/10.1073/pnas.1533501100
Spooner D, Alvarez N, Peralta I, Clausen A (2016) Taxonomy of wild potatoes and their relatives in Southern South America (Solanum sect. Petota and Etuberosum). Syst Bot Monogr 100:240
Stefańczyk E, Sobkowiak S, Brylińska M, Śliwka J (2017) Expression of the potato late blight resistance gene Rpi-phu1 and Phytophthora infestans effectors in the compatible and incompatible interactions in potato. Phytopathology 107:740–748. https://doi.org/10.1094/phyto-09-16-0328-r
Stefańczyk E, Plich J, Janiszewska M, Smyda-Dajmund P, Sobkowiak S, Śliwka J (2020) Marker-assisted pyramiding of potato late blight resistance genes Rpi-rzc1 and Rpi-phu1 on di- and tetraploid levels. Mol Breed 40:89. https://doi.org/10.1007/s11032-020-01169-x
Storck T, Böhme T, Schultheiss H (2012) Status and perspectives of GM approaches to fight late blight. In: Schepers HTAM (ed) Thirteenth EuroBlight workshop. PPO Publications, St. Petersburg, pp 45–48
Su Y, Viquez-Zamora M, den Uil D, Sinnige J, Kruyt H, Vossen J, Lindhout P, van Heusden S (2020) Introgression of genes for resistance against Phytophthora infestans in diploid potato. Am J Potato Res 97:33–42. https://doi.org/10.1007/s12230-019-09741-8
Tomczyńska I, Stefańczyk E, Chmielarz M, Karasiewicz B, Kamiński P, Jones JDG, Lees AK, Śliwka J (2014) A locus conferring effective late blight resistance in potato cultivar Sárpo Mira maps to chromosome XI. Theor Appl Genet 127:647–657. https://doi.org/10.1007/s00122-013-2248-9
Trognitz BR, Trognitz FC (2007) Occurrence of the R1 allele conferring resistance to late blight in potato R-gene differentials and commercial cultivars. Plant Pathol 56:150–155. https://doi.org/10.1111/j.1365-3059.2006.01489.x
Turnbull D, Wang H, Breen S, Malec M, Naqvi S, Yang L, Welsh L, Hemsley P, Zhendong T, Brunner F, Gilroy EM, Birch PRJ (2019) AVR2 targets BSL family members, which act as susceptibility factors to suppress host immunity. Plant Physiol 180:571–581. https://doi.org/10.1104/pp.18.01143
Van der Lee T, Robold A, Testa A, Van’t Klooster JW, Govers F (2001) Mapping of avirulence genes in Phytophthora infestans with amplified fragment length polymorphism markers selected by bulked segregant analysis. Genetics 157:949–956. https://doi.org/10.1093/genetics/157.3.949
Van der Vossen EAG, Sikkema A, Hekkert B, Gros J, Stevens P, Muskens M, Wouters D, Pereira A, Stiekema W, Allefs S (2003) An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J 36:867–882. https://doi.org/10.1046/j.1365-313x.2003.01934.x
Van der Vossen EAG, Gros J, Sikkema A, Muskens M, Wouters D, Wolters P, Pereira A, Allefs S (2005) The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J 44:208–222. https://doi.org/10.1111/j.1365-313X.2005.02527.x
Van Doorn B (2020) Editing of inactive late blight resistance genes in Solanum tuberosum. PhD thesis, Wageningen University.
Van Esse HP, Reuber TL, Van der Does D (2020) Genetic modification to improve disease resistance in crops. New Phytol 225:70–86. https://doi.org/10.1111/nph.15967
Van Poppel PM, Guo J, Van de Vondervoort PJ, Jung MW, Birch PR, Whisson SC, Govers F (2008) The Phytophthora infestans avirulence gene Avr4 encodes an RXLR-dEER effector. Mol Plant Microbe Interact 21:1460–1470. https://doi.org/10.1094/mpmi-21-11-1460
Verzaux E, Van Arkel G, Vleeshouwers VGAA, Van der Vossen EAG, Niks RE, Jacobsen E, Vossen J, Visser RGF (2012) High-resolution mapping of two broad-spectrum late blight resistance genes from two wild species of the Solanum circaeifolium group. Potato Res 55:109–123. https://doi.org/10.1007/s11540-012-9213-x
Verzaux E (2010) Resistance and susceptibility to late blight in Solanum: gene mapping, cloning and stacking. Doctoral thesis, Wageningen Universiteit
Villamon FG, Spooner DM, Orrillo M, Mihovilovich E, Pérez W, Bonierbale M (2005) Late blight resistance linkages in a novel cross of the wild potato species Solanum paucissectum (series Piurana). Theor Appl Genet 111:1201–1214. https://doi.org/10.1007/s00122-005-0053-9
Vleeshouwers VGAA, Rietman H, Krenek P, Champouret N, Young C, Oh SK, Wang M, Bouwmeester K, Vosman B, Visser RGF, Jacobsen E, Govers F, Kamoun S, van der Vossen EAG (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE 3:e2875. https://doi.org/10.1371/journal.pone.0002875
Vleeshouwers VG, Raffaele S, Vossen JH, Champouret N, Oliva R, Segretin ME, Rietman H, Cano LM, Lokossou A, Kessel G, Pel MA, Kamoun S (2011) Understanding and exploiting late blight resistance in the age of effectors. Annu Rev Phytopathol 49:507–531. https://doi.org/10.1146/annurev-phyto-072910-095326
Vossen JH, Van Arkel G, Bergervoet M, Jo KR, Jacobsen E, Visser RG (2016) The Solanum demissum R8 late blight resistance gene is an Sw-5 homologue that has been deployed worldwide in late blight resistant varieties. Theor Appl Genet 129:1785–1796. https://doi.org/10.1007/s00122-016-2740-0
Wang M, Allefs S, Van den Berg RG, Vleeshouwers VG, Van der Vossen EA, Vosman B (2008) Allele mining in Solanum: conserved homologues of Rpi-blb1 are identified in Solanum stoloniferum. Theor Appl Genet 116:933–943. https://doi.org/10.1007/s00122-008-0725-3
Wang J, Hu M, Wang J, Qi J, Han Z, Wang G, Qi Y, Wang HW, Zhou JM, Chai J (2019) Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364:eavv5870. https://doi.org/10.1126/science.aav5870
Webi EN, Kariuki D, Kinyua J, Njoroge A, Ghislain M, Magembe E (2019) Extreme resistance to late blight disease by transferring R3 genes from wild relatives into African farmer-preferred potato varieties. Afr J Biotechnol 18:845–856
Witek K, Jupe F, Witek AI, Baker D, Clark MD, Jones JD (2016) Accelerated cloning of a potato late blight-resistance gene using RenSeq and SMRT sequencing. Nat Biotechnol 34:656–660. https://doi.org/10.1038/nbt.3540
Witek K, Lin X, Karki HS et al (2021) A complex resistance locus in Solanum americanum recognizes a conserved Phytophthora effector. Nat Plants 7:198–208. https://doi.org/10.1038/s41477-021-00854-9
Yang L, Wang D, Xu Y, Zhao H, Wang L, Cao X, Chen Y, Chen Q (2017) A new resistance gene against potato late blight originating from Solanum pinnatisectum located on potato chromosome 7. Front Plant Sci 8:1729. https://doi.org/10.3389/fpls.2017.01729
Yin J, Gu B, Huang G, Tian Y, Quan J, Lindqvist-Kreuze H, Shan W (2017) Conserved RXLR effector genes of Phytophthora infestans expressed at the early stage of potato infection are suppressive to host defense. Front Plant Sci 8:2155. https://doi.org/10.3389/fpls.2017.02155
Zhang C, Liu L, Wang X, Vossen J, Li G, Li T, Zheng Z, Gao J, Guo Y, Visser RGF, Li J, Bai Y, Du Y (2014) The Ph-3 gene from Solanum pimpinellifolium encodes CC-NBS-LRR protein conferring resistance to Phytophthora infestans. Theor Appl Genet 127:1353–1364. https://doi.org/10.1007/s00122-014-2303-1
Zheng J, Duan S, Armstrong MR, Duan Y, Xu J, Chen X, Hein I, Jin L, Li G (2020) New findings on the resistance mechanism of an elite diploid wild potato species JAM1-4 in response to a super race strain of Phytophthora infestans. Phytopathology 110:1375–1387. https://doi.org/10.1094/phyto-09-19-0331-r
Zhu S, Vossen JH, Bergervoet M, Nijenhuis M, Kodde L, Kessel GJ, Vleeshouwers V, Visser RG, Jacobsen E (2015) An updated conventional-and a novel GM potato late blight R gene differential set for virulence monitoring of Phytophthora infestans. Euphytica 202:219–234. https://doi.org/10.1007/s10681-014-1276-0
Zoteyeva NM (2020) Late blight resistance of wild potato species under field conditions in the Northwest of Russia. Proc Appl Bot Genet Breed 180:159–169. https://doi.org/10.30901/2227-8834-2019-4-159-169
Zoteyeva NM, Chrzanowska M, Flis B, Zimnoch-Guzowska E (2012) Resistance to pathogens of the potato accessions from the collection of N. I. Vavilov Institute of Plant Industry (VIR). Am J Potato Res 89:277–293. https://doi.org/10.1007/s12230-012-9252-5
Funding
The research leading to these results has received funding from the Norwegian Financial Mechanism 2014–2021, project DivGene: UMO-2019/34/H/NZ9/00559. The work was supported by a statutory subvention from Polish Ministry of Education and Science for Plant Breeding and Acclimatization Institute-National Research Institute. In particular, PP was supported by project for young scientists 1-1-00-1-04 MN.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Communicated by Gerhard Leubner.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Paluchowska, P., Śliwka, J. & Yin, Z. Late blight resistance genes in potato breeding. Planta 255, 127 (2022). https://doi.org/10.1007/s00425-022-03910-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-022-03910-6