Abstract
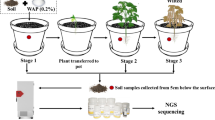
Climate change contributes to drought stress and subsequently affects crop growth, development, and yield. The microbial community, such as fungi and bacteria in the rhizosphere, is of special importance to plant productivity. In this study, soil collected from a cotton research field was used to grow cotton plants (Gossypium hirsutum cv. Jin668) under controlled environment conditions. Drought stress was applied at flowering stage, while control plants were regularly watered. At the same time, the soil without plants was also subjected to drought, while control pots were regularly watered. The soil was collected in sterilized tubes and microbial DNA was isolated and high-throughput sequencing of 16S rRNA genes was carried out. The alpha diversity of bacteria community significantly increased in the soil with cotton plants compared to the soil without cotton plants. Taxonomic analysis revealed that the bacterial community structure of the cotton rhizosphere predominantly consisted of the phyla Proteobacteria (31.7%), Actinobacteria (29.6%), Gemmatimonadetes (9.8%), Chloroflexi (9%), Cyanobacteria (5.6%), and Acidobacteria. In the drought-treated rhizosphere, Chloroflexi and Gemmatimonadetes were the dominant phyla. This study reveals that the cotton rhizosphere has a rich pool of bacterial communities even under drought stress, and which may improve drought tolerance in plants. These data will underpin future improvement of drought tolerance of cotton via the soil microbial community.






Similar content being viewed by others
References
Ullah A, Sun H, Munis MFH, Fahad S, Yang X (2015) Phytoremediation of heavy metals assisted by plant growth promoting (PGP) bacteria: a review. Environ. Exp. Bot. 117:28–40
Yu LH, Wu SJ, Peng YS, Liu RN, Chen X, Zhao P, Xu P, Zhu JB, Jiao GL, Pei Y, Xiang CB (2016) Arabidopsis EDT1/HDG11 improves drought and salt tolerance in cotton and poplar and increases cotton yield in the field. Plant Biotechnol. J. 14:72–84
Comas LH, Becker SR, Cruz VMV, Byrne PF, Dierig DA (2013) Root traits contributing to plant productivity under drought. Front. Plant Sci. 4:442
Ullah A, Sun H, Yang X, Zhang X (2017) Drought coping strategies in cotton: increased crop per drop. Plant Biotechnol. J. 15:281–284
Zhang F, Li S, Yang S, Wang L, Guo W (2015) Overexpression of a cotton annexing gene, GhAnn1, enhances drought and salt stress tolerance in transgenic cotton. Plant Mol. Biol. 87:47–67
Dawn news (2016) http://www.dawn.com/news/1240448
Hejnák V, Tatar Ö, Atasoy GD, Martinková J, Çelen AE, Hnilička F, Skalický M (2015) Growth and photosynthesis of Upland and Pima cotton: response to drought and heat stress. Plant Soil Environ. 61:507–514
Lareen A, Burton F, Schäfer P (2016) Plant root-microbe communication in shaping root microbiomes. Plant Mol. Biol. 90:575–587
Mendes R, Garbeva P, Raaijmakers JM (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37:634–663
Bai Y, Müller DV, Srinivas G, Garrido-Oter R, Potthoff E, Rott M et al (2015) Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528:364–369
Finkel OM, Castrillo G, Paredes SH, González IS, Dangl JL (2017) Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant Biol. 38:155–163
Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90
Glick BR (2014) Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169:30–39
Bartram AK, Lynch MDJ, Stearns JC, Moreno-Hagelsieb G, Neufeld JD (2011) Generation of multi-million 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl. Environ. Microbiol. 77:3846–3852
Masella AP, Bartram AK, Brown DG, Truszkowski JM, Neufeld JD (2012) PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 13(31):31
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336
Blaxter M, Mann J, Chapman T, Thomas F, Whitton C, Floyd R, Abebe E (2005) Defining operational taxonomic units using DNA barcode data. Philos Trans R Soc Lond B Biol Sci 360:1935–1943
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596
Kemp PF, Aller JY (2004) Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol. Ecol. 47:161–177
Chao A, Shen TJ (2004) Nonparametric prediction in species sampling. J. Agric. Biol. Environ. Stat. 9:253–269
Ramette A (2007) Multivariates analyses in microbial ecology. FEMS Microbiol. Ecol. 62:142–160
Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31:814–821
Yan N, Marschner P, Cao W, Zuo C, Qin W (2015) Influence of salinity and water content on soil microorganisms. ISWCR 3:316–323
Verbon EH, Liberman LM (2016) Beneficial microbes affect endogenous mechanisms controlling root development. Trends Plant Sci 21:218–229
Qiao Q, Wang F, Zhang J, Chen Y, Zhang C, Liu G, Zhang H, Ma C, Zhang J (2017) The variation in the rhizosphere microbiome of cotton with soil type, genotype and developmental stage. Sci. Rep. 7:3940
Yamada T, Sekiguchi Y, Imachi H, Kamagata Y, Ohashi A, Harada H (2005) Diversity, localization, and physiological properties of filamentous microbes belonging to Chloroflexi subphylum I in mesophilic and thermophilic methanogenic sludge granules. Appl. Environ. Microbiol. 71:7493–7503
DeBruyn JM, Nixon LT, Fawaz MN, Johnson AM, Radosevich M (2011) Global biogeography and quantitative seasonal dynamics of Gemmatimonadetes in soil. Appl. Environ. Microbiol. 77:6295–6300
Nunoura T, Hirai M, Miyazaki M, Kazama H, Makita H, Hirayama H, Furushima Y, Yamamoto H, Imachi H, Takai K (2013) Isolation and characterization of a thermophilic, obligately anaerobic and heterotrophic marine Chloroflexi bacterium from a Chloroflexi-dominated microbial community associated with a Japanese shallow hydrothermal system, and proposal for Thermomarinilinea lacunofontalis gen. nov., sp. nov. Microbes Environ. 28:228–235
Ward LM, Hemp J, Shih PM, McGlynn SE, Fischer WW (2018) Evolution of phototrophy in the Chloroflexi phylum driven by horizontal gene transfer. Front. Microbiol. 9:260
Yandigeri MS, Meena KK, Singh D, Malviya N, Singh DP, Solanki MK, Yadav AK, Arora DK (2012) Drought-tolerant endophytic actinobacteria promote growth of wheat (Triticum aestivum) under water stress conditions. Plant Growth Regul. 68:411–420
Viaene T, Langendries S, Beirinckx S, Maes M, Goormachtig S (2016) Streptomyces as a plant’s best friend? FEMS Microbiol. Ecol. 92:fiw119
de Jesus Sousa JA, Olivares FL (2016) Plant growth promotion by streptomycetes: ecophysiology, mechanisms and applications. CBTA 3:24
Naylor D, DeGraaf S, Purdom E, Coleman-Derr D (2017) Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J 11:2691–2704
Bouskill NJ, Wood TE, Baran R, Ye Z, Bowen BP, Lim H et al (2016) Belowground response to drought in a tropical forest soil. I. Changes in microbial functional potential and metabolism. Front. Microbiol. 7:525
Vurukonda SKP, Vardharajula S, Shrivastava M, Ali SKZ (2016) Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 184:13–24
Naylor D, Coleman-Derr D (2018) Drought stress and root-associated bacterial communities. Front. Plant Sci. 8:2223
Santana-Vieira DDS, Freschi L, Almeida LAD, Moraes DHS, Neves DM, Santos LM et al (2016) Survival strategies of citrus rootstocks subjected to drought. Sci. Rep. 6:38775
Numan M, Bashir S, Khan Y, Mumtaza R, Shinwari ZK, Khan AL et al (2018) Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: a review. Microbiol. Res. 209:21–32
Santos-Medellín C, Edwards J, Liechty Z, Nguyen B, Sundaresan V (2017) Drought stress results in a compartment-specific restructuring of the rice root-associated microbiomes. mBio 8:e00764–e00717
Funding
Funding was provided by the National Key Project of Research and Development Plan (2016YFD0101006) and National R&D Project of Transgenic Crops of Ministry of Science and Technology of China (2016ZX08005-004-002). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Xiyan Yang designed the project; Abid Ullah and Adnan Akbar conducted the experiments and wrote the paper; Qingqing Luo, Hakim, and Muhammad Shaban helped Abid Ullah in the experiments; Aamir Hamid Khan helped Abid Ullah in writing the paper; and Xiyan Yang critically reviewed the paper.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Electronic supplementary material
ESM 1
(PDF 66 kb)
Fig. S1
The abundance distribution of the top 20 taxa with the most significant differences among samples, Phylum (A) and Genus (B). The abscissa is the most significant first 20 taxa, and the ordinate is the sequence amount of each taxon in each sample (group). In the case of no grouping of samples, they are displayed in the form of scatter plots; in the case of grouping of samples, they are displayed in the form of a violin chart combined with a box plot: Among them, the violin plot visually display the distribution characteristics of the data, “violin”. The “skinny” reflects the density of the sample data distribution (the greater the width, the more samples corresponding to the sequence); the box plot borders represent the Interquartile range (IQR), horizontal lines Representing the median value, the upper and lower whiskers represent the 1.5-fold IQR range outside the upper and lower quartiles respectively, and the symbol “•” indicates the extreme value that exceeds the range. (PDF 590 kb)
Fig. S2
Affiliation network diagram. The nodes represent the dominant genus and are identified by different colors. The connection between the nodes indicates that there is a correlation between the two genera, the red line indicates a positive correlation, and the green line indicates a negative correlation. The more connections that pass through a node, the more associations the genus has with other members of the flora. (PDF 22 kb)
Fig. S3
PICRUSt predicted KEGG second-level distribution map. (A-D) Metabolic pathways divided into five categories, including metabolism, genetic information processing, environmental information processing, cellular processes, and organism systems. In the figure, the abscissa is the KEGG second-class functional group, and the ordinate is the relative abundance of each functional group within each sample (group). In the case of no grouping of samples, they are displayed in the form of a bar graph; in the case of grouping of samples, they are displayed in the form of violin plots combined with box plots: among them, violin plots can visually display the distribution characteristics of data, “violin”. The “skinny” reflects the density of the distribution of the sample data (the greater the width, the more the corresponding sample under the abundance); the box plot border represents the Interquartile range (IQR), horizontal line representing the median value, the upper and lower whiskers represent the 1.5-fold IQR range outside the upper and lower quartiles respectively, and the symbol “•” indicates the extreme value that exceeds the range. (PDF 98 kb)
Rights and permissions
About this article
Cite this article
Ullah, A., Akbar, A., Luo, Q. et al. Microbiome Diversity in Cotton Rhizosphere Under Normal and Drought Conditions. Microb Ecol 77, 429–439 (2019). https://doi.org/10.1007/s00248-018-1260-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-018-1260-7