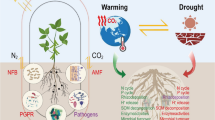
Abstract
Cotton root microbiomes were investigated in two long-term rotation systems established in 2000, a bahiagrass (Paspalum notatum Flugge)-bahiagrass-peanut (Arachis hypogaea L.)-cotton (Gossypium hirsutum L.) rotation (sod-based rotation, SBR) and a peanut-cotton-cotton rotation (conventional rotation, ConR), from 2017 to 2019. Our results demonstrate that bacterial communities were primarily structured by interannual variability, while fungal alpha and beta diversity were significantly affected by both rotation and interannual variability, with greater fungal diversity and distinct fungal communities in SBR compared to ConR across three sampling years. Cotton roots in SBR also harbored more complex and stable microbial networks. These increased resistance to environmental changes driven by interannual variability, such as temperature and precipitation. Beneficial microbial communities (e.g., Opitutaceae, Pseudonocardiaceae, Rhizobiaceae, Bacillaceae, Comamonadaceae, Serendipitaceae, and Glomeraceae) that may promote plant growth, improve tolerance to abiotic stress, and enhance pathogen defense were associated with cotton roots in SBR, along with fewer pathogenic microbes. These beneficial microbial communities (core microbiomes) together with complex and stable microbial networks were significantly and positively correlated with cotton yield across three sampling years, suggesting that long-term conversion to SBR shaped root microbiomes in a way that increased cotton productivity. This study improves our understanding of the microbial mechanisms that underlie the agronomic and economic benefits observed when integrating perennial grasses to diversify the conventional peanut-cotton rotation.







Similar content being viewed by others
References
Acosta-Martínez V, Burow G (2010) Soil microbial communities and function in alternative systems to continuous cotton. Soil Sci Soc Am J 74:1181–1192
Aliche EB, Talsma W, Munnik T (2021) Characterization of maize root microbiome in two different soils by minimizing plant DNA contamination in metabarcoding analysis. Biol Fertil Soils 57:731–737
Allard-Massicotte R, Tessier L, Lécuyer F, Lakshmanan V, Lucier JF, Garneau D, Caudwell L, Vlamakis H, Bais HP, Beauregard PB (2016) Bacillus subtilis early colonization of Arabidopsis thaliana roots involves multiple chemotaxis receptors. Mbio 7:01664–01716
Anthony VM, Ferroni M (2012) Agricultural biotechnology and smallholder farmers in developing countries. Curr Opin Biotechnol 23:278–285
Backer R, Rokem JS, Ilangumaran G, Lamont J, Praslickova D, Ricci E, Subramanian S, Smith DL (2018) Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front Plant Sci 9:1473
Bahulikar RA, Chaluvadi SR, Torres-Jerez I, Mosali J, Bennetzen JL, Udvardi M (2020) Nitrogen fertilization reduces nitrogen fixation activity of diverse diazotrophs in switchgrass roots. Phytobiomes J 5:80–87
Banerjee S, Walder F, Büchi L, Meyer M, Held AY, Gattinger A, Keller T, Charles R, van der Heijden MG (2019) Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J 13:1722–1736
Barnes CJ, van der Gast CJ, Burns CA, McNamara NP, Bending GD (2016) Temporally variable geographical distance effects contribute to the assembly of root-associated fungal communities. Front Microbiol 7:195
Bastian M, Heymann S, Jacomy M (2009) Gephi: An Open Source Software for Exploring and Manipulating Networks. ICWSM 3
Belk A, Xu ZZ, Carter DO, Lynne A, Bucheli S, Knight R, Metcalf JL (2018) Microbiome data accurately predicts the postmortem interval using random forest regression models. Genes 9:104
Bell TH, El-Din Hassan S, Lauron-Moreau A, Al-Otaibi F, Hijri M, Yergeau E, St-Arnaud M (2014) Linkage between bacterial and fungal rhizosphere communities in hydrocarbon-contaminated soils is related to plant phylogeny. ISME J 8:331–343
Beneduzi A, Ambrosini A, Passaglia LMP (2012) Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genet Mol Biol 35:1044–1051
Berg G, Smalla K (2009) Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68:1–13
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857
Bonfante P, Genre A (2010) Mechanisms underlying beneficial plant–fungus interactions in mycorrhizal symbiosis. Nat Commun 1:1–11
Brabcová V, Nováková M, Davidová A, Baldrian P (2016) Dead fungal mycelium in forest soil represents a decomposition hotspot and a habitat for a specific microbial community. New Phytol 210:1369–1381
Bulgarelli D, Rott M, Schlaeppi K, van Themaat EV, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488:91–95
Busby PE, Soman C, Wagner MR, Friesen ML, Kremer J, Bennett A, Morsy M, Eisen JA, Leach JE, Dangl JL (2017) Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol 15:e2001793
Carlisle L, De Wit MM, DeLonge MS, Calo A, Getz C, Ory J, Munden-Dixon K, Galt R, Melone B, Knox R, Iles A (2019) Securing the future of US agriculture: the case for investing in new entry sustainable farmers. Elem Sci Anth 7:17
Cassán F, Coniglio A, López G, Molina R, Nievas S, de Carlan CL, Donadio F, Torres D, Rosas S, Pedrosa FO, de Souza E (2020) Everything you must know about Azospirillum and its impact on agriculture and beyond. Biol Fertil Soils 56:461–479
Chen K-H, Longley R, Bonito G, Liao H-L (2021) A two-step PCR protocol enabling flexible primer choice and high sequencing yield for Illumina MiSeq meta-barcoding. Agronomy 11:1274
D’Acunto L, Andrade JF, Poggio SL, Semmartin M (2018) Diversifying crop rotation increased metabolic soil diversity and activity of the microbial community. Agr Ecosyst Environ 257:159–164
de-Bashan LE, Nannipieri P, Antoun H, Lindermann RG, (2020) Application of beneficial microorganisms and their effects on soil, plants, and the environment: the scientific legacy of Professor Yoav Bashan. Biol Fertil Soils 56:439–442
Delaux P-M, Schornack S (2021) Plant evolution driven by interactions with symbiotic and pathogenic microbes. Science 371: eaba6605
Dourte D, Bartel RL, George S, Marois JJ, Wright D (2016) A sod-based cropping system for irrigation reductions. Renew Agric Food Syst 31:485–494
Durán P, Thiergart T, Garrido-Oter R, Agler M, Kemen E, Schulze-Lefert P, Hacquard S (2018) Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 175:973-983.e14
Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci U S A 112:E911–E920
Emmett BD, Buckley DH, Drinkwater LE (2020) Plant growth rate and nitrogen uptake shape rhizosphere bacterial community composition and activity in an agricultural field. New Phytol 225:960–973
Faust K, Raes J (2012) Microbial interactions: from networks to models. Nat Rev Microbiol 10:538–550
Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C (2012) Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 8:e1002606
Fitzpatrick CR, Copeland J, Wang PW, Guttman DS, Kotanen PM, Johnson MT (2018) Assembly and ecological function of the root microbiome across angiosperm plant species. Proc Natl Acad Sci U S A 115:E1157–E1165
Frøslev TG, Kjøller R, Bruun HH, Ejrnæs R, Brunbjerg AK, Pietroni C, Hansen AJ (2017) Algorithm for post-clustering curation of DNA amplicon data yields reliable biodiversity estimates. Nat Commun 8:1188
Gaiero JR, McCall CA, Thompson KA, Day NJ, Best AS, Dunfield KE (2013) Inside the root microbiome: bacterial root endophytes and plant growth promotion. Am J Bot 100:1738–1750
Glaeser SP, Kämpfer P (2014) The family sphingomonadaceae. The Prokaryotes: Alphaproteobacteria and Betaproteobacteria. Springer, Berlin, pp 641–707
Glick BR (2012) Plant growth-promoting bacteria: mechanisms and applications. Scientifica 2012:1–15
Gupta VVSR, Zhang B, Penton CR, Yu J, Tiedje JM (2019) Diazotroph diversity and nitrogen fixation in summer active perennial grasses in a Mediterranean region agricultural soil. Front Mol Biosci 6:115
Hannula SE, Kielak AM, Steinauer K, Huberty M, Jongen R, De Long JR, Heinen R, Bezemer TM (2019) Time after time: temporal variation in the effects of grass and forb species on soil bacterial and fungal communities. MBio 10.: https://doi.org/10.1128/mBio.02635-19
Hannula SE, Ma H-K, Pérez-Jaramillo JE, Pineda A, Bezemer TM (2020) Structure and ecological function of the soil microbiome affecting plant-soil feedbacks in the presence of a soil-borne pathogen. Environ Microbiol 22:660–676
Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, Döring M, Sessitsch A (2015) The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev 79:293–320
Hardoim PR, van Overbeek LS, van Elsas JD (2008) Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol 16:463–471
Hartman K, van der Heijden MGA, Wittwer RA, Banerjee S, Walser JC, Schlaeppi K (2018) Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 6:14
Haskett TL, Tkacz A, Poole PS (2020) Engineering rhizobacteria for sustainable agriculture. ISME J 15:949–964
Hector A (2015) The New Statistics with R: An Introduction for Biologists. Oxford University Press
Hernandez DJ, David AS, Menges ES, Searcy CA, Afkhami ME (2021) Environmental stress destabilizes microbial networks. ISME J 15:1722–1734
Hernández-Restrepo M, Bezerra JDP, Tan YP, Wiederhold N, Crous PW, Guarro J, Gené J (2019) Re-evaluation of Mycoleptodiscus species and morphologically similar fungi. Persoonia 42:205–227
Hontoria C, García-González I, Quemada M, Roldán A, Alguacil MM (2019) The cover crop determines the AMF community composition in soil and in roots of maize after a ten-year continuous crop rotation. Sci Total Environ 660:913–922
Huang LF, Song LX, Xia XJ, Mao WH, Shi K, Zhou YH, Yu JQ (2013) Plant-soil feedbacks and soil sickness: from mechanisms to application in agriculture. J Chem Ecol 39:232–242
Hugoni M, Luis P, Guyonnet J, Haichar FEZ (2018) Plant host habitat and root exudates shape fungal diversity. Mycorrhiza 28:451–463
Isbell F, Craven D, Connolly J, Loreau M, Schmid B, Beierkuhnlein C, Bezemer TM, Bonin C, Bruelheide H, De Luca E, Ebeling A (2015) Biodiversity increases the resistance of ecosystem productivity to climate extremes. Nature 526:574–577
Jach-Smith LC, Jackson RD (2018) N addition undermines N supplied by arbuscular mycorrhizal fungi to native perennial grasses. Soil Biol Biochem 116:148–157
da Jesus E, C, Liang C, Quensen JF, Susilawati E, Jackson RD, Balser TC, Tiedje JM, (2016) Influence of corn, switchgrass, and prairie cropping systems on soil microbial communities in the upper Midwest of the United States. Glob Change Biol Bioenergy 8:481–494
Johnson AW, Minton NA, Brenneman TB (1999) Bahiagrass, corn, cotton rotations, and pesticides for managing nematodes, diseases, and insects on peanut. J Nematol 31:191
Kaminsky LM, Trexler RV, Malik RJ, Hockett KL, Bell TH (2019) The inherent conflicts in developing soil microbial inoculants. Trends Biotechnol 37:140–151
Katsvairo TW, Wright DL, Marois JJ (2007a) Transition from conventional farming to organic farming using bahiagrass. J Therm Sci 87:2751–2756
Katsvairo TW, Wright DL, Marois JJ (2007b) Performance of peanut and cotton in a bahiagrass cropping system. Agron J 99:1245–1251
Katsvairo TW, Wright DL, Marois JJ (2007c) Cotton roots, earthworms, and infiltration characteristics in sod–peanut–cotton cropping systems. Agron J 99:390–398
Katsvairo TW, Wright DL, Marois JJ, Rich JR (2009) Comparative plant growth and development in two cotton rotations under irrigated and non-irrigated conditions. Crop Sci 49:2233–2245
Kumari B, Mallick MA, Solanki MK, Solanki AC, Hora A, Guo W (2019) Plant Growth Promoting Rhizobacteria (PGPR): Modern Prospects for Sustainable Agriculture. In: Ansari RA, Mahmood I (eds) Plant Health Under Biotic Stress, vol 2. Microbial Interactions. Springer Singapore, Singapore, pp 109–127
Kumar R, Bhatia R, Kukreja K, Behl RK, Dudeja SS, Narula N (2007) Establishment of Azotobacter on plant roots: chemotactic response, development and analysis of root exudates of cotton (Gossypium hirsutum L.) and wheat (Triticum aestivum L.). J Basic Microbiol 47:436–439
Li X, Zhang Y, Ding C, Jia Z, He Z, Zhang T, Wang X (2015) Declined soil suppressiveness to Fusarium oxysporum by rhizosphere microflora of cotton in soil sickness. Biol Fertil Soils 51:935–946
Liao H-L, Chen Y, Bruns TD, Peay KG, Taylor JW, Branco S, Talbot JM, Vilgalys R (2014) Metatranscriptomic analysis of ectomycorrhizal roots reveals genes associated with Piloderma-Pinus symbiosis: improved methodologies for assessing gene expression in situ. Environ Microbiol 16:3730–3742
Liu J, Yu Z, Yao Q, Hu X, Zhang W, Mi G, Chen X, Wang G (2017) Distinct soil bacterial communities in response to the cropping system in a Mollisol of northeast China. App Soil Ecol 119:407–416
Louca S, Polz MF, Mazel F, Albright MB, Huber JA, O’Connor MI, Ackermann M, Hahn AS, Srivastava DS, Crowe SA, Doebeli M (2018) Function and functional redundancy in microbial systems. Nat Ecol Evol 2:936–943
Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, Del Rio TG, Edgar RC (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90
Ma B, Wang H, Dsouza M, Lou J, He Y, Dai Z, Brookes PC, Xu J, Gilbert JA (2016) Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J 10:1891–1901
Ma B, Wang Y, Ye S, Liu S, Stirling E, Gilbert JA, Faust K, Knight R, Jansson JK, Cardona C, Röttjers L (2020) Earth microbial co-occurrence network reveals interconnection pattern across microbiomes. Microbiome 8:82
Mavrodi DV, Mavrodi OV, Elbourne LDH, Tetu S, Bonsall RF, Parejko J, Yang M, Paulsen IT, Weller DM (2018) Long-term irrigation affects the dynamics and activity of the wheat rhizosphere microbiome. Front Plant Sci 9:345
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P (2012) An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618
Naylor D, Coleman-Derr D (2017) Drought stress and root-associated bacterial communities. Front Plant Sci 8:2223
Pascale A, Proietti S, Pantelides IS, Stringlis IA (2019) Modulation of the root microbiome by plant molecules: the basis for targeted disease suppression and plant growth promotion. Front Plant Sci 10:1741
Pausch J, Kuzyakov Y (2018) Carbon input by roots into the soil: quantification of rhizodeposition from root to ecosystem scale. Glob Chang Biol 24:1–12
Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci U S A 110:6548–6553
Platas G, Morón R, González I, Collado J, Genilloud O, Peláez F, Diez MT (1998) Production of antibacterial activities by members of the family Pseudonocardiaceae: influence of nutrients. World J Microbiol Biotechnol 14:521–527
Põlme S, Abarenkov K, Henrik Nilsson R, Lindahl BD, Clemmensen KE, Kauserud H, Nguyen N, Kjøller R, Bates ST, Baldrian P, Frøslev TG (2020) FungalTraits: a user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers 105:1–16
Qiao Q, Wang F, Zhang J, Chen Y, Zhang C, Liu G, Zhang H, Ma C, Zhang J (2017) The variation in the rhizosphere microbiome of cotton with soil type, genotype and developmental stage. Sci Rep 7:3940
Ray P, Guo Y, Chi MH, Krom N, Saha MC, Craven KD (2020) Serendipita bescii promotes winter wheat growth and modulates the host root transcriptome under phosphorus and nitrogen starvation. Environ Microbiol 23:1876–1888
Rodrigues JLM, Isanapong J (2014) The Family Opitutaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The Prokaryotes. Springer, Berlin, pp 751–756
Rosseel Y (2012) lavaan: AnRPackage for Structural Equation Modeling. J Stat Softw 48:1–36
Santolini M, Barabási A-L (2018) Predicting perturbation patterns from the topology of biological networks. Proc Natl Acad Sci U S A 115:E6375–E6383
Schumacher LA, Grabau ZJ, Wright DL, Small IM, Liao HL (2020) Nematicide influence on cotton yield and plant-parasitic nematodes in conventional and sod-based crop rotation. J Nematol 52:1–14
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Singh BK, Trivedi P, Egidi E, Macdonald CA, Delgado-Baquerizo M (2020) Crop microbiome and sustainable agriculture. Nat Rev Microbiol 18:601–602
Smith SE, Read DJ (2010) Mycorrhizal Symbiosis. Academic Press
Strobel T, Al-Dilaimi A, Blom J, Gessner A, Kalinowski J, Luzhetska M, Pühler A, Szczepanowski R, Bechthold A, Rückert C (2012) Complete genome sequence of Saccharothrix espanaensis DSM 44229 T and comparison to the other completely sequenced Pseudonocardiaceae. BMC Genomics 13:1–13
Sunilkumar G, Campbell LM, Puckhaber L, Stipanovic RD, Rathore KS (2006) Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc Natl Acad Sci U S A 103:18054–18059
Tackmann J, Arora N, Schmidt TSB, Rodrigues JF, von Mering C (2018) Ecologically informed microbial biomarkers and accurate classification of mixed and unmixed samples in an extensive cross-study of human body sites. Microbiome 6:192
Thiergart T, Durán P, Ellis T, Vannier N, Garrido-Oter R, Kemen E, Roux F, Alonso-Blanco C, Ågren J, Schulze-Lefert P, Hacquard S (2020) Root microbiota assembly and adaptive differentiation among European Arabidopsis populations. Nat Ecol Evol 4:122–131
Trivedi P, Delgado-Baquerizo M, Trivedi C, Hu H, Anderson IC, Jeffries TC, Zhou J, Singh BK (2016) Microbial regulation of the soil carbon cycle: evidence from gene–enzyme relationships. ISME J 10:2593–2604
Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK (2020) Plant-microbiome interactions: from community assembly to plant health. Nat Rev Microbiol 18:607–621
Tsigbey FK, Rich JR, Marois JJ, Wright DL (2009) Effect of bahiagrass (Paspalum notatum Fluegge) on nematode populations in the field and their behavior under greenhouse and laboratory conditions. Nematropica 111–120
Ullah A, Akbar A, Luo Q, Khan AH, Manghwar H, Shaban M, Yang X (2019) Microbiome diversity in cotton rhizosphere under normal and drought conditions. Microb Ecol 77:429–439
van der Heijden MGA, Hartmann M (2016) Networking in the plant microbiome. PLoS Biol. 14:e1002378
Větrovský T, Morais D, Kohout P, Lepinay C, Algora C, Hollá SA, Bahnmann BD, Bílohnědá K, Brabcová V, D’Alò F, Human ZR (2020) GlobalFungi, a global database of fungal occurrences from high-throughput-sequencing metabarcoding studies. Sci Data 7:228
Vohník M, Pánek M, Fehrer J, Selosse M-A (2016) Experimental evidence of ericoid mycorrhizal potential within Serendipitaceae (Sebacinales). Mycorrhiza 26:831–846
Vuko M, Cania B, Vogel C, Kublik S (2020) Shifts in reclamation management strategies shape the role of exopolysaccharide and lipopolysaccharide-producing bacteria during soil formation. Microb Biotechnol 13:584–598
Wei F, Zhao L, Xu X, Feng H, Shi Y, Deakin G, Feng Z, Zhu H (2019) Cultivar-dependent variation of the cotton rhizosphere and endosphere microbiome under field conditions. Front Plant Sci 10:1659
Weiß M, Waller F, Zuccaro A, Selosse MA (2016) Sebacinales–one thousand and one interactions with land plants. New Phytol 211:20–40
West SG, Taylor AB, Wu W (2012) Model fit and model selection in structural equation modeling. Handbook of structural equation modeling. Guilford Press, New York, pp 209–231
Wu L, Wu H, Chen J, Wang J, Lin W (2016) Microbial community structure and its temporal changes in Rehmannia glutinosa rhizospheric soils monocultured for different years. Eur J Soil Biol 72:1–5
Xia Y, Sun J, Chen DG (2018) Statistical analysis of microbiome data with R. Springer, Singapore
Xiong C, Singh BK, He JZ, Han YL, Li PP, Wan LH, Meng GZ, Liu SY, Wang JT, Wu CF, Ge AH, Zhang LM (2021) Plant developmental stage drives the differentiation in ecological role of the maize microbiome. Microbiome 9:179
Xiong C, Zhu YG, Wang JT, Singh B, Han LL, Shen JP, Li PP, Wang GB, Wu CF, Ge AH, Zhang LM (2020) Host selection shapes crop microbiome assembly and network complexity. New Phytol 229:1091–1104
Yadav V, Kumar M, Deep DK, Kumar H, Sharma R, Tripathi T, Tuteja N, Saxena AK, Johri AK (2010) A phosphate transporter from the root endophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. J Biol Chem 285:26532–26544
Yu P, Wang C, Baldauf JA, Tai H, Gutjahr C, Hochholdinger F, Li C (2018) Root type and soil phosphate determine the taxonomic landscape of colonizing fungi and the transcriptome of field-grown maize roots. New Phytol 217:1240–1253
Zhang K, Bonito G, Hsu CM, Hameed K, Vilgalys R, Liao HL (2020a) Mortierella elongata increases plant biomass among non-leguminous crop species. Agronomy 10:754
Zhang K, Chen L, Li Y, Brookes PC, Xu J, Luo Y (2017) The effects of combinations of biochar, lime, and organic fertilizer on nitrification and nitrifiers. Biol Fertil Soils 53:77–87
Zhang K, Chen L, Li Y, Brookes PC, Xu J, Luo Y (2020b) Interactive effects of soil pH and substrate quality on microbial utilization. Eur J Soil Biol 96:103151
Zhang K, Maltais-Landry G, Liao HL (2021) How soil biota regulate C cycling and soil C pools in diversified crop rotations. Soil Biol Biochem 156:108219
Zhang K, Schumacher L, Maltais-Landry G, Grabau ZJ, George S, Wright D, Small IM, Liao HL (2022) Integrating perennial bahiagrass into the conventional rotation of cotton and peanut enhances interactions between microbial and nematode communities. Appl Soil Ecol 170:104254
Zhao D, Wright DL, Marois JJ, Mackowiak CL (2010) Improved growth and nutrient status of an oat cover crop in sod-based versus conventional peanut-cotton rotations. Agron Sustain Dev 30:497–504
Zhao Z-B, He JZ, Geisen S, Han LL, Wang JT, Shen JP, Wei WX, Fang YT, Li PP, Zhang LM (2019) Protist communities are more sensitive to nitrogen fertilization than other microorganisms in diverse agricultural soils. Microbiome 7:33
Zhou X, Liu J, Wu F (2017) Soil microbial communities in cucumber monoculture and rotation systems and their feedback effects on cucumber seedling growth. Plant Soil 415:507–520
Acknowledgements
We thank Chih-Ming Hsu, Neetika Thakur and Lesley Schumacher for their help in collecting root samples and performing DNA extraction for the cotton root samples. We also thank the staff at the North Florida Research and Education Center for maintenance of the long-term field site at Quincy.
Funding
This work was financially supported by a United States Department of Agriculture (USDA)-Southern Sustainable Agriculture Research and Education (SSARE) grant award (2017–38640-26914) to Z. Grabau, I. Small, D. Wright and H.-L. Liao, a USDA-NIFA (2019–67013-29107) award to H.-L. Liao, a USDA-SSARE (2019–38640-29878, SUB00002463) grant award to H.-L. Liao and K. Zhang, and a UF Graduate School Funding Award to K. Zhang.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors have approved the manuscript in its entirety and agreed for its publication.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, K., Maltais-Landry, G., George, S. et al. Long-term sod-based rotation promotes beneficial root microbiomes and increases crop productivity. Biol Fertil Soils 58, 403–419 (2022). https://doi.org/10.1007/s00374-022-01626-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-022-01626-z