Abstract
Lignin is a heterogeneous aromatic biopolymer and a major constituent of lignocellulosic biomass, such as wood and agricultural residues. Despite the high amount of aromatic carbon present, the severe recalcitrance of the lignin macromolecule makes it difficult to convert into value-added products. In nature, lignin and lignin-derived aromatic compounds are catabolized by a consortia of microbes specialized at breaking down the natural lignin and its constituents. In an attempt to bridge the gap between the fundamental knowledge on microbial lignin catabolism, and the recently emerging field of applied biotechnology for lignin biovalorization, we have developed the eLignin Microbial Database (www.elignindatabase.com), an openly available database that indexes data from the lignin bibliome, such as microorganisms, aromatic substrates, and metabolic pathways. In the present contribution, we introduce the eLignin database, use its dataset to map the reported ecological and biochemical diversity of the lignin microbial niches, and discuss the findings.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lignin is one of the three main components in lignocellulosic biomass and the most abundant terrestrial aromatic macromolecule and is as such a potentially great source of renewable aromatic compounds (Holladay et al. 2007). It is found in the cell walls of lignocellulosic plants (Fig. 1), where it is intertwined with the other two main polymers (cellulose and hemicellulose), and confers structural strength, impermeability, and water transport in the cell wall (Ayyachamy et al. 2013). The main characteristic traits of the lignin macropolymer are its highly amorphous structure—caused by the high heterogeneity of its aromatic building blocks (in turn directly depending on the plant species) (Gellerstedt and Henriksson 2008; Lewis and Yamamoto 1990; Vanholme et al. 2010)—and its severe recalcitrance to chemical and microbial depolymerization (Ruiz-Dueñas and Martínez 2009). Various types of lignin streams (here called technical lignins) are produced in high amounts in the pulp and paper industry and are today primarily used to generate process steam and electricity by incineration (Li and Takkellapati 2018; Naqvi et al. 2012). These lignin streams are therefore a largely untapped resource for sustainable production of platform chemicals and have the potential to become a key feedstock in a future expanded biorefinery concepts (Beckham et al. 2016).
Schematic representation of the lignin microbial niche. In this model of the niche, lignin is mineralized by two subgroups: lignolytic species and aromatic degrading species. Some species degrade or modify lignin to access the hemi-/cellulose on which they grow (subgroup 1), and other species catabolize the aromatic lignin fragments that result from the enzymatic depolymerization (subgroup 2). There is also some overlap between the subgroups, with species capable of both lignolysis and aromatic degradation. Yellow circles represent the different origins of isolation reported for this niche. The poplar lignin structure was adapted from Vanholme et al. (2010)
Microbial lignin degradation in nature has been studied for decades, with the scientific literature stretching back to at least the 1960s and studies on, e.g., Pseudomonas putida (Ornston and Stanier 1966). Due to the high diversity of the lignin heteropolymer, the microbial modes of lignin catabolism are also diverse (Bugg et al. 2011b; Durante-Rodríguez et al. 2018; Fuchs et al. 2011). Lignin degraders are typically bacteria and fungi: among the former, the species mostly belong to the Actinobacteria and Proteobacteria phyla (Bugg et al. 2011b; Tian et al. 2014); as for the fungi, the common degraders are of the white rot fungi, filamentous fungi, and yeast taxa (Durham et al. 1984; Guillén et al. 2005; Martins et al. 2015). Furthermore, the lignin recalcitrance often prevents one single species from fully degrading the lignin polymer, and instead a symbiosis where rot-type fungi and bacteria are working together is needed to achieve a complete degradation (Cragg et al. 2015; de Boer et al. 2005), thus generating a specific niche (Fig. 1) that selects for a small set of microbial genera.
On the applied side, chemical depolymerization of natural or technical lignins is required to establish a biotechnological value chain from mono- or oligoaromatics. The lignin streams, e.g., from the pulp and paper industry, must be depolymerized to yield mono- and oligomeric aromatic compounds (Ragauskas et al. 2014; Zakzeski et al. 2010) that are then fed to suitable microbes (natural or engineered) for bioconversion into value-added products. However, most knowledge on the microbial side of this process comes from natural degraders, and little is currently known about microbial growth and utilization on the cocktail of aromatic compounds found in depolymerized technical lignin. Furthermore, although different lignocellulosic feedstocks (e.g., softwood, hardwood, agricultural residues) are known to contain different amounts and types of aromatic building blocks (Gellerstedt and Henriksson 2008; Ragauskas et al. 2014), it is very challenging to predict the chemical composition of the mixture resulting from a depolymerization process, especially for technical lignins (Abdelaziz et al. 2016). Consequently, it is difficult to a priori select a suitable microbial host until chemical analysis has been performed on the depolymerized (low molecular weight) lignin stream.
The literature on microbial lignin catabolism is vast and combines fundamental microbiology and applied studies that have in particular seen a surge in popularity during the last decade. However, there has been little effort yet to facilitate an overview of the large amount of publications in this field, especially regarding intracellular microbial events. For this reason, we have created a new database named The eLignin Microbial Database (www.elignindatabase.com) for collection of data from scientific literature on the catabolism of lignin and lignin-derived aromatic compound by microorganisms. The eLignin database was launched online in March 2017 and aims to bring together the bibliome of this field in one self-contained searchable platform, and thus fill a gap presently not covered by other online biological databases, as well as to demonstrate the high diversity of this microbial niche (Fig. 1). As the database primarily focuses on intracellular conversion steps, information on extracellular enzymes with lignolytic activities are currently not covered and the readers are redirected to, e.g., the following reviews (Janusz et al. 2017; Sigoillot et al. 2012).
The present minireview will introduce the design philosophy of the eLignin database and present our outcome of the diversity analysis with prime focus on intracellular microbial events. What sets this paper apart from other recent reviews discussing the diversity of microbial lignin degradation (Bugg et al. 2011a; Tian et al. 2014) is that we have been able to use the established content of the database (Table 1) to make pattern recognitions over the indexed publications in eLignin (for instance using relational SQL queries and Python scripts).
Scope and design of the eLignin database
The eLignin database was created because there is, to our knowledge, no currently available database dedicated to microbial lignin catabolism. A literature survey showed that there have been published databases on lignin biochemistry in the past, but they are, at the time of writing, all unavailable and/or discontinued: FOLy, a database on fungal oxidoreductases for lignin catabolism (Levasseur et al. 2008); LD2L, a database similar in scope as eLignin (Arumugam et al. 2014); and an NMR database for lignin structures (Ralph et al. 2004), with the latter not treating microbial catabolism. The objective of eLignin is to collect data on strains of microorganisms (bacteria, yeasts, and fungi) known to degrade and/or catabolize lignin and lignin-derived aromatic compounds. Specifically, the database content includes microorganisms, substrates, pathways, genes, metabolic reactions, and enzymes related to the topic (Table 1). So far, its prime focus has been on collecting data on microbial diversity and intracellular events; however, the database can later be expanded with extracellular enzymes and reactions (such as laccases and peroxidases), as these play an important role in microbial degradation of native lignin and can be applied for enzymatic depolymerzation of technical lignins (Bourbonnais et al. 1995; Pardo et al. 2018; Zhao et al. 2016).
In practice, the data in eLignin is retrieved from scientific literature (peer-reviewed articles, reviews, and books), manually curated and supplemented with links to relevant entries in other well-established biological and chemical databases (e.g., GenBank (Benson et al. 2012), KEGG (Kanehisa and Goto 2000), PubChem (Kim et al. 2015), and ChEBI (Hastings et al. 2012)). The initial dataset was collected by performing a systematic literature review according to the Kitchenham protocol (Kitchenham 2004), where 561 articles (title, abstract, and keywords) were screened and analyzed for their inclusion in the database bibliome. Since the eLignin dataset originates from scientific literature, users are encouraged to read the primary references for any data of interest, since there will be aspects of the data that are not indexed or reviewed by eLignin (such as experimental conditions). Due to the nature of the data collection for eLignin (scientific publications), there will be some overlap with other biological databases such as MetaCyc (Caspi et al. 2015), GenBank (Benson et al. 2012), or UniProt (UniProtConsortium 2017), when it comes to information on pathways, genes, and enzymes. As we do not strive to master features that already established databases already do, eLignin entries are annotated with links to specialized databases where possible.
Two major entry points were considered for eLignin: a microorganism- and a substrate-oriented search (Fig. 2). This design choice was made in order to cater to what we foresee are the two most common information needs both in fundamental and applied lignin microbial conversion: (i) What substrates can my microbe of choice breakdown and/or utilize?; (ii) What microorganism can I use to consume the lignin and lignin-derived aromatics in my substrate stream? Using these entry points, we will now describe the current state of the bibliome and use eLignin content to map and discuss the presently known diversity of the lignin microbial niche.
The microbial diversity in the lignin niche, as reported in the eLignin bibliome
Lignocellulose degradation through cellulolytic activity has been found to be distributed in a wide range of genera within the Bacteria, Archaea, Fungi, and Animalia kingdoms (Cragg et al. 2015). However, the known lignin-degrading subset of lignocellulose degraders is so far limited to a few bacterial and fungal phyla (Janusz et al. 2017; Tian et al. 2014). Mineralization of the lignin requires two main steps: (1) breakdown of the lignin macropolymer to yield smaller aromatic compounds and (2) ring fission of the resulting aromatic compounds (Tuor et al. 1995). The first step is carried out by microbes able to secrete extracellular enzymes with lignolytic and/or lignin-modifying activities such as laccases and peroxidases—typically wood-decaying fungi and certain bacterial species (Bugg et al. 2011b; Janusz et al. 2017; Sigoillot et al. 2012) (Fig. 1). The resulting heterogeneous mixture of aromatic breakdown products is then metabolized by the lignolytic secreters themselves or by other microorganisms in the vicinity capable of aromatic catabolism (Cragg et al. 2015). This leads to the establishment of a microbial niche that favors microbes with matching substrate specificity for the resulting aromatic compounds and with tolerance to the often inhibitory or toxic nature of the aromatics (Díaz et al. 2013; Krell et al. 2012; Schweigert et al. 2001). During catabolism, the aromatic breakdown products are typically shunted through a number of reactions that are collectively referred to as funneling pathways (Harwood and Parales 1996)—or sometimes upper pathways (Linger et al. 2014)—that eventually converge on a couple of conserved ring fission pathways where the aromatic rings are cleaved and the subsequent metabolites enter the central carbon metabolism (Fuchs et al. 2011). Because of these two main steps (depolymerization and ring fission), the lignin microbial niche can be said to contain two main groups of microbes: lignin macropolymer degraders and degraders of lignin-derived aromatic compounds (with the former often being capable of the latter (Nakamura et al. 2012)), from here on referred to as niche subgroups 1 and 2 (Fig. 1). The eLignin database aims to index both, and for the remainder of the minireview, the concept of the lignin microbial niche will be used to refer to all microbes that are capable of degrading lignin and lignin-derived aromatic compounds. Subgroup 2 is of importance for applied studies aiming to, e.g., valorize chemically depolymerized lignin, or to perform in situ bioremediation, and thus, an extra effort has been put on this group in the eLignin database.
Within the applied side of lignin bioconversion, a quick survey of the recent literature shows that a substantial amount of research articles focus on a few commonly used model organisms such as Pseudomonas putida (Linger et al. 2014), Sphingobium sp. (Masai et al. 1999), Rhodococcus jostii (Sainsbury et al. 2013), and Rhodococcus opacus (Kosa and Ragauskas 2012). Reviews on microbial lignin degradation that often include large tables with important isolates (Abdelaziz et al. 2016; Bugg et al. 2011a; Tian et al. 2014) are seldom listing more than ~ 50 different microbes. Still, over 250 microorganisms with lignin and lignin-derived aromatics catabolic activity are currently mapped in the eLignin bibliome (Tables 1, 2, and 3), which indicates its usefulness for meta-analysis of the field.
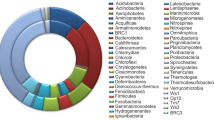
The listed species in the current dataset of eLignin are distributed over 90 different genera, which in turn can be classified into six bacterial phyla (Acidobacteria, Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria, and Spirochaetes) and two fungal phyla (Ascomycota and Basidiomycota) (see Tables 2 and 3). However, the majority of the microbes belong to five of the eight observed phyla: Proteobacteria (114 species/strains), Basidiomycota (58 species/strains), Actinobacteria (31 species/strains), Ascomycota (27 species/strains), and Firmicutes (22 species/strains) (Tables 2 and 3). Evidence of some aromatic-degrading archaea (of the kingdom of Euryarchaeota) is also beginning to emerge (Emerson et al. 1994; Erdoğmuş et al. 2013; Khemili-Talbi et al. 2015). Overall, the large occurrence of Proteobacteria is noteworthy, and the species of this phylum are indeed enriched in studies of isolates found from lignin-rich environments and selected on growth on lignin and aromatic compounds (Jimenez et al. 2002; Jurková and Wurst 1993; Kuhnigk and Konig 1997; Narbad and Gasson 1998; Overhage et al. 1999; Perestelo et al. 1996; Ravi et al. 2017). Likewise, when the same organisms were analyzed for their origin of isolation, it was clear that a majority originated from soil and from the forest ground layer (Table 4), which is probably the most expected ecosystem for the lignin microbial niche (Cragg et al. 2015; Harwood and Parales 1996) given the abundance of lignocellulose in different states of decay found in there.
The following subsections will discuss the outcome of the analysis of the database content in terms of fungal, bacterial, and archaeal diversity. Also, in order to complement the pure isolate approach of the database, the last subsection will discuss microbial communities.
Fungal diversity
The fungi listed in the database are either of the wood rot-type or yeasts. Wood-decaying, or wood-rot, fungi are found within the Basidiomycota and Ascomycota phyla and can be divided into three different types that all have lignin-modifying activities to various extent: soft-rot, brown-rot, and white-rot fungi (Hatakka 2005; Janusz et al. 2017). Soft-rot fungi tend to prefer hardwood and seem to only weakly affect lignin (Sigoillot et al. 2012), but a few species have been reported to exhibit white-rot–like activity toward the end of the wood decay (Pildain et al. 2005). Brown-rot fungi, which are mainly found in the Basidiomycota phylum, selectively attack hemicellulose and cellulose and leave a modified (e.g., dealkylated, demethoxylated, and/or demethylated) lignin signified by its brown color (hence the name of this group of wood degraders); they are primarily found in softwood ecosystems (Hatakka 2005; Sigoillot et al. 2012). Finally, white-rot fungi can degrade all three main components of lignocellulose, i.e., hemicellulose, cellulose, and lignin, and leave a decayed wood with a bleached color (Blanchette 1984; Eriksson et al. 1980; Sigoillot et al. 2012). White-rots are the only wood-decaying fungi that can completely degrade lignin to CO2 and H2O; however, it has been proposed that lignin cannot be used as the sole carbon source by white-rots; rather, the lignin degradation is probably a process that the fungi use to access the cellulose and hemicellulose (ten Have and Teunissen 2001). Like brown-rot fungi, white-rot fungi mostly belong to the Basidiomycota phylum and to a smaller extent to the Ascomycota (Sigoillot et al. 2012).
Both brown-rot and white-rot fungi invade the wood cell lumen by hyphal growth and secrete their lignocellolytic enzymes (Kirk and Farrell 1987; Leonowicz et al. 1999). The lignolytic mechanisms of white-rot fungi secretome have been thoroughly studied (Leonowicz et al. 1999; ten Have and Teunissen 2001). The known lignolytic enzymes (e.g., lignin peroxidases, manganese peroxidases, versatile peroxidases, and laccases (Janusz et al. 2017)) work by nonspecific oxidation, and although nucleophilic cleavage can be used for chemical depolymerization of lignin (e.g., in kraft pulping), the highly variable tertiary structure of lignin could explain why no nucleophilic lignolytic enzymes have been described (Hammel and Cullen 2008). The level and patterns of decay vary between different fungal species and the type of wood (Worrall et al. 1997) as well as the state of decay of the wood. Fukasawa and colleagues subjected beech wood in varying levels of decay to different fungal species and were able to demonstrate that the Basidiomycota caused its highest weight loss in nondecayed wood, whereas the assayed Ascomycota caused more weight loss in predecayed wood (Fukasawa et al. 2011), which implicates that there is a sequential order within the lignin microbial niche with different types of fungi taking turns for degrading the (residual) wood. A very thorough indexing of microbes (primarily fungi) that secrete lignolytic enzymes can be found in a supplemental table of the review of Janusz et al. (2017).
The 85 eukaryotes (fungi and yeasts from 43 different genera) currently listed in eLignin are distributed between Ascomycota and Basidiomycota (Table 3) and were primarily isolated from soil and forest environments (Table 4), which is in accordance with other reviews of the ecological occurrence (Janusz et al. 2017). The clinical isolates reported in Table 4 are mainly different species of Candida yeasts, which aside from their opportunistic pathogenicity in humans, are known degraders of lignocellulose-derived compounds such as xylose and different aromatics (Gérecová et al. 2015; Holesova et al. 2011; Jeffries 1981; Krug et al. 1985). In general, the yeasts species in the database are aromatic degraders and not lignin degraders (Bergauer et al. 2005; Holesova et al. 2011; Middelhoven 1993; Yaegashi et al. 2017) and, therefore, play a role in the niche as degraders of lignin breakdown products. Three species in the dataset have, however, been reported to have activity on lignin: Rhodotorula sp. R2 modified wheat straw and Sarkanda grass (Hainal et al. 2012), whereas Geotrichum klebahnii CCY 74-6-2 and Trichosporon pullulans CCY 30-1-10 acted on beechwood lignin fractionated from the prehydrolysis step of kraft pulping (Sláviková and Košíková 2001; Sláviková et al. 2002).
When it comes to lignin-degrading activity, fungi tend to be more studied than bacteria because of their higher prevalence of lignolytic secretomes (Janusz et al. 2017). However, if the system boundaries are expanded to include the whole lignin aromatic niche, i.e., the species that lack delignification activities but grow on the lignin-derived aromatic compounds (Fig. 1), the ratio between fungi and bacteria could be rather different. In eLignin, which was built on this niche principle, there are about two times as many bacterial isolates listed as fungal ones (Tables 1, 2, and 3). We cannot determine if this is a bias in the literature, comes from the database boundaries (which were initially created with a focus on intracellular events, and not on secreted enzymes), or if the “true” diversity holds less fungal species than bacterial. The number of wood-rotting Basidiomycetes has been estimated to up to 1700 species in North America only, but the number of lignolytic fungi is unknown (Gilbertson 1980; Janusz et al. 2017).
Bacterial diversity
By using the holistic ecological approach to list both degraders of lignin and lignin-derived aromatic compounds, 171 different bacteria distributed over 63 different genera have been indexed in eLignin at the time of writing (Table 2). As mentioned above, three main phyla encompasses the bulk of the dataset (Proteobacteria, Actinobacteria, and Firmicutes), with Proteobacteria dominating the list with its 114 entries (Table 2). Within these Proteobacteria, γ-Proteobacteria was the main class (66 species/strains), followed by β-Proteobacteria (27 species/strains), α-Proteobacteria (18 species/strains), and δ-Proteobacteria (3 species/strains), again highlighting that certain types of microbes are greatly enriched in the eLignin bibliome. It can also be noted that many of the organisms in this particular niche have undergone one or several taxonomical reclassifications since they were first isolated and described (see, e.g., Cupriavidus necator which was previously known as, e.g., Ralstonia eutropha and Wautersia eutropha (Vandamme and Coenye 2004)), meaning that the binomial names in articles from the 1960–1980s may be different from the currently prevailing names. Therefore, the organism entry in the database has, when possible, been harmonized with links to the corresponding entry in the NCBI Taxonomy Database (https://www.ncbi.nlm.nih.gov/taxonomy; Acland et al. 2014).
The Gram stain distribution tends to follow the phyla and, thus, is dominated by Gram-negative bacteria (121 species/strains), with the remainder being Gram-positive (46 species/strains) and unknown/Gram-indeterminate (4 species/strains). This may have implication on studies focusing on, e.g., transport of compounds over membranes (discussed in a separate section below), or when expanding a species’ substrate range by metabolic engineering. In the latter case, the difference in total GC content in the genome that is in general seen between Gram-positives and Gram-negatives (Muto and Osawa 1987) will affect the feasibility of heterologous expression if using traditional PCR-based cloning.
Although fungi are known as the main degraders of the lignin macropolymer (as described in the previous subsection), there are a substantial number of studies that describe delignifying bacteria. Tian et al. reviewed the topic and performed phylogeny on 57 lignin-degrading and 463 laccase-encoding prokaryotes that led them to propose that screening for laccases genes may be a good way to detect new lignin-degrading species (Tian et al. 2014). Furthermore, the authors suggest that aromatic metabolism is a prerequisite for but not a proof of lignolytic activity (Tian et al. 2014), which is in line with our division of the lignin bacterial niche into subgroups 1 and 2 that specialize in different aspects of the full lignin catabolism (Fig. 1). The metabolism of the resulting lignin breakdown products, which mainly takes place intracellularly, will be discussed in the “Distribution of metabolic pathways and substrate specificities” section below.
Soil is absolutely the most common origin of isolation mapped in the database (Table 4), which also reflects how popular this environment has been for studies on isolation of lignin and aromatic degraders. Other than soil, termite guts are a main origin of isolation. There seems to be no clear evidence that the termites themselves are able to degrade lignin (instead they live of the hydrolysis products of hemicellulose and cellulose) (Brune and Ohkuma 2010). The lignin barrier is overcome by the termites by a symbiotic relationship with a diverse microbial community, e.g., by exosymbiotic fungi and endosymbiotic gut flora (Maurice and Erdei 2018). Examples of aromatic degrading bacteria isolated from the gut flora include Proteobacteria (Harazono et al. 2003; Kuhnigk and Konig 1997; Suman et al. 2016; Tsegaye et al. 2018; Van Dexter and Boopathy 2018), Actinobacteria (Chung et al. 1994; Kuhnigk and Konig 1997; Watanabe et al. 2003), and Firmicutes (Kuhnigk and Konig 1997), as well as the only Spirochaetes entry in the database (Lucey and Leadbetter 2014). Another enrichment reported in Table 4 for bacteria is the isolates from different man-made environments. One example is pulp and paper mill effluents that contain residual lignins and aromatics and have been a source of many isolates (Chandra et al. 2007; Duan et al. 2016b; González et al. 1997; Hooda et al. 2015; Mathews et al. 2014; Nishikawa et al. 1998; Ravi et al. 2018); likewise, sludge from waste water treatment plants has been a source of a number of isolates, some of which are strictly anaerobic (Gorny et al. 1992; Mechichi et al. 1999, 2005; Ni et al. 2013; Traunecker et al. 1991; Tschech and Fuchs 1987).
Anaerobic aromatic degrading bacteria are in a minority compared to the aerobic fission bacteria and were even for a long time believed to be impossible (Kirk and Farrell 1987). However, with recent advances in the field, the molecular biology of these pathways has begun to be understood (Durante-Rodríguez et al. 2018). Some examples found in the database include, e.g., Pelobacter acidigallici Ma Gal2 (Schink and Pfennig 1982), Desulfobacterium phenolicum Ph01 (Bak and Widdel 1986), Rhodopseudomonas palustris CGA001 (Harwood and Gibson 1988), Clostridium thermoaceticum ATCC 39073 (Daniel et al. 1988), and Dysgonomonas sp. WJDL-Y1 (Duan et al. 2016b); Holophaga foetida TMBS4 is also worthy of mention as it the only observed species in the Acidobacteria phylum reported in the database, and it grows anaerobically on a couple of typically lignin-derived aromatics such as ferulic acid and syringic acid (Bak et al. 1992).
Archaeal diversity
Of the three domains in the Woeseian system (Woese et al. 1990), archaea is the most underrepresented in the lignin microbial niche. To our knowledge, there are no reported archaeal single culture isolates with lignolytic capacity at the time of writing. Recently, by enrichment cultures from estuarine sediment, it was possible to infer growth of Bathyarchaeota on alkali lignin by the increase in gene-copy number and the incorporation of inorganic carbon in the archaeal lipids over 11 months (Yu et al. 2018). Likewise, putative laccase genes have been reported in some archaeal species (Ausec et al. 2011; Sharma and Kuhad 2009; Tian et al. 2014). A laccase from Haloferax volcanii DS70 has been purified with activity on model compounds such as syringaldazine and ABTS (Uthandi et al. 2010). However, to our understanding, the in vivo lignolytic activity of these putative and purified laccases remains to be assayed.
Five archaeal isolates—classified in niche subgroup 2 (growth on aromatics; Fig. 1)—have so far been indexed in eLignin, all of them being halophiles, i.e., extremophiles that prefer high salt concentration. Haloferax sp. D1227 was isolated from soil and grew on benzoic, cinnamic, and phenylpropanoic acid (Emerson et al. 1994). Haloferax sp. C-24, Halorubrum ezzemoulense C-46, and Haloarcula sp. D1 were isolated from high-saline samples and grew on, e.g., 4-hydroxybenzoic acid (Erdoğmuş et al. 2013; Fairley et al. 2002). Natrialba sp. C21 degraded phenol (Khemili-Talbi et al. 2015). The halophilic nature of these isolates and the lack of known lignolytic activity seem to suggest that they contribute with the degradation of aromatic breakdown products that have ended up in saltwater environments, which could be speculated to be a downstream (or downriver) extension of the lignin microbial niche.
The communities of the lignin microbial niche
Lignin degradation is a community effort and is in itself often a subpart of a lignocellulose-degrading niche (de Boer et al. 2005). Microbial communities—organisms that live and interact within a contiguous environment (Konopka 2009)—are in a way what we are illustrating by looking at the isolates from the point of the niche subgroups (Fig. 1). It has been proposed that lignin degradation is more rapid with consortia than single isolates due to synergism (Wang et al. 2013). Furthermore, studies on fungal–bacterial interactions in the lignin microbial niche have reported examples of commensalism as well as amensalism between certain species: some bacteria have been reported to promote growth of a white-rot fungi when co-cultivated (Harry-asobara and Kamei 2018), and there is a report showing two different white-rot species outcompeting opportunistic bacteria (Folman et al. 2008). At the moment, consortia are not mapped in eLignin but are nevertheless important for the understanding of the lignin microbiology.
Many studies have reported physiological characterization of a community with unknown or partly known composition, either because it was not possible to isolate single cultures with the desired phenotype—for instance, 99% of the bacteria in soil have been estimated to be unculturable (Pham and Kim 2012)—or because the aim was to study the community effort. Examples include communities capable of degrading lignin (DeAngelis et al. 2011; Wang et al. 2013; Wu and He 2013), syringic acid (Kaiser and Hanselmann 1982; Phelps and Young 1997), resorcinol and catechol (Milligan and Häggblom 1998), coniferyl alcohol (Grbić-Galić 1983), and plant lignin–soil community studies (Bennett et al. 2015; Bradley et al. 2007), to name a few. Many of these studies were reported under anaerobic conditions.
Another approach to analyze microbial communities is to consider the makeup of the metagenome as a unique property of a given community (Konopka 2009). 16S rRNA sequencing can be used to taxonomically identify members of a community (González et al. 1996). A common methodology is to divide the results of the 16S rRNA sequencing of a metagenome into operational taxonomic units (OTUs) to attempt to resolve, e.g., phylum level abundances (Moraes et al. 2018); this is similar to what is done here with the eLignin database using single isolates (Tables 2 and 3). In addition to taxonomical metagenomics, Moraes and colleagues reconstructed draft bacterial genomes from a lignin-degrading consortium and could identify conserved domains related to lignin degradation in their metagenome (Moraes et al. 2018).
Distribution of metabolic pathways and substrate specificities
The lignin macropolymer is primarily depolymerized by extracellular enzymes secreted by lignolytic microbes. Due to its heterogeneity, the resulting depolymerization products are commonly a mixture of different mono- and di- and oligoaromatic compounds (Bugg et al. 2011b). This has led to the evolution of a panel of intracellular funneling pathways, i.e., metabolic routes that connect substituted aromatic compounds with a ring fission pathway leading to the central carbon metabolism, often (but not always) via acetyl-CoA (Fig. 3). In this section, the eLignin database was used to assess the diversity of substrates and metabolic routes within the lignin microbial niche.
Schematic distribution of the known pathways for aromatic catabolism currently indexed in the eLignin database. Please note that this representation should be seen as a hypothetical map of the existing possibilities within aromatic catabolism, and not as a map of a “superbug.” Funneling pathways refer to routes that reduce larger/more substituted aromatic compounds down to the different catabolic nodes from where ring fission occurs (here called fission pathways). The three routes that funnel compounds derived from the primary monolignols (S, H, G) are indicated in dotted boxes: the sinapyl (S), p-coumaryl (H), and coniferyl (G) branches
Reported substrate specificities
Similar to how fundamental and applied studies on lignin focus on a few model organisms, many studies use a few common model aromatic model compounds that represent different funneling pathways (e.g., 4-hydroxybenzoic acid, vanillic acid, ferulic acid, p-coumaric acid, and benzoic acid) to evaluate the physiology of the microbial niche (see, e.g., Fischer et al. 2008; González et al. 1997; Kosa and Ragauskas 2012; Ravi et al. 2017; Vardon et al. 2015). However, from browsing eLignin, there appears to be a much higher substrate diversity in this niche than just these model compounds. This is illustrated in Fig. 4a, showing a meta-analysis of the “most popular” substrates in the eLignin bibliome in terms of the number of different microbes that have been reported in the literature to degrade them. Evidently, the model aromatics are in the top, which both suggest that they indeed are good model compounds for the different funneling pathways and that they have been popular choices for the experimental work that has been published on this topic. In addition, some natural and technical lignins (corn stover, kraft, Klason, and alkaline lignin), “synthetic” oligoaromatics (dehydropolymerisate), and dimers (biphenol, benzylvanillin) are among these top 32 substrates (Fig. 4a). The number of microbes in the database that have been reported to degrade natural and technical lignins and di-/oligoaromatics is presented in Fig. 4b. The results show that fungi are the most prevalent degraders of natural lignins, which is reasonable given the high diversity of lignolytic fungi. The reported technical lignins include chemically modified lignin polymers as well as chemically depolymerized lignin (i.e., a mixture of both high (polymeric) and low molecular weight lignins (mono- and oligomers)) which explains the high number of bacteria that have been reported to grow on technical lignins. Di- and oligoaromatic compounds were primarily reported in Proteobacteria in eLignin, but this is likely a literature bias since (model) monoaromatic compounds tend to be more commonly studied across all phyla. Note that there are no Acidobacteria or Spirocheates in the eLignin bibliome that have been reported to degrade natural/technical lignins and di-/oligoaromatics.
a Substrates that can be utilized by > 10 organisms listed in the database; the numbers represent the number of strains in the database that utilize each compound. Total number of substrates that satisfied the > 10 cutoff—32; total number of substrates in dataset—141. b Number of species that can degrade natural and technical lignins, and di- and oligomeric aromatic compounds, sorted by phylum. To distinguish the bacteria from the representatives of the other two kingdoms, the fungal phyla are presented with stripes and the only archaeal phylum is in solid black
It is equally important to know the substrates that cannot be used by a given organism, as this will give the limitations of its metabolism. In fact, many isolation papers both list substrates that can and that cannot support growth (for a few examples, see Bache and Pfennig 1981; Defnoun et al. 2000; Harwood and Gibson 1988; Song 2009) and thereby present a valuable hint to which funneling pathways can and cannot be expected in the organism. At the moment, the indexing in eLignin has been focused on the substrates that can be used, but a logical next step for the database development is to also include substrates that an organism cannot use.
Prediction of funneling pathway distributions
Lignin consists of three primary building blocks known as monolignols that plants produce from the amino acid phenylalanine: sinapyl alcohol (called syringyl, or S, unit when incorporated in the lignin polymer), coniferyl alcohol (guaiacyl unit; G), and p-coumaryl alcohol (p-hydroxyphenyl unit; H) (Vanholme et al. 2010). The ratio of units in the polymer differs depending on the lignin source, with softwood consisting of mainly G units with a small fraction of H units, hardwood having a combination of almost exclusively S and G, and monocots all three (Gellerstedt and Henriksson 2008; Gosselink et al. 2010). Recent reports have also shown that a caffeyl alcohol homopolymer (caffeyl unit; C) can be found in seed coats of, e.g., vanilla orchard and some cacti species (Barsberg et al. 2018; Chen et al. 2012a). Consequently, the composition of aromatics in the depolymerized lignin will differ greatly between different lignocellulose feedstocks.
Following the S, G, and H types, three main funneling pathways for monoaromatic catabolism have been defined, based on which of the main lignin units (or derivatives thereof) they catabolize: the sinapyl branch (two methoxy groups), coniferyl branch (one methoxy group), and the p-coumaryl branch (no methoxy groups) (see Fig. 3). Within eLignin, these branches were further divided into one or more sequential pathways in order to better specify which reactions a species have been characterized with, i.e., a bacteria with a vanillin degradation pathway will not necessarily have the pathway for ferulic acid, although these pathways are sequential in the coniferyl branch. Microbial aromatic catabolism is also not limited to the S, G, and H funneling branches, meaning that there is a need for naming of other routes as well, including aromatics that are derived from other origins than lignins (e.g., other plant matter). Some examples include the caffeic acid, benzoyl, resorcinol, and cresol pathways (Fig. 3). Funneling pathways for di- and oligomeric aromatics, the study of which has started emerging in certain species (Bugg et al. 2011b; Kamimura et al. 2017), is another example of essential catabolic routes.
In a lot of bibliome studies, the substrate specificity of a species is presented without going into the intracellular conversion mechanisms nor reporting evidence of a specific funneling pathway. Therefore, in order to be able to use the eLignin dataset to look at pathway diversity, we developed a prediction algorithm to infer funneling branches from reported substrates from the literature. This is possible since many of the funneling branches are linear, e.g., ferulic acid is degraded via vanillin, and any species that have been reported to grow on these compounds and their intermediates can then be theoretically inferred to have the coniferyl branch (Fig. 3). Cinnamic acid is reported to be catabolized by 18 organisms (Fig. 4a), but due to the alternate metabolic routes for its degradation—e.g., via benzoic acid, 3-phenylpropionic acid, or styrene (Chamkha et al. 2001; Defnoun et al. 2000; Monisha et al. 2018; Mäkelä et al. 2015)—it was omitted from the prediction model. Also, according to current knowledge, anaerobic aromatic catabolism frequently (but not exclusively) relies on pathways that converge on benzoyl-CoA, that is further subjected to ATP-dependent hydrolysis to open the aromatic ring (Durante-Rodríguez et al. 2018; Fuchs et al. 2011); but since the exact mechanisms are largely unknown for the species in the dataset, all anaerobic microbes have been put in an “anaerobic branch (es)” cluster (Fig. 5).
Distribution of putative funneling pathway branches in the eLignin bibliome, inferred from reported substrates, sorted by phylum. Because of the overall linear nature of the aerobic funneling pathways (Fig. 3), it is possible to use the substrates reported in the literature for a given organism and correlate that to a funneling pathway branch (i.e., a collection of funneling pathways). The small group of species that has been reported to degrade aromatics anaerobically has all been clustered in the anaerobic branch in order not to generate false positives in the other branches. To distinguish the bacteria from the representatives of the other two kingdoms, the fungal phyla are presented with stripes and the only archaeal phylum is in solid black. A detailed outcome of the prediction for each species with links to the different references is found online at www.elignindatabase.com under each organism entry page. Please note that the results are theoretical and it is up to everyone to assess the probability of these inferences, e.g., by reading the primary references for each organism
The result of the theoretical prediction is presented in Fig. 5. The main conclusion is that, of the three main funneling branches (S, G, H), the coniferyl (G) and p-coumaric (H) branches seem by far to be the most abundant in niche 2. This might be correlated to the number of methoxy groups (none in the H unit, one in the G unit, two in the S unit; Fig. 3), as ring fission usually seems to occur after the methoxy groups have been demethylated to hydroxyl groups (Gupta et al. 1986; Nishikawa et al. 1998; Sampaio 1999). As the demethylation often requires a cofactor such as tetrahydrofolic acid (Masai et al. 2004) and NADH and FAD (Mallinson et al. 2018), the degradation of methylated aromatics may be limited by the rate of cofactor recycling. Furthermore, it is noteworthy that there is no caffeic acid degrading Actinobacteria yet in the eLignin database, despite the fact that they are the prokaryotic phylum that is commonly the second most abundant for most branches in the dataset (Fig. 5). Another observation is that metabolites of the resorcinol branch (Fig. 3) seem to be degraded by fungi to a larger extent than the other branches according to the current data (Fig. 5). Resorcinols are part of the phenolics in plants and soil humic acids (Burges et al. 1964; Kluge et al. 1990) and do not seem to be derived from lignin per se, which would put this compound within niche subgroup 2.
Predicting organisms that can catabolize a given depolymerization mix
Many lignin valorization studies apply chemical depolymerization since microbial enzymatic breakdown of lignin is a very slow process taking many weeks (Fackler et al. 2006; Hedges et al. 1988; Liers et al. 2011). Therefore, from an applied point-of-view, it would be of interest to run the prediction model “backwards” in order to identify which organism(s) would be likely to grow on the mixture of aromatic monomers resulting from chemical depolymerization. The outcome of the depolymerization is highly dependent on process conditions and lignin source (Sun et al. 2018), and predicting the monomeric yield is beyond the scope of this review. However, the distribution of H, G, and S units in a given lignin might be indicative of the possible monomeric composition in the depolymerisate. Using this assumption, depolymerized softwood lignin would need microbes with funneling pathways for coniferyl- (G) and p-coumaric (H)-derived monomers. Spruce lignosulfonate has for instance been reported to yield vanillin, guaiacol, acetovanillone, and vanillic acid (Pérez and Tuck 2018). Some examples of organisms that can catabolize both vanillic acid and guaiacol include Amycolatopsis sp. ATCC 39116 (Pometto III et al. 1981), Comamonas sp. B-9 (Chen et al. 2012c), and Rhodotorula rubra IFO 889 (Huang et al. 1993). Hardwood depolymerisates would require species that can handle monomers derived from S and G units, and therefore, organisms with the syringyl (S) and coniferyl (G) branches would be needed, such as Sphingobium sp. SYK-6 (Katayama et al. 1988), Acetobacterium woodii NZva16 (Bache and Pfennig 1981), or Rhizobium sp. YS-1r (Jackson et al. 2017). Species that seem able to degrade compounds from all the S, G, and H branches, which would be representative of grass lignins, would include Oceanimonas doudoroffii JCM21046T (Numata and Morisaki 2015) and Exophiala jeanselmei CBS 658.76 (Middelhoven 1993). Please note that these predictions do not take culture and process conditions into account, meaning that some of these species might be better suited for process applications than others.
Transport proteins
Although the chemical structure of many aromatic compounds allow them to passively diffuse through the lipid bilayers of biological membranes (Engelke et al. 1996), many microorganisms have dedicated transport channels or proteins for aromatic compounds—reviewed, e.g., by Parales and by Kamimura and their colleagues (Kamimura et al. 2017; Parales and Ditty 2017; Parales et al. 2008). In fact, transporter genes are commonly found within the catabolic operons for aromatic acids (Parales et al. 2008) which could suggest that the natural diffusion rate of certain aromatics is too limited for growth on aromatics as a sole carbon source. Transporters are of interest for metabolic engineering purposes, as a part of uptake optimization and/or expansion of the substrate range of a given strain. As more and more of the metabolic pathways for aromatic degradation are now elucidated, there seems to be an emerging effort within the fundamental molecular biology studies on lignin degradation to look into transport proteins. We have begun indexing transport proteins as part of the organism pages in eLignin, and we anticipate that this section will grow as this field expands.
Current knowledge on bacterial aromatic transporters mostly focuses on Gram-negative bacteria which have a cell envelope with two lipid bilayers separated by a periplasmic space: the outer and the inner membrane (Nikaido 2003). Some Gram-negatives have been reported to have substrate-specific diffusion channels for aromatic compounds on the outer membrane (Hearn et al. 2008; Nikaido 2003). Inner membrane transport of aromatic acids seems to be achieved by active transporters and not by diffusion in many species. This may be explained by the fact that these compounds are commonly protonated at neutral pH and—due to the hydrophobic charge—can partition into the membrane and damage the structure (Kamimura et al. 2017; Parales and Ditty 2017). Gram-negative bacteria with reported aromatic transporters include Acinetobacter baylyi ADP1 (Collier et al. 1997; D’Argenio et al. 1999), Bradyrhizobium japonicum USDA110 (Michalska et al. 2012), Klebsiella pneumoniae M5a1 (Xu et al. 2012), Pseudomonas putida KT2440 (Nishikawa et al. 2008) and PRS2000 (Nichols and Harwood 1997), Rhodopseudomonas palustris CGA009 (Giuliani et al. 2011; Michalska et al. 2012), Sinorhizobium meliloti 1024 (Michalska et al. 2012), and Sphingobium sp. SYK-6 (Mori et al. 2018). Gram-positives, on the other hand, only have a single cell membrane in their envelope: the cytoplasmic membrane (Parales and Ditty 2017). There seem to be less studies on Gram-positive than Gram-negative species with regard to aromatic transport. Some examples include Corynebacterium glutamicum ATCC 13032 (Chaudhry et al. 2007; Xu et al. 2006), Lactobacillus plantarum WCFS1 (Reverón et al. 2017), and Rhodococcus jostii RHA1 (Otani et al. 2014). It is also worthwhile to note that in addition to the mechanisms for transport of aromatics into the cell, many species also have efflux pumps in order to cope with the often cytotoxic properties of aromatic compounds (Parales and Ditty 2017), or as a means to excrete detoxified compounds. Ravi and colleagues have for instance described a Pseudomonas isolate that excreted vanillyl alcohol during growth on vanillin as a tolerance mechanism to handle excess vanillin that was not catabolized to vanillic acid fast enough, but the mechanism by which vanillyl alcohol was transported out of the cell has not been elucidated yet (Ravi et al. 2018).
Conclusions and outlook
The interest for lignin as an underexploited carbon source has markedly increased during the last two decades, as evidenced by the exponential increase in published papers on lignin valorization (Abejón et al. 2018). In this minireview, we used our recently created resource, the eLignin database, to analyze the diversity of the lignin microbial niche, which we have defined as all microbes that can either degrade lignin or lignin-derived aromatic compounds. It should, however, be kept in mind that the data in eLignin encompasses the diversity in the bibliome, meaning that it reflects what people have reported in the literature. The papers that are indexed in the database concern microbial isolates, i.e., species that were cultivable. It is, therefore, inevitable that this approach does not represent the overall diversity of the lignin microbial niche, as there are many species within the niche community that cannot be detected and sustained with the common isolation methodologies. Although the aim of this minireview is to show the diversity of the niche, it also reveals the diversity and fashions within the scientific community, which may or may not correlate with the biological diversity. We can also conclude that the literature is enriched with physiological characterization, i.e., aromatic substrate specificities of different organisms are rather well known. The molecular biology of specific metabolic routes is, in contrast, less well elucidated, which will be an important next step both for the fundamental understanding of the biology and for the many projects that apply microbes in a value chain for lignin valorization. The prediction algorithm for aromatic pathways presented in this review can hopefully generate new hypotheses on the molecular biology of the niche and pave the way for future studies.
The microbiological aspects of lignin and aromatics degradation have a long history with a vast bibliome, and the need for resources such as the eLignin database will continue to grow as the field expands. In the future, we expect to further implement in eLignin a number of discussed features including improved prediction algorithms, lignolytic communities, and substrates that cannot be converted by a given organism. Economically feasible lignin valorization will require advanced metabolic engineering and thorough knowledge on microbial physiology. In that context, the objective of eLignin is not only to generate new overviews of the field but also to fuel new research ideas and engineering strategies and thus become an operational tool for studies on the microbiological aspect of lignin degradation, catabolism, and valorization.
References
Abdelaziz OY, Brink DP, Prothmann J, Ravi K, Sun M, García-Hidalgo J, Sandahl M, Hulteberg CP, Turner C, Lidén G, Gorwa-Grauslund MF (2016) Biological valorization of low molecular weight lignin. Biotechnol Adv 34(8):1318–1346
Abejón R, Pérez-Acebo H, Clavijo L (2018) Alternatives for chemical and biochemical lignin valorization: hot topics from a bibliometric analysis of the research published during the 2000–2016 period. Processes 6(8):98
Acevedo F, Pizzul L, Castillo MD, Cuevas R, Diez MC (2011) Degradation of polycyclic aromatic hydrocarbons by the Chilean white-rot fungus Anthracophyllum discolor. J Hazard Mater 185(1):212–219. https://doi.org/10.1016/j.jhazmat.2010.09.020
Acland A, Agarwala R, Barrett T, Beck J, Benson DA, Bollin C, Bolton E, Bryant SH, Canese K, Church DM (2014) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 42(Database issue):D7
Alexieva Z, Yemendzhiev H, Zlateva P (2010) Cresols utilization by Trametes versicolor and substrate interactions in the mixture with phenol. Biodegradation 21(4):625–635
Antai SP, Crawford DL (1981) Degradation of softwood, hardwood, and grass lignocelluloses by two Streptomyces strains. Appl Environ Microbiol 42(2):378–380
Aoyama A, Yamada K, Suzuki Y, Kato Y, Nagai K, Kurane R (2014) Newly-isolated laccase high productivity Streptomyces sp. grown in cedar powder as the sole carbon source. Int J Waste Resour 4(2):1–5
Arora D, Sandhu D (1985) Laccase production and wood degradation by a white-rot fungus Daedalea flavida. Enzym Microb Technol 7(8):405–408
Arumugam N, Kalavathi P, Mahalingam P (2014) Lignin database for diversity of lignin degrading microbial enzymes (LD2L). Research in Biotechnology 5(1):13–18
Ausec L, Zakrzewski M, Goesmann A, Schlüter A, Mandic-Mulec I (2011) Bioinformatic analysis reveals high diversity of bacterial genes for laccase-like enzymes. PLoS One 6(10):e25724
Ayyachamy M, Cliffe FE, Coyne JM, Collier J, Tuohy MG (2013) Lignin: untapped biopolymers in biomass conversion technologies. Biomass Convers Biorefin 3(3):255–269
Bache R, Pfennig N (1981) Selective isolation of Acetobacterium woodii on methoxylated aromatic acids and determination of growth yields. Arch Microbiol 130(3):255–261
Bak F, Widdel F (1986) Anaerobic degradation of phenol and phenol derivatives by Desulfobacterium phenolicum sp. nov. Arch Microbiol 146(2):177–180
Bak F, Finster K, Rothfuß F (1992) Formation of dimethylsulfide and methanethiol from methoxylated aromatic compounds and inorganic sulfide by newly isolated anaerobic bacteria. Arch Microbiol 157(6):529–534
Balkwill DL, Drake GR, Reeves RH, Fredrickson JK, White DC, Ringelberg DB, Chandler DP, Romine MF, Kennedy DW, Spadoni CM (1997) Taxonomic study of aromatic-degrading bacteria from deep-terrestrial-subsurface sediments and description of Sphingomonas aromaticivorans sp. nov., Sphingomonas subterranea sp. nov., and Sphingomonas stygia sp. nov. Int J Syst Evol Microbiol 47(1):191–201
Bandounas L, Wierckx NJP, de Winde JH, Ruijssenaars HJ (2011) Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential. BMC Biotechnol 11: Artn 94. doi:https://doi.org/10.1186/1472-6750-11-94
Barapatre A, Jha H (2017) Degradation of alkali lignin by two ascomycetes and free radical scavenging activity of the products. Biocatal Biotransformation 35(4):269–286
Barsberg ST, Lee Y-I, Rasmussen HN (2018) Development of C-lignin with G/S-lignin and lipids in orchid seed coats–an unexpected diversity exposed by ATR-FT-IR spectroscopy. Seed Sci Res 28(1):41–51
Beckham GT, Johnson CW, Karp EM, Salvachúa D, Vardon DR (2016) Opportunities and challenges in biological lignin valorization. Curr Opin Biotechnol 42:40–53
Benjamin S, Kamimura N, Takahashi K, Masai E (2016) Achromobacter denitrificans SP1 efficiently utilizes 16 phthalate diesters and their downstream products through protocatechuate 3, 4-cleavage pathway. Ecotoxicol Environ Saf 134:172–178
Bennett AE, Grussu D, Kam J, Caul S, Halpin C (2015) Plant lignin content altered by soil microbial community. New Phytol 206(1):166–174
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2012) GenBank. Nucleic Acids Res 41(D1):D36–D42
Bergauer P, Fonteyne P-A, Nolard N, Schinner F, Margesin R (2005) Biodegradation of phenol and phenol-related compounds by psychrophilic and cold-tolerant alpine yeasts. Chemosphere 59(7):909–918
Bi R, Lawoko M, Henriksson G (2016) Phoma herbarum, a soil fungus able to grow on natural lignin and synthetic lignin (DHP) as sole carbon source and cause lignin degradation. J Ind Microbiol Biotechnol 43(8):1175–1182
Billings AF, Fortney JL, Hazen TC, Simmons B, Davenport KW, Goodwin L, Ivanova N, Kyrpides NC, Mavromatis K, Woyke T (2015) Genome sequence and description of the anaerobic lignin-degrading bacterium Tolumonas lignolytica sp. nov. Stand Genomic Sci 10(1):106
Blanchette RA (1984) Screening wood decayed by white rot fungi for preferential lignin degradation. Appl Environ Microbiol 48(3):647–653
Bourbonnais R, Paice M, Reid I, Lanthier P, Yaguchi M (1995) Lignin oxidation by laccase isozymes from Trametes versicolor and role of the mediator 2, 2′-azinobis (3-ethylbenzthiazoline-6-sulfonate) in kraft lignin depolymerization. Appl Environ Microbiol 61(5):1876–1880
Bradley KL, Hancock JE, Giardina CP, Pregitzer KS (2007) Soil microbial community responses to altered lignin biosynthesis in Populus tremuloides vary among three distinct soils. Plant Soil 294(1–2):185–201
Brune A, Ohkuma M (2010) Role of the termite gut microbiota in symbiotic digestion. In: Biology of termites: a modern synthesis. Springer, pp 439–475
Bugg TD, Ahmad M, Hardiman EM, Singh R (2011a) The emerging role for bacteria in lignin degradation and bio-product formation. Curr Opin Biotechnol 22(3):394–400
Bugg TDH, Ahmad M, Hardiman EM, Rahmanpour R (2011b) Pathways for degradation of lignin in bacteria and fungi. Nat Prod Rep 28(12):1883–1896. https://doi.org/10.1039/C1np00042j
Burges N, Hurst H, Walkden B (1964) The phenolic constituents of humic acid and their relation to the lignin of the plant cover. Geochim Cosmochim Acta 28(10–11):1547–1554
Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA (2015) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 44(D1):D471–D480
Chamkha M, Garcia J-L, Labat M (2001) Metabolism of cinnamic acids by some Clostridiales and emendation of the descriptions of Clostridium aerotolerans, Clostridium celerecrescens and Clostridium xylanolyticum. Int J Syst Evol Microbiol 51(6):2105–2111
Chandra R, Bharagava RN (2013) Bacterial degradation of synthetic and kraft lignin by axenic and mixed culture and their metabolic products. J Environ Biol 34(6):991–999
Chandra R, Raj A, Purohit HJ, Kapley A (2007) Characterisation and optimisation of three potential aerobic bacterial strains for kraft lignin degradation from pulp paper waste. Chemosphere 67(4):839–846. https://doi.org/10.1016/j.chemosphere.2006.10.011
Chang AJ, Fan J, Wen X (2012) Screening of fungi capable of highly selective degradation of lignin in rice straw. Int Biodeterior Biodegrad 72:26–30
Chang Y-C, Choi D, Takamizawa K, Kikuchi S (2014) Isolation of Bacillus sp. strains capable of decomposing alkali lignin and their application in combination with lactic acid bacteria for enhancing cellulase performance. Bioresour Technol 152:429–436
Chapman PJ, Ribbons DW (1976) Metabolism of resorcinylic compounds by bacteria: alternative pathways for resorcinol catabolism in Pseudomonas putida. J Bacteriol 125(3):985–998
Chaudhry MT, Huang Y, Shen X-H, Poetsch A, Jiang C-Y, Liu S-J (2007) Genome-wide investigation of aromatic acid transporters in Corynebacterium glutamicum. Microbiology 153(3):857–865
Chen F, Tobimatsu Y, Havkin-Frenkel D, Dixon RA, Ralph J (2012a) A polymer of caffeyl alcohol in plant seeds. Proc Natl Acad Sci 109(5):1772–1777
Chen Y, Chai L, Tang C, Yang Z, Zheng Y, Shi Y, Zhang H (2012b) Kraft lignin biodegradation by Novosphingobium sp. B-7 and analysis of the degradation process. Bioresour Technol 123:682–685
Chen YH, Chai LY, Zhu YH, Yang ZH, Zheng Y, Zhang H (2012c) Biodegradation of kraft lignin by a bacterial strain Comamonas sp B-9 isolated from eroded bamboo slips. J Appl Microbiol 112(5):900–906. https://doi.org/10.1111/j.1365-2672.2012.05275.x
Chong G-G, Huang X-J, Di J-H, Xu D-Z, He Y-C, Pei Y-N, Tang Y-J, Ma C-L (2018) Biodegradation of alkali lignin by a newly isolated Rhodococcus pyridinivorans CCZU-B16. Bioprocess Biosyst Eng 41(4):501–510
Chow KT, Pope MK, Davies J (1999) Characterization of a vanillic acid non-oxidative decarboxylation gene cluster from Streptomyces sp. D7. Microbiology 145(9):2393–2403
Chowdhury SP, Khanna S, Verma S, Tripathi A (2004) Molecular diversity of tannic acid degrading bacteria isolated from tannery soil. J Appl Microbiol 97(6):1210–1219
Chung S-Y, Maeda M, Song E, Horikoshij K, Kudo T (1994) A Gram-positive polychlorinated biphenyl-degrading bacterium, Rhodococcus erythropolis strain TA421, isolated from a termite ecosystem. Biosci Biotechnol Biochem 58(11):2111–2113
Collier LS, Nichols NN, Neidle EL (1997) benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobacter sp. strain ADP1. J Bacteriol 179(18):5943–5946
Cragg SM, Beckham GT, Bruce NC, Bugg TD, Distel DL, Dupree P, Etxabe AG, Goodell BS, Jellison J, McGeehan JE (2015) Lignocellulose degradation mechanisms across the tree of life. Curr Opin Chem Biol 29:108–119
Crawford R, McCoy E, Harkin J, Kirk T, Obst J (1973) Degradation of methoxylated benzoic acids by a Nocardia from a lignin-rich environment: significance to lignin degradation and effect of chloro substituents. Appl Microbiol 26(2):176–184
Crawford RL, Bromley JW, Perkins-Olson PE (1979) Catabolism of protocatechuate by Bacillus macerans. Appl Environ Microbiol 37(3):614–618
Cronin CN, Kim J, Fuller JH, Zhang X, Mcintire WS (1999) Organization and sequences of p-hydroxybenzaldehyde dehydrogenase and other plasmid-encoded genes for early enzymes of the p-cresol degradative pathway in Pseudomonas putida NCIMB 9866 and 9869. DNA Seq 10(1):7–17
D’Argenio DA, Segura A, Coco WM, Bünz PV, Ornston LN (1999) The physiological contribution of Acinetobacter PcaK, a transport system that acts upon protocatechuate, can be masked by the overlapping specificity of VanK. J Bacteriol 181(11):3505–3515
Daniel SL, Wu Z, Drake HL (1988) Growth of thermophilic acetogenic bacteria on methoxylated aromatic acids. FEMS Microbiol Lett 52(1–2):25–28
Davis JR, Sello JK (2010) Regulation of genes in Streptomyces bacteria required for catabolism of lignin-derived aromatic compounds. Appl Microbiol Biotechnol 86(3):921–929. https://doi.org/10.1007/s00253-009-2358-0
de Boer W, Folman LB, Summerbell RC, Boddy L (2005) Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev 29(4):795–811. https://doi.org/10.1016/j.femsre.2004.11.005
DeAngelis KM, Allgaier M, Chavarria Y, Fortney JL, Hugenholtz P, Simmons B, Sublette K, Silver WL, Hazen TC (2011) Characterization of trapped lignin-degrading microbes in tropical forest soil. PLoS One 6(4):e19306
DeAngelis KM, Sharma D, Varney R, Simmons B, Isern NG, Markilllie LM, Nicora C, Norbeck AD, Taylor RC, Aldrich JT, Robinson EW (2013) Evidence supporting dissimilatory and assimilatory lignin degradation in Enterobacter lignolyticus SCF1. Front Microbiol 4:Artn 280. doi:https://doi.org/10.3389/Fmicb.2013.00280
Defnoun S, Labat M, Ambrosio M, Garcia J-L, Patel B (2000) Papillibacter cinnamivorans gen. nov., sp. nov., a cinnamate-transforming bacterium from a shea cake digester. Int J Syst Evol Microbiol 50(3):1221–1228
Delneri D, Degrassi G, Rizzo R, Bruschi CV (1995) Degradation of trans-ferulic and p-coumaric acid by Acinetobacter calcoaceticus DSM 586. Biochim Biophys Acta Gen Subj 1244(2–3):363–367
Deschamps AM, Mahoudeau G, Lebeault JM (1980) Fast degradation of kraft lignin by bacteria. Eur J Appl Microbiol 9(1):45–51. https://doi.org/10.1007/Bf00500001
Díaz E, Jiménez JI, Nogales J (2013) Aerobic degradation of aromatic compounds. Curr Opin Biotechnol 24(3):431–442
Duan J, Huo X, Du W, Liang J, Wang D, Yang S (2016a) Biodegradation of kraft lignin by a newly isolated anaerobic bacterial strain, Acetoanaerobium sp. WJDL-Y2. Lett Appl Microbiol 62(1):55–62
Duan J, Liang J, Wang Y, Du W, Wang D (2016b) Kraft lignin biodegradation by Dysgonomonas sp. WJDL-Y1, a new anaerobic bacterial strain isolated from sludge of a pulp and paper mill. J Microbiol Biotechnol 26(10):1765–1773
Durante-Rodríguez G, Gómez-Álvarez H, Blázquez B, Fernández-Llamosas H, Martín-Moldes Z, Sanz D, Nogales J, Carmona M, Díaz E (2018) Anaerobic pathways for the catabolism of aromatic compounds lignin valorization. pp 333–390
Durham DR, Mcnamee CG, Stewart DB (1984) Dissimilation of aromatic compounds in Rhodotorula graminis: biochemical characterization of pleiotropically negative mutants. J Bacteriol 160(2):771–777
Edlin DA, Narbad A, Dickinson JR, Lloyd D (1995) The biotransformation of simple phenolic compounds by Brettanomyces anomalus. FEMS Microbiol Lett 125(2–3):311–315
Eggert C, Temp U, Eriksson K-E (1996) The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: purification and characterization of the laccase. Appl Environ Microbiol 62(4):1151–1158
Emerson D, Chauhan S, Oriel P, Breznak JA (1994) Haloferax sp. D1227, a halophilic Archaeon capable of growth on aromatic compounds. Arch Microbiol 161(6):445–452
Engelke M, Tähti H, Vaalavirta L (1996) Perturbation of artificial and biological membranes by organic compounds of aliphatic, alicyclic and aromatic structure. Toxicol in Vitro 10(2):111–115
Erdoğmuş SF, Mutlu B, Korcan SE, Güven K, Konuk M (2013) Aromatic hydrocarbon degradation by halophilic archaea isolated from Çamaltı Saltern, Turkey. Water Air Soil Pollut 224(3):1449
Eriksson KE, Grünewald A, Vallander L (1980) Studies of growth conditions in wood for three white-rot fungi and their cellulaseless mutants. Biotechnol Bioeng 22(2):363–376
Eriksson K-E, Johnsrud SC, Vallander L (1983) Degradation of lignin and lignin model compounds by various mutants of the white-rot fungus Sporotrichum pulverulentum. Arch Microbiol 135(3):161–168
Eriksson K-E, Gupta JK, Nishida A, Rao M (1984) Syringic acid metabolism by some white-rot, soft-rot and brown-rot fungi. Microbiology 130(10):2457–2464
Eulberg D, Golovleva LA, Schlömann M (1997) Characterization of catechol catabolic genes from Rhodococcus erythropolis 1CP. J Bacteriol 179(2):370–381
Fackler K, Gradinger C, Hinterstoisser B, Messner K, Schwanninger M (2006) Lignin degradation by white rot fungi on spruce wood shavings during short-time solid-state fermentations monitored by near infrared spectroscopy. Enzym Microb Technol 39(7):1476–1483
Fairley D, Boyd D, Sharma N, Allen C, Morgan P, Larkin M (2002) Aerobic metabolism of 4-hydroxybenzoic acid in Archaea via an unusual pathway involving an intramolecular migration (NIH shift). Appl Environ Microbiol 68(12):6246–6255
Falcon M, Rodriguez A, Carnicero A, Regalado V, Perestelo F, Milstein O, De la Fuente G (1995) Isolation of microorganisms with lignin transformation potential from soil of Tenerife Island. Soil Biol Biochem 27(2):121–126
Fialova A, Boschke E, Bley T (2004) Rapid monitoring of the biodegradation of phenol-like compounds by the yeast Candida maltosa using BOD measurements. Int Biodeterior Biodegradation 54(1):69–76
Fischer R, Bleichrodt FS, Gerischer UC (2008) Aromatic degradative pathways in Acinetobacter baylyi underlie carbon catabolite repression. Microbiology 154(10):3095–3103
Folman LB, Klein Gunnewiek PJ, Boddy L, De Boer W (2008) Impact of white-rot fungi on numbers and community composition of bacteria colonizing beech wood from forest soil. FEMS Microbiol Ecol 63(2):181–191
Fuchs G, Boll M, Heider J (2011) Microbial degradation of aromatic compounds—from one strategy to four. Nat Rev Microbiol 9(11):803–816. https://doi.org/10.1038/nrmicro2652
Fukasawa Y, Osono T, Takeda H (2011) Wood decomposing abilities of diverse lignicolous fungi on nondecayed and decayed beech wood. Mycologia 103(3):474–482
Gao X, Tan CL, Yeo CC, Poh CL (2005) Molecular and biochemical characterization of the xlnD-encoded 3-hydroxybenzoate 6-hydroxylase involved in the degradation of 2, 5-xylenol via the gentisate pathway in Pseudomonas alcaligenes NCIMB 9867. J Bacteriol 187(22):7696–7702
Gellerstedt G, Henriksson G (2008) Lignins: major sources, structure and properties. Elsevier, Amsterdam
Gérecová G, Neboháčová M, Zeman I, Pryszcz LP, Tomáška Ľ, Gabaldón T, Nosek J (2015) Metabolic gene clusters encoding the enzymes of two branches of the 3-oxoadipate pathway in the pathogenic yeast Candida albicans. FEMS Yeast Res 15(3):fov006
Gilbertson RL (1980) Wood-rotting fungi of North America. Mycologia 72(1):1–49
Giroux H, Vidal P, Bouchard J, Lamy F (1988) Degradation of kraft indulin lignin by Streptomyces viridosporus and Streptomyces badius. Appl Environ Microbiol 54(12):3064–3070
Giuliani SE, Frank AM, Corgliano DM, Seifert C, Hauser L, Collart FR (2011) Environment sensing and response mediated by ABC transporters. BMC Genomics 12(1):S8
González B, Acevedo C, Brezny R, Joyce T (1993) Metabolism of chlorinated guaiacols by a guaiacol-degrading Acinetobacter junii strain. Appl Environ Microbiol 59(10):3424–3429
González JM, Whitman WB, Hodson RE, Moran MA (1996) Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture. Appl Environ Microbiol 62(12):4433–4440
González J, Mayer F, Moran M, Hodson R, Whitman W (1997) Microbulbifer hydrolyticus gen. nov., sp. nov., and Marinobacterium georgiense gen. nov., sp. nov., two marine bacteria from a lignin-rich pulp mill waste enrichment community. Int J Syst Evol Microbiol 47(2):369–376
Gonzalez JM, Mayer F, Moran MA, Hodson RE, Whitman WB (1997) Sagittula stellata gen. nov, sp. nov, a lignin-transforming bacterium from a coastal environment. Int J Syst Bacteriol 47(3):773–780
Gorny N, Wahl G, Brune A, Schink B (1992) A strictly anaerobic nitrate-reducing bacterium growing with resorcinol and other aromatic compounds. Arch Microbiol 158(1):48–53
Gosselink RJ, van Dam JE, de Jong E, Scott EL, Sanders JP, Li J, Gellerstedt G (2010) Fractionation, analysis, and PCA modeling of properties of four technical lignins for prediction of their application potential in binders. Holzforschung 64(2):193–200
Grbić-Galić D (1983) Anaerobic degradation of coniferyl alcohol by methanogenic consortia. Appl Environ Microbiol 46(6):1442–1446
Grbić-Galić D (1985) Fermentative and oxidative transformation of ferulate by a facultatively anaerobic bacterium isolated from sewage sludge. Appl Environ Microbiol 50(4):1052–1057
Groseclose EE, Ribbons DW (1981) Metabolism of resorcinylic compounds by bacteria: new pathway for resorcinol catabolism in Azotob acter vinelandii. J Bacteriol 146(2):460–466
Guillén F, Martínez MJ, Gutiérrez A, Del Rio J (2005) Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int Microbiol 8:195–204
Gupta JK, Jebsen C, Kneifel H (1986) Sinapic acid degradation by the yeast Rhodotorula glutinis. Microbiology 132(10):2793–2799
Gupta VK, Minocha AK, Jain N (2001) Batch and continuous studies on treatment of pulp mill wastewater by Aeromonas formicans. Journal of Chemical Technology & Biotechnology: International Research in Process, Environmental & Clean Technology 76(6):547–552
Hainal AR, Capraru AM, Volf I, Popa VI (2012) Lignin as a carbon source for the cultivation of some Rhodotorula species. Cellul Chem Technol 46:87–96
Hammel KE, Cullen D (2008) Role of fungal peroxidases in biological ligninolysis. Curr Opin Plant Biol 11(3):349–355
Hamzah R, Al-Baharna B (1994) Catechol ring-cleavage in Pseudomonas cepacia: the simultaneous induction of ortho and meta pathways. Appl Microbiol Biotechnol 41(2):250–256
Haq I, Kumar S, Kumari V, Singh SK, Raj A (2016) Evaluation of bioremediation potentiality of ligninolytic Serratia liquefaciens for detoxification of pulp and paper mill effluent. J Hazard Mater 305:190–199
Harazono K, Yamashita N, Shinzato N, Watanabe Y, Fukatsu T, Kurane R (2003) Isolation and characterization of aromatics-degrading microorganisms from the gut of the lower termite Coptotermes formosanus. Biosci Biotechnol Biochem 67(4):889–892
Harry-asobara JL, Kamei I (2018) Bacterial strains isolated from cedar wood improve the mycelial growth and morphology of white rot fungus Phlebia brevispora on agar and liquid medium. J Wood Sci 64(4):444–450
Harwood CS, Gibson J (1988) Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris. Appl Environ Microbiol 54(3):712–717
Harwood CS, Parales RE (1996) The beta-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol 50:553–590. https://doi.org/10.1146/annurev.micro.50.1.553
Hastings J, de Matos P, Dekker A, Ennis M, Harsha B, Kale N, Muthukrishnan V, Owen G, Turner S, Williams M (2012) The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic Acids Res 41(D1):D456–D463
Hatakka A (2005) Biodegradation of lignin. Biopolymers Online
Hearn EM, Patel DR, Van den Berg B (2008) Outer-membrane transport of aromatic hydrocarbons as a first step in biodegradation. Proc Natl Acad Sci 105(25):8601–8606
Hedges JI, Blanchette RA, Weliky K, Devol AH (1988) Effects of fungal degradation on the CuO oxidation products of lignin: a controlled laboratory study. Geochim Cosmochim Acta 52(11):2717–2726
Henson WR, Campbell T, DeLorenzo DM, Gao Y, Berla B, Kim SJ, Foston M, Moon TS, Dantas G (2018) Multi-omic elucidation of aromatic catabolism in adaptively evolved Rhodococcus opacus. Metab Eng 49:69–83
Hiratsuka N, Oyadomari M, Shinohara H, Tanaka H, Wariishi H (2005) Metabolic mechanisms involved in hydroxylation reactions of diphenyl compounds by the lignin-degrading basidiomycete Phanerochaete chrysosporium. Biochem Eng J 23(3):241–246. https://doi.org/10.1016/j.bej.2005.01.008
Hirose J, Nagayoshi A, Yamanaka N, Araki Y, Yokoi H (2013) Isolation and characterization of bacteria capable of metabolizing lignin-derived low molecular weight compounds. Biotechnol Bioprocess Eng 18(4):736–741
Hirose J, Tsuda N, Miyatake M, Yokoi H, Shimodaira J (2018) Draft genome sequence of Pseudomonas sp. strain LLC-1 (NBRC 111237), capable of metabolizing lignin-derived low-molecular-weight compounds. Genome Announc 6(16):e00308–e00318
Hofrichter M, Vares T, Kalsi M, Galkin S, Scheibner K, Fritsche W, Hatakka A (1999) Production of manganese peroxidase and organic acids and mineralization of 14C-labelled lignin (14C-DHP) during solid-state fermentation of wheat straw with the white rot fungus Nematoloma frowardii. Appl Environ Microbiol 65(5):1864–1870
Holesova Z, Jakubkova M, Zavadiakova I, Zeman I, Tomaska L, Nosek J (2011) Gentisate and 3-oxoadipate pathways in the yeast Candida parapsilosis: identification and functional analysis of the genes coding for 3-hydroxybenzoate 6-hydroxylase and 4-hydroxybenzoate 1-hydroxylase. Microbiology 157:2152–2163. https://doi.org/10.1099/mic.0.048215-0
Holladay JE, White JF, Bozell JJ, Johnson D (2007) Top value added chemicals from biomass—volume II, results of screening for potential candidates from biorefinery lignin. Pacific Northwest National Lab. (PNNL), Richland, WA (United States); National Renewable Energy Laboratory (NREL), Golden, CO (United States)
Hooda R, Bhardwaj NK, Singh P (2015) Screening and identification of ligninolytic bacteria for the treatment of pulp and paper mill effluent. Water Air Soil Pollut 226(9):305
Hooda R, Bhardwaj NK, Singh P (2018) Brevibacillus parabrevis MTCC 12105: a potential bacterium for pulp and paper effluent degradation. World J Microbiol Biotechnol 34(2):31
Huang Z, Dostal L, Rosazza J (1993) Mechanisms of ferulic acid conversions to vanillic acid and guaiacol by Rhodotorula rubra. J Biol Chem 268(32):23954–23958
Huang XF, Santhanam N, Badri DV, Hunter WJ, Manter DK, Decker SR, Vivanco JM, Reardon KF (2013) Isolation and characterization of lignin-degrading bacteria from rainforest soils. Biotechnol Bioeng 110(6):1616–1626. https://doi.org/10.1002/Bit.24833
Hughes E, Bayly RC (1983) Control of catechol meta-cleavage pathway in Alcaligenes eutrophus. J Bacteriol 154(3):1363–1370
Ishiyama D, Vujaklija D, Davies J (2004) Novel pathway of salicylate degradation by Streptomyces sp. strain WA46. Appl Environ Microbiol 70(3):1297–1306
Iwabuchi N, Takiguchi H, Hamaguchi T, Takihara H, Sunairi M, Matsufuji H (2015) Transformation of lignin-derived aromatics into nonaromatic polymeric substances with fluorescent activities (NAPSFA) by Pseudomonas sp. ITH-SA-1. ACS Sustain Chem Eng 3(11):2678–2685
Jackson C, Couger M, Prabhakaran M, Ramachandriya K, Canaan P, Fathepure B (2017) Isolation and characterization of Rhizobium sp. strain YS-1r that degrades lignin in plant biomass. J Appl Microbiol 122(4):940–952
Janusz G, Pawlik A, Sulej J, Świderska-Burek U, Jarosz-Wilkołazka A, Paszczyński A (2017) Lignin degradation: microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol Rev 41(6):941–962
Jeffries TW (1981) Conversion of xylose to ethanol under aerobic conditions by Candida tropicalis. Biotechnol Lett 3(5):213–218
Jimenez JI, Minambres B, Garcia JL, Diaz E (2002) Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ Microbiol 4(12):824–841. https://doi.org/10.1046/j.1462-2920.2002.00370.x
Jones DC, Cooper RA (1990) Catabolism of 3-hydroxybenzoate by the gentisate pathway in Klebsiella pneumoniae M5a1. Arch Microbiol 154(5):489–495
Jurková M, Wurst M (1993) Biodegradation of aromatic carboxylic acids by Pseudomonas mira. FEMS Microbiol Lett 111(2–3):245–250
Kaiser J-P, Hanselmann KW (1982) Fermentative metabolism of substituted monoaromatic compounds by a bacterial community from anaerobic sediments. Arch Microbiol 133(3):185–194
Kamimura N, Aoyama T, Yoshida R, Takahashi K, Kasai D, Abe T, Mase K, Katayama Y, Fukuda M, Masai E (2010) Characterization of the protocatechuate 4, 5-cleavage pathway operon in Comamonas sp. strain E6 and discovery of a novel pathway gene. Appl Environ Microbiol 76(24):8093–8101
Kamimura N, Takahashi K, Mori K, Araki T, Fujita M, Higuchi Y, Masai E (2017) Bacterial catabolism of lignin-derived aromatics: new findings in a recent decade: update on bacterial lignin catabolism. Environ Microbiol Rep 9(6):679–705
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28(1):27–30
Karlson U, Dwyer D, Hooper S, Moore E, Timmis K, Eltis L (1993) Two independently regulated cytochromes P-450 in a Rhodococcus rhodochrous strain that degrades 2-ethoxyphenol and 4-methoxybenzoate. J Bacteriol 175(5):1467–1474
Katayama Y, Nishikawa S, Murayama A, Yamasaki M, Morohoshi N, Haraguchi T (1988) The metabolism of biphenyl structures in lignin by the soil bacterium (Pseudomonas paucimobilis SYK-6). FEBS Lett 233(1):129–133
Kato K, Kozaki S, Sakuranaga M (1998) Degradation of lignin compounds by bacteria from termite guts. Biotechnol Lett 20(5):459–462
Kaufmann F, Wohlfarth G, Diekert G (1998) O-demethylase from Acetobacterium dehalogenans: cloning, sequencing, and active expression of the gene encoding the corrinoid protein. Eur J Biochem 257(2):515–521
Kerr TJ, Kerr RD, Benner R (1983) Isolation of a bacterium capable of degrading peanut hull lignin. Appl Environ Microbiol 46(5):1201–1206
Khemili-Talbi S, Kebbouche-Gana S, Akmoussi-Toumi S, Angar Y, Gana ML (2015) Isolation of an extremely halophilic arhaeon Natrialba sp. C21 able to degrade aromatic compounds and to produce stable biosurfactant at high salinity. Extremophiles 19(6):1109–1120
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA (2015) PubChem substance and compound databases. Nucleic Acids Res 44(D1):D1202–D1213
Kirk TK, Farrell RL (1987) Enzymatic “combustion”: the microbial degradation of lignin. Annu Rev Microbiol 41(1):465–501
Kitchenham B (2004) Procedures for performing systematic reviews. Keele, UK, Keele University 33(2004):1–26
Kluge C, Tschech A, Fuchs G (1990) Anaerobic metabolism of resorcyclic acids (m-dihydroxybenzoic acids) and resorcinol (1, 3-benzenediol) in a fermenting and in a denitrifying bacterium. Arch Microbiol 155(1):68–74
Knežević A, Stajić M, Milovanović I, Vukojević J (2018) Wheat straw degradation by Trametes gibbosa: the effect of calcium ions. Waste Biomass Valorization 9(10):1903–1908
Konopka A (2009) What is microbial community ecology? ISME J 3(11):1223
Korniłłowicz-Kowalska T, Rybczyńska K (2015) Screening of microscopic fungi and their enzyme activities for decolorization and biotransformation of some aromatic compounds. Int J Environ Sci Technol 12(8):2673–2686
Kosa M, Ragauskas AJ (2012) Bioconversion of lignin model compounds with oleaginous Rhodococci. Appl Microbiol Biotechnol 93(2):891–900. https://doi.org/10.1007/s00253-011-3743-z
Krell T, Lacal J, Guazzaroni ME, Busch A, Silva-Jiménez H, Fillet S, Reyes-Darías JA, Muñoz-Martínez F, Rico-Jiménez M, García-Fontana C (2012) Responses of Pseudomonas putida to toxic aromatic carbon sources. J Biotechnol 160(1–2):25–32
Krug M, Ziegler H, Straube G (1985) Degradation of phenolic-compounds by the yeast Candida tropicalis Hp-15. 1. Physiology of growth and substrate utilization. J Basic Microbiol 25(2):103–110. https://doi.org/10.1002/jobm.3620250206
Kuhnigk T, Konig H (1997) Degradation of dimeric lignin model compounds by aerobic bacteria isolated from the hindgut of xylophagous termites. J Basic Microbiol 37(3):205–211. https://doi.org/10.1002/jobm.3620370309
Kumar M, Singh J, Singh MK, Singhal A, Thakur IS (2015) Investigating the degradation process of kraft lignin by β-proteobacterium, Pandoraea sp. ISTKB. Environ Sci Pollut Res 22(20):15690–15702
Leonowicz A, Matuszewska A, Luterek J, Ziegenhagen D, Wojtaś-Wasilewska M, Cho N-S, Hofrichter M, Rogalski J (1999) Biodegradation of lignin by white rot fungi. Fungal Genet Biol 27(2–3):175–185
Levasseur A, Piumi F, Coutinho PM, Rancurel C, Asther M, Delattre M, Henrissat B, Pontarotti P, Asther M, Record E (2008) FOLy: an integrated database for the classification and functional annotation of fungal oxidoreductases potentially involved in the degradation of lignin and related aromatic compounds. Fungal Genet Biol 45(5):638–645
Lewis NG, Yamamoto E (1990) Lignin: occurrence, biogenesis and biodegradation. Annu Rev Plant Biol 41(1):455–496
Li T, Takkellapati S (2018) The current and emerging sources of technical lignins and their applications. Biofuels Bioprod Biorefin 12(5):756–787
Li DH, Yan YL, Ping SZ, Chen M, Zhang W, Li L, Lin WN, Geng LZ, Liu W, Lu W, Lin M (2010) Genome-wide investigation and functional characterization of the beta-ketoadipate pathway in the nitrogen-fixing and root-associated bacterium Pseudomonas stutzeri A1501. BMC Microbiol 10: Artn 36. https://doi.org/10.1186/1471-2180-10-36
Liers C, Arnstadt T, Ullrich R, Hofrichter M (2011) Patterns of lignin degradation and oxidative enzyme secretion by different wood- and litter-colonizing basidiomycetes and ascomycetes grown on beech-wood. FEMS Microbiol Ecol 78(1):91–102. https://doi.org/10.1111/j.1574-6941.2011.01144.x
Linger JG, Vardon DR, Guarnieri MT, Karp EM, Hunsinger GB, Franden MA, Johnson CW, Chupka G, Strathmann TJ, Pienkos PT, Beckham GT (2014) Lignin valorization through integrated biological funneling and chemical catalysis. Proc Natl Acad Sci USA 111(33):12013–12018. https://doi.org/10.1073/pnas.1410657111
Liu Z-P, Wang B-J, Liu Y-H, Liu S-J (2005) Novosphingobium taihuense sp. nov., a novel aromatic-compound-degrading bacterium isolated from Taihu Lake, China. Int J Syst Evol Microbiol 55(3):1229–1232
Lucey KS, Leadbetter JR (2014) Catechol 2,3-dioxygenase and other meta-cleavage catabolic pathway genes in the ‘anaerobic’ termite gut spirochete Treponema primitia. Mol Ecol 23(6):1531–1543. https://doi.org/10.1111/mec.12598
MacLean AM, MacPherson G, Aneja P, Finan TM (2006) Characterization of the β-ketoadipate pathway in Sinorhizobium meliloti. Appl Environ Microbiol 72(8):5403–5413
Mahiudddin M, Fakhruddin A (2012) Degradation of phenol via meta cleavage pathway by Pseudomonas fluorescens PU1. ISRN Microbiology 2012
Mäkelä MR, Marinović M, Nousiainen P, Liwanag AJ, Benoit I, Sipilä J, Hatakka A, de Vries RP, Hilden KS (2015) Aromatic metabolism of filamentous fungi in relation to the presence of aromatic compounds in plant biomass. Adv Appl Microbiol. Vol 91. Elsevier, pp 63–137
Mallinson SJ, Machovina MM, Silveira RL, Garcia-Borràs M, Gallup N, Johnson CW, Allen MD, Skaf MS, Crowley MF, Neidle EL (2018) A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion. Nat Commun 9(1):2487
Martins TM, Hartmann DO, Planchon S, Martins I, Renaut J, Pereira CS (2015) The old 3-oxoadipate pathway revisited: new insights in the catabolism of aromatics in the saprophytic fungus Aspergillus nidulans. Fungal Genet Biol 74:32–44. https://doi.org/10.1016/j.fgb.2014.11.002
Maruyama K, Shibayama T, Ichikawa A, Sakou Y, Yamada S, Sugisaki H (2004) Cloning and characterization of the genes encoding enzymes for the protocatechuate meta-degradation pathway of Pseudomonas ochraceae NGJ1. Biosci Biotechnol Biochem 68(7):1434–1441
Masai E, Katayama Y, Nishikawa S, Fukuda M (1999) Characterization of Sphingomonas paucimobilis SYK-6 genes involved in degradation of lignin-related compounds. J Ind Microbiol Biotechnol 23(4–5):364–373. https://doi.org/10.1038/sj.jim.2900747
Masai E, Sasaki M, Minakawa Y, Abe T, Sonoki T, Miyauchi K, Katayama Y, Fukuda M (2004) A novel tetrahydrofolate-dependent O-demethylase gene is essential for growth of Sphingomonas paucimobilis SYK-6 with syringate. J Bacteriol 186(9):2757–2765
Masai E, Katayama Y, Fukuda M (2007) Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci Biotechnol Biochem 71(1):1–15. https://doi.org/10.1271/Bbb.60437
Mathews SL, Pawlak JJ, Grunden AM (2014) Isolation of Paenibacillus glucanolyticus from pulp mill sources with potential to deconstruct pulping waste. Bioresource Technol 164:100–105. https://doi.org/10.1016/j.biortech.2014.04.093
Maurice N, Erdei L (2018) Termite gut microbiome termites and sustainable management. Springer, pp 69–99
Mazzoli R, Pessione E, Giuffrida MG, Fattori P, Barello C, Giunta C, Lindley ND (2007) Degradation of aromatic compounds by Acinetobacter radioresistens S13: growth characteristics on single substrates and mixtures. Arch Microbiol 188(1):55–68
McCarthy AJ, Broda P (1984) Screening for lignin-degrading actinomycetes and characterization of their activity against [14C] lignin-labelled wheat lignocellulose. Microbiology 130(11):2905–2913
Mechichi T, Labat M, Patel BK, Woo TH, Thomas P, Garcia J-L (1999) Clostridium methoxybenzovorans sp. nov., a new aromatic o-demethylating homoacetogen from an olive mill wastewater treatment digester. Int J Syst Evol Microbiol 49(3):1201–1209
Mechichi T, Patel BK, Sayadi S (2005) Anaerobic degradation of methoxylated aromatic compounds by Clostridium methoxybenzovorans and a nitrate-reducing bacterium Thauera sp. strain Cin3, 4. Int Biodeterior Biodegradation 56(4):224–230
Michalska K, Chang C, Mack JC, Zerbs S, Joachimiak A, Collart FR (2012) Characterization of transport proteins for aromatic compounds derived from lignin: benzoate derivative binding proteins. J Mol Biol 423(4):555–575
Michielse CB, Reijnen L, Olivain C, Alabouvette C, Rep M (2012) Degradation of aromatic compounds through the beta-ketoadipate pathway is required for pathogenicity of the tomato wilt pathogen Fusarium oxysporum f. sp. lycopersici. Mol Plant Pathol 13(9):1089–1100. https://doi.org/10.1111/j.1364-3703.2012.00818.x
Middelhoven WJ (1993) Catabolism of benzene compounds by ascomycetous and basidiomycetous yeasts and yeastlike fungi. Antonie Van Leeuwenhoek 63(2):125–144
Milligan PW, Häggblom MM (1998) Biodegradation of resorcinol and catechol by denitrifying enrichment cultures. Environ Toxicol Chem 17(8):1456–1461
Monisha T, Ismailsab M, Masarbo R, Nayak AS, Karegoudar T (2018) Degradation of cinnamic acid by a newly isolated bacterium Stenotrophomonas sp. TRMK2. 3 Biotech 8(8):368
Moraes EC, Alvarez TM, Persinoti GF, Tomazetto G, Brenelli LB, Paixão DA, Ematsu GC, Aricetti JA, Caldana C, Dixon N (2018) Lignolytic-consortium omics analyses reveal novel genomes and pathways involved in lignin modification and valorization. Biotechnol Biofuels 11(1):75
Mori K, Kamimura N, Masai E (2018) Identification of the protocatechuate transporter gene in Sphingobium sp. strain SYK-6 and effects of overexpression on production of a value-added metabolite. Appl Microbiol Biotechnol 102(11):4807–4816
Murray K, Duggleby CJ, Williams PA, Sala-Trepat JM (1972) The metabolism of benzoate and methylbenzoates via the meta-cleavage pathway by Pseudomonas arvilla mt-2. Eur J Biochem 28(3):301–310
Muto A, Osawa S (1987) The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci 84(1):166–169
Nakamura T, Ichinose H, Wariishi H (2012) Flavin-containing monooxygenases from Phanerochaete chrysosporium responsible for fungal metabolism of phenolic compounds. Biodegradation 23(3):343–350. https://doi.org/10.1007/s10532-011-9521-x
Naqvi M, Yan J, Dahlquist E (2012) Bio-refinery system in a pulp mill for methanol production with comparison of pressurized black liquor gasification and dry gasification using direct causticization. Appl Energy 90(1):24–31
Narbad A, Gasson MJ (1998) Metabolism of ferulic acid via vanillin using a novel CoA-dependent pathway in a newly-isolated strain of Pseudomonas fluorescens. Microbiology 144:1397–1405
Ni B, Zhang Y, Chen D-W, Wang B-J, Liu S-J (2013) Assimilation of aromatic compounds by Comamonas testosteroni: characterization and spreadability of protocatechuate 4, 5-cleavage pathway in bacteria. Appl Microbiol Biotechnol 97(13):6031–6041
Nichols NN, Harwood CS (1997) PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J Bacteriol 179(16):5056–5061
Nikaido H (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67(4):593–656
Nikodem P, Hecht V, Schlömann M, Pieper DH (2003) New bacterial pathway for 4- and 5-chlorosalicylate degradation via 4-chlorocatechol and maleylacetate in Pseudomonas sp. strain MT1. J Bacteriol 185(23):6790–6800
Nishikawa S, Sonoki T, Kasahara T, Obi T, Kubota S, Kawai S, Morohoshi N, Katayama Y (1998) Cloning and sequencing of the Sphingomonas (Pseudomonas) paucimobilis gene essential for the O demethylation of vanillate and syringate. Appl Environ Microbiol 64(3):836–842
Nishikawa Y, Yasumi Y, Noguchi S, Sakamoto H, J-i N (2008) Functional analyses of Pseudomonas putida benzoate transporters expressed in the yeast Saccharomyces cerevisiae. Biosci Biotechnol Biochem 72(8):2034–2038
Numata K, Morisaki K (2015) Screening of marine bacteria to synthesize polyhydroxyalkanoate from lignin: contribution of lignin derivatives to biosynthesis by Oceanimonas doudoroffii. ACS Sustain Chem Eng 3(4):569–573. https://doi.org/10.1021/acssuschemeng.5b00031
Ornston L, Parke D (1976) Properties of an inducible uptake system for beta-ketoadipate in Pseudomonas putida. J Bacteriol 125(2):475–488
Ornston LN, Stanier RY (1966) Conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. I. Biochemistry. J Biol Chem 241(16):3776–&
Otani H, Lee Y-E, Casabon I, Eltis LD (2014) Characterization of p-hydroxycinnamate catabolism in a soil Actinobacterium. J Bacteriol. https://doi.org/10.1128/JB.02247-14
Overhage J, Priefert H, Steinbüchel A (1999) Biochemical and genetic analyses of ferulic acid catabolism in Pseudomonas sp. strain HR199. Appl Environ Microbiol 65(11):4837–4847
Parales RE, Ditty JL (2017) Substrate transport. In: Krell T (ed) Cellular ecophysiology of microbe. Springer International Publishing, Cham, pp 1–16
Parales RE, Ju K-S, Rollefson JB, Ditty J (2008) Bioavailability, chemotaxis, and transport of organic pollutants. Caister Academic Press, Madrid
Pardo I, Rodríguez-Escribano D, Aza P, de Salas F, Martínez AT, Camarero S (2018) A highly stable laccase obtained by swapping the second cupredoxin domain. Sci Rep 8(1):15669
Parke D (1997) Acquisition, reorganization, and merger of genes: novel management of the β-ketoadipate pathway in Agrobacterium tumefaciens. FEMS Microbiol Lett 146(1):3–12
Perestelo F, Falcon MA, Perez ML, Roig EC, Martin GD (1989) Bioalteration of kraft pine lignin by Bacillus megaterium isolated from compost piles. J Ferment Bioeng 68(2):151–153. https://doi.org/10.1016/0922-338x(89)90066-4
Perestelo F, Falcon M, de la Fuente G (1990) Biotransformation of kraft lignin fractions by Serratia marcescens. Lett Appl Microbiol 10(2):61–64
Perestelo F, Rodriguez A, Perez R, Carnicero A, De La Fuente G, Falcon M (1996) Isolation of a bacterium capable of limited degradation of industrial and labelled, natural and synthetic lignins. World J Microbiol Biotechnol 12(1):111–112
Pérez E, Tuck CO (2018) Quantitative analysis of products from lignin depolymerisation in high-temperature water. Eur Polym J 99:38–48
Perez-Pantoja D, De la Lglesia R, Pieper DH, Gonzalez B (2008) Metabolic reconstruction of aromatic compounds degradation from the genome of the amazing pollutant-degrading bacterium Cupriavidus necator JMP134. FEMS Microbiol Rev 32(5):736–794. https://doi.org/10.1111/j.1574-6976.2008.00122.x
Périé FH, Gold MH (1991) Manganese regulation of manganese peroxidase expression and lignin degradation by the white rot fungus Dichomitus squalens. Appl Environ Microbiol 57(8):2240–2245
Pham VH, Kim J (2012) Cultivation of unculturable soil bacteria. Trends Biotechnol 30(9):475–484
Phelps C, Young L (1997) Microbial metabolism of the plant phenolic compounds ferulic and syringic acids under three anaerobic conditions. Microb Ecol 33(3):206–215
Pildain M, Novas M, Carmarán C (2005) Evaluation of anamorphic state, wood decay and production of lignin-modifying enzymes for diatrypaceous fungi from Argentina. Journal of Agricultural Technology 1(1):81–96
Pometto AL III, Sutherland JB, Crawford DL (1981) Streptomyces setonii: catabolism of vanillic acid via guaiacol and catechol. Can J Microbiol 27(6):636–638
Providenti MA, O’Brien JM, Ruff J, Cook AM, Lambert IB (2006) Metabolism of isovanillate, vanillate, and veratrate by Comamonas testosteroni strain BR6020. J Bacteriol 188(11):3862–3869
Qi SW, Chaudhry MT, Zhang Y, Meng B, Huang Y, Zhao KX, Poetsch A, Jiang CY, Liu S, Liu SJ (2007) Comparative proteomes of Corynebacterium glutamicum grown on aromatic compounds revealed novel proteins involved in aromatic degradation and a clear link between aromatic catabolism and gluconeogenesis via fructose-1, 6-bisphosphatase. Proteomics 7(20):3775–3787
Rabus R, Widdel F (1995) Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch Microbiol 163(2):96–103
Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M (2014) Lignin valorization: improving lignin processing in the biorefinery. Science 344(6185):1246843
Raj A, Chandra R, Reddy MMK, Purohit HJ, Kapley A (2007) Biodegradation of kraft lignin by a newly isolated bacterial strain, Aneurinibacillus aneurinilyticus from the sludge of a pulp paper mill. World J Microbiol Biotechnol 23(6):793–799. https://doi.org/10.1007/s11274-006-9299-x
Ralph SA, Ralph J, Landucci L, Landucci L (2004) NMR database of lignin and cell wall model compounds. US Forest Prod Lab, Madison (http://ars.usda.gov/Services/docs.htm)
Ravi K, García-Hidalgo J, Gorwa-Grauslund MF, Lidén G (2017) Conversion of lignin model compounds by Pseudomonas putida KT2440 and isolates from compost. Appl Microbiol Biotechnol 101(12):5059–5070
Ravi K, García-Hidalgo J, Nöbel M, Gorwa-Grauslund MF, Lidén G (2018) Biological conversion of aromatic monolignol compounds by a Pseudomonas isolate from sediments of the Baltic Sea. AMB Express 8(1):32
Reverón I, Jiménez N, Curiel JA, Peñas E, de Felipe FL, de las Rivas B, Muñoz R (2017) Differential gene expression on Lactobacillus plantarum WCFS1 in response to phenolic compounds unravels new genes involved in tannin degradation. Appl Environ Microb:AEM. 03387-16
Rhoads TL, Mikell AT Jr, Eley MH (1995) Investigation of the lignin-degrading activity of Serratia marcescens: biochemical screening and ultrastructural evidence. Can J Microbiol 41(7):592–600
Rodriguez A, Carnicero A, Perestelo F, Delafuente G, Milstein O, Falcon MA (1994) Effect of Penicillium chrysogenum on lignin transformation. Appl Environ Microbiol 60(8):2971–2976
Ruiz-Dueñas FJ, Martínez ÁT (2009) Microbial degradation of lignin: how a bulky recalcitrant polymer is efficiently recycled in nature and how we can take advantage of this. Microb Biotechnol 2(2):164–177
Rüttimann-Johnson C, Salas L, Vicuña R, Kirk TK (1993) Extracellular enzyme production and synthetic lignin mineralization by Ceriporiopsis subvermispora. Appl Environ Microbiol 59(6):1792–1797
Ryazanova T, Chuprova N, Luneva T (2015) Effect of Trichoderma fungi on lignin from tree species barks. Catal Ind 7(1):82–89
Saha BC, Qureshi N, Kennedy GJ, Cotta MA (2016) Biological pretreatment of corn stover with white-rot fungus for improved enzymatic hydrolysis. Int Biodeterior Biodegradation 109:29–35
Sainsbury PD, Hardiman EM, Ahmad M, Otani H, Seghezzi N, Eltis LD, Bugg TDH (2013) Breaking down lignin to high-value chemicals: the conversion of lignocellulose to vanillin in a gene deletion mutant of Rhodococcus jostii RHA1. ACS Chem Biol 8(10):2151–2156. https://doi.org/10.1021/Cb400505a
Saito Y, Tsuchida H, Matsumoto T, Makita Y, Kawashima M, Kikuchi J, Matsui M (2018) Screening of fungi for decomposition of lignin-derived products from Japanese cedar. J Biosci Bioeng
Salmon RC, Cliff MJ, Rafferty JB, Kelly DJ (2013) The CouPSTU and TarPQM transporters in Rhodopseudomonas palustris: redundant, promiscuous uptake systems for lignin-derived aromatic substrates. PLoS One 8(3): ARTN e59844. doi:https://doi.org/10.1371/journal.pone.0059844
Sampaio J (1999) Utilization of low molecular weight aromatic compounds by heterobasidiomycetous yeasts: taxonomic implications. Can J Microbiol 45(6):491–512
Sato Y, Nishihara H, Yoshida M, Watanabe M, Rondal JD, Concepcion RN, Ohta H (2006) Cupriavidus pinatubonensis sp. nov. and Cupriavidus laharis sp. nov., novel hydrogen-oxidizing, facultatively chemolithotrophic bacteria isolated from volcanic mudflow deposits from Mt. Pinatubo in the Philippines. Int J Syst Evol Microbiol 56(5):973–978
Schink B, Pfennig N (1982) Fermentation of trihydroxybenzenes by Pelobacter acidigallici gen. nov. sp. nov., a new strictly anaerobic, non-sporeforming bacterium. Arch Microbiol 133(3):195–201
Schweigert N, Zehnder AJB, Eggen RIL (2001) Chemical properties of catechols and their molecular modes of toxic action in cells, from microorganisms to mammals. Environ Microbiol 3(2):81–91. https://doi.org/10.1046/j.1462-2920.2001.00176.x
Sethuraman A, Akin D, Eriksson K-E (1999) Production of ligninolytic enzymes and synthetic lignin mineralization by the bird’s nest fungus Cyathus stercoreus. Appl Microbiol Biotechnol 52(5):689–697
Sharma KK, Kuhad RC (2009) An evidence of laccases in archaea. Indian J Microbiol 49(2):142–150
Shettigar M, Balotra S, Cahill D, Warden AC, Lacey MJ, Kohler H-PE, Rentsch D, Oakeshott JG, Pandey G (2018) Isolation of the (+)-pinoresinol-mineralizing Pseudomonas sp. strain SG-MS2 and elucidation of its catabolic pathway. Appl Environ Microbiol 84(4):e02531–e02517
Shi Y, Chai L, Tang C, Yang Z, Zhang H, Chen R, Chen Y, Zheng Y (2013a) Characterization and genomic analysis of kraft lignin biodegradation by the beta-proteobacterium Cupriavidus basilensis B-8. Biotechnol Biofuels 6(1):1
Shi Y, Chai L, Tang C, Yang Z, Zheng Y, Chen Y, Jing Q (2013b) Biochemical investigation of kraft lignin degradation by Pandoraea sp. B-6 isolated from bamboo slips. Bioprocess Biosyst Eng 36(12):1957–1965
Sietmann R, Hammer E, Specht M, Cerniglia CE, Schauer F (2001) Novel ring cleavage products in the biotransformation of biphenyl by the yeast Trichosporon mucoides. Appl Environ Microbiol 67(9):4158–4165
Sigoillot J-C, Berrin J-G, Bey M, Lesage-Meessen L, Levasseur A, Lomascolo A, Record E, Uzan-Boukhris E (2012) Fungal strategies for lignin degradation Adv Bot Res. Vol 61. Elsevier, pp 263–308
Sláviková E, Košíková B (2001) Modification of lignin by Geotrichum klebahnii. World J Microbiol Biotechnol 17(1):1–3
Sláviková E, Košíková B, Mikulášová M (2002) Biotransformation of waste lignin products by the soil-inhabiting yeast Trichosporon pullulans. Can J Microbiol 48(3):200–203
Song Y-J (2009) Characterization of aromatic hydrocarbon degrading bacteria isolated from pine litter. Kor J Microbiol Biotechnol 37:333–339
Sperfeld M, Diekert G, Studenik S (2018) Anaerobic aromatic compound degradation in Sulfuritalea hydrogenivorans sk43H. FEMS Microbiol Ecol 95(1):fiy199
Sudtachat N, Ito N, Itakura M, Masuda S, Eda S, Mitsui H, Kawaharada Y, Minamisawa K (2009) Aerobic vanillate degradation and C1 compound metabolism in Bradyrhizobium japonicum. Appl Environ Microbiol 75(15):5012–5017
Suman SK, Dhawaria M, Tripathi D, Raturi V, Adhikari DK, Kanaujia PK (2016) Investigation of lignin biodegradation by Trabulsiella sp. isolated from termite gut. Int Biodeterior Biodegradation 112:12–17
Sun Z, Fridrich B, de Santi A, Elangovan S, Barta K (2018) Bright side of lignin depolymerization: toward new platform chemicals. Chem Rev 118(2):614–678
Sutherland JB (1986) Demethylation of veratrole by cytochrome P-450 in Streptomyces setonii. Appl Environ Microbiol 52(1):98–100
Szewzyk R, Pfennig N (1987) Complete oxidation of catechol by the strictly anaerobic sulfate-reducing Desulfobacterium catecholicum sp. nov. Arch Microbiol 147(2):163–168
Taylor CR, Hardiman EM, Ahmad M, Sainsbury PD, Norris PR, Bugg TDH (2012) Isolation of bacterial strains able to metabolize lignin from screening of environmental samples. J Appl Microbiol 113(3):521–530. https://doi.org/10.1111/j.1365-2672.2012.05352.x
ten Have R, Teunissen PJ (2001) Oxidative mechanisms involved in lignin degradation by white-rot fungi. Chem Rev 101(11):3397–3414
Tian JH, Pourcher AM, Bouchez T, Gelhaye E, Peu P (2014) Occurrence of lignin degradation genotypes and phenotypes among prokaryotes. Appl Microbiol Biotechnol 98(23):9527–9544. https://doi.org/10.1007/s00253-014-6142-4
Tian JH, Pourcher AM, Peu P (2016) Isolation of bacterial strains able to metabolize lignin and lignin-related compounds. Lett Appl Microbiol 63(1):30–37
Traunecker J, Preuß A, Diekert G (1991) Isolation and characterization of a methyl chloride utilizing, strictly anaerobic bacterium. Arch Microbiol 156(5):416–421
Tschech A, Fuchs G (1987) Anaerobic degradation of phenol by pure cultures of newly isolated denitrifying pseudomonads. Arch Microbiol 148(3):213–217
Tsegaye B, Balomajumder C, Roy P (2018) Biodelignification and hydrolysis of rice straw by novel bacteria isolated from wood feeding termite. 3 Biotech 8(10):447
Tuor U, Winterhalter K, Fiechter A (1995) Enzymes of white-rot fungi involved in lignin degradation and ecological determinants for wood decay. J Biotechnol 41(1):1–17
UniProtConsortium (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res 45(D1):D158–D169
Uthandi S, Saad B, Humbard MA, Maupin-Furlow JA (2010) LccA, an archaeal laccase secreted as a highly stable glycoprotein into the extracellular medium by Haloferax volcanii. Appl Environ Microbiol 76(3):733–743
Van Dexter S, Boopathy R (2018) Biodegradation of phenol by Acinetobacter tandoii isolated from the gut of the termite. Environ Sci Pollut Res: 1–6
Vandamme P, Coenye T (2004) Taxonomy of the genus Cupriavidus: a tale of lost and found. Int J Syst Evol Microbiol 54(6):2285–2289
Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. Plant Physiol 153(3):895–905
Vardon DR, Franden MA, Johnson CW, Karp EM, Guarnieri MT, Linger JG, Salm MJ, Strathmann TJ, Beckham GT (2015) Adipic acid production from lignin. Energy Environ Sci 8(2):617–628. https://doi.org/10.1039/c4ee03230f
Vares T, Niemenmaa O, Hatakka A (1994) Secretion of ligninolytic enzymes and mineralization of 14C-ring-labelled synthetic lignin by three Phlebia tremellosa strains. Appl Environ Microbiol 60(2):569–575
Vasudevan N, Mahadevan A (1992) Utilization of complex phenolic compounds by Acinetobacter sp. Appl Microbiol Biotechnol 37(3):404–407
Wang Y, Liu Q, Yan L, Gao Y, Wang Y, Wang W (2013) A novel lignin degradation bacterial consortium for efficient pulping. Bioresour Technol 139:113–119
Watanabe Y, Shinzato N, Fukatsu T (2003) Isolation of actinomycetes from termites' guts. Biosci Biotechnol Biochem 67(8):1797–1801
Woese CR, Kandler O, Wheelis ML (1990) Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci 87(12):4576–4579
Woo HL, Ballor NR, Hazen TC, Fortney JL, Simmons B, Davenport KW, Goodwin L, Ivanova N, Kyrpides NC, Mavromatis K (2014a) Complete genome sequence of the lignin-degrading bacterium Klebsiella sp. strain BRL6-2. Stand Genomic Sci 9(1):19
Woo HL, Utturkar S, Klingeman D, Simmons BA, DeAngelis KM, Brown SD, Hazen TC (2014b) Draft genome sequence of the lignin-degrading Burkholderia sp. strain LIG30, isolated from wet tropical forest soil. Genome Announc 2(3):e00637–e00614
Worrall JJ, Anagnost SE, Zabel RA (1997) Comparison of wood decay among diverse lignicolous fungi. Mycologia:199–219
Wu Y-R, He J (2013) Characterization of anaerobic consortia coupled lignin depolymerization with biomethane generation. Bioresour Technol 139:5–12
Xiong X, Liao H, Ma J, Liu X, Zhang L, Shi X, Yang X, Lu X, Zhu Y (2014) Isolation of a rice endophytic bacterium, Pantoea sp. S d-1, with ligninolytic activity and characterization of its rice straw degradation ability. Lett Appl Microbiol 58(2):123–129
Xu Y, Yan D-Z, Zhou N-Y (2006) Heterologous expression and localization of gentisate transporter Ncg12922 from Corynebacterium glutamicum ATCC 13032. Biochem Biophys Res Commun 346(2):555–561
Xu C, Ma F, Zhang X (2009) Lignocellulose degradation and enzyme production by Irpex lacteus CD2 during solid-state fermentation of corn stover. J Biosci Bioeng 108(5):372–375
Xu Y, Gao X, Wang S-H, Liu H, Williams PA, Zhou N-Y (2012) MhbT is a specific transporter for 3-hydroxybenzoate uptake by Gram-negative bacteria. Appl Environ Microbiol. 01511-12
Xu Z, Qin L, Cai M, Hua W, Jin M (2018) Biodegradation of kraft lignin by newly isolated Klebsiella pneumoniae, Pseudomonas putida, and Ochrobactrum tritici strains. Environ Sci Pollut Res 25(14):14171–14181
Yaegashi J, Kirby J, Ito M, Sun J, Dutta T, Mirsiaghi M, Sundstrom ER, Rodriguez A, Baidoo E, Tanjore D (2017) Rhodosporidium toruloides: a new platform organism for conversion of lignocellulose into terpene biofuels and bioproducts. Biotechnol Biofuels 10(1):241
Yaguchi A, Robinson A, Mihealsick E, Blenner M (2017) Metabolism of aromatics by Trichosporon oleaginosus while remaining oleaginous. Microb Cell Factories 16(1):206
Yang Y, Zhou J, Lu H, Yuan Y, Zhao L (2011) Isolation and characterization of a fungus Aspergillus sp. strain F-3 capable of degrading alkali lignin. Biodegradation 22(5):1017–1027
Yang YS, Zhou JT, Lu H, Yuan YL, Zhao LH (2012) Isolation and characterization of Streptomyces spp. strains F-6 and F-7 capable of decomposing alkali lignin. Environ Technol 33(23):2603–2609. https://doi.org/10.1080/09593330.2012.672473
Yang CX, Wang T, Gao LN, Yin HJ, Lü X (2017) Isolation, identification and characterization of lignin-degrading bacteria from Qinling, China. J Appl Microbiol 123(6):1447–1460
Yoshida T, Inami Y, Matsui T, Nagasawa T (2010) Regioselective carboxylation of catechol by 3, 4-dihydroxybenzoate decarboxylase of Enterobacter cloacae P. Biotechnol Lett 32(5):701–705
Yu T, Wu W, Liang W, Lever MA, Hinrichs K-U, Wang F (2018) Growth of sedimentary Bathyarchaeota on lignin as an energy source. Proc Natl Acad Sci :201718854
Zakzeski J, Bruijnincx PC, Jongerius AL, Weckhuysen BM (2010) The catalytic valorization of lignin for the production of renewable chemicals. Chem Rev 110(6):3552–3599
Zeida M, Wieser M, Yoshida T, Sugio T, Nagasawa T (1998) Purification and characterization of gallic acid decarboxylase from Pantoea agglomerans T71. Appl Environ Microbiol 64(12):4743–4747
Zeng J, Singh D, Laskar DD, Chen S (2013) Degradation of native wheat straw lignin by Streptomyces viridosporus T7A. Int J Environ Sci Technol 10(1):165–174. https://doi.org/10.1007/s13762-012-0085-z
Zhao C, Xie S, Pu Y, Zhang R, Huang F, Ragauskas AJ, Yuan JS (2016) Synergistic enzymatic and microbial lignin conversion. Green Chem 18(5):1306–1312
Zhu D, Zhang P, Xie C, Zhang W, Sun J, Qian W-J, Yang B (2017) Biodegradation of alkaline lignin by Bacillus ligniniphilus L1. Biotechnol Biofuels 10(1):44
Acknowledgements
The authors would like to thank Javier García-Hidalgo for his contributions to the initial data mining performed before the construction of the database.
Availability of data and materials
The database is available online at www.elignindatabase.com, with no restrictions for academic or non-academic use. The eLignin Database: Copyright 2016–-2019 Daniel Brink, Applied Microbiology, Department of Chemistry, Lund University, Sweden. We welcome contributions to the database; instructions are available at the homepage. All contributions will undergo manual curation.
Funding
This work was financed by the Swedish Foundation for Strategic Research through the grant contract RBP14-0052.
Author information
Authors and Affiliations
Contributions
DB designed the study, designed and programmed the database and web interface, performed the data mining, wrote the manuscript, and curated the database content. KR assisted in the data mining and curation. GL and MGG conceived the study and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Brink, D.P., Ravi, K., Lidén, G. et al. Mapping the diversity of microbial lignin catabolism: experiences from the eLignin database. Appl Microbiol Biotechnol 103, 3979–4002 (2019). https://doi.org/10.1007/s00253-019-09692-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-09692-4