Abstract
Key message
A cytogenetic map of wheat was constructed using FISH with cDNA probes. FISH markers detected homoeology and chromosomal rearrangements of wild relatives, an important source of genes for wheat improvement.
Abstract
To transfer agronomically important genes from wild relatives to bread wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD) by induced homoeologous recombination, it is important to know the chromosomal relationships of the species involved. Fluorescence in situ hybridization (FISH) can be used to study chromosome structure. The genomes of allohexaploid bread wheat and other species from the Triticeae tribe are colinear to some extent, i.e., composed of homoeoloci at similar positions along the chromosomes, and with genic regions being highly conserved. To develop cytogenetic markers specific for genic regions of wheat homoeologs, we selected more than 60 full-length wheat cDNAs using BLAST against mapped expressed sequence tags and used them as FISH probes. Most probes produced signals on all three homoeologous chromosomes at the expected positions. We developed a wheat physical map with several cDNA markers located on each of the 14 homoeologous chromosome arms. The FISH markers confirmed chromosome rearrangements within wheat genomes and were successfully used to study chromosome structure and homoeology in wild Triticeae species. FISH analysis detected 1U-6U chromosome translocation in the genome of Aegilops umbellulata, showed colinearity between chromosome A of Ae. caudata and group-1 wheat chromosomes, and between chromosome arm 7S#3L of Thinopyrum intermedium and the long arm of the group-7 wheat chromosomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A wide variety of important genes were transferred to bread wheat, Triticum aestivum L., (2n = 6x = 42, genome AABBDD) from Triticeae species, but only few of them are exploited in wheat cultivars (reviewed by Friebe et al. 1996; Kilian et al. 2011). To select an optimal gene transfer strategy resulting in the introgression of only a small part of an alien chromosome with the gene of interest, but without undesirable traits and compensating for the replaced wheat chromatin, it is necessary to know the evolutionary distance, homoeologous relationships and degree of colinearity between wheat and alien chromosomes.
Several approaches were used to study the homoeology between chromosomes of hexaploid wheat and other Triticeae species. Gametophytic compensation test is based on non-viability of wheat male gametes with substituting alien non-compensating chromosomes. The analysis uses cytological or phenotypic screening of the progeny from crosses between wheat and set of wheat-alien double monosomic lines. The sporophytic compensation ability of alien chromosomes can be tested by measuring the fertility of wheat-alien substitution lines. Because of variation in the ability to substitute chromosomes from A, B and D genomes, this analysis requires the development of wheat-alien substitution lines for all three wheat genomes (Sears 1952; Dvorak 1980; Friebe et al. 1993).
Analysis of chromosome pairing in metaphase I of meiosis can also be used to determine the homoeology of wheat and alien chromosomes (Yang et al. 1996).
A comparative genetic analysis of alien chromosomes with wheat is another approach, which employs molecular or morphological markers. Only a few morphological, isozymic or seed storage protein markers are available for defining the homoeology of an alien chromosome, which limit the ability of the approach to reveal chromosomal rearrangements. (Yang et al. 1996; Zhang et al. 1998; Friebe et al. 1999; Qi et al. 2007; McArthur et al. 2012). Restriction fragment length polymorphisms (RFLPs) are the most informative wheat molecular markers (Qi et al. 2007). RFLPs were successfully used to develop high-resolution wheat genetic and physical maps (Qi et al. 2003, 2004), define homoeology of alien chromosomes and reveal their rearrangements relative to wheat (Devos et al. 1993a; Zhang et al. 1998; McArthur et al. 2012). Southern hybridization, however, is not a high-throughput technique and it employs radioisotopes (Qi et al. 2007). To perform genetic mapping of molecular markers on alien chromosomes it is necessary to develop and analyze segregating populations.
Fluorescence in situ hybridization (FISH) is a useful tool for physical mapping of chromosomes and for studying evolutionary chromosome rearrangements. FISH labeling of tandem repeats and microsatellites allows the identification of individual wheat chromosomes and chromosomes of related species (Mukai et al. 1993; Pedersen et al. 1996; Pedersen and Langridge 1997; Cuadrado and Schwarzacher 1998; Cuadrado et al. 2008a, b; Komuro et al. 2013). Genic sequences are highly conserved among Triticeae species (Feuillet and Keller 2002; Akhunov et al. 2003) and free of repeats, which makes them homoeolog-specific wheat FISH markers and potentially universal Triticeae markers. Single genes with size larger than 3 kb can be routinely localized on mitotic plant chromosomes by FISH, as was shown on maize, barley and wheat (Lamb et al. 2007; Danilova and Birchler 2008; Ma et al. 2010; Karafiátová et al. 2013). In our previous study, we used wheat cDNA of cytosolic acetyl-CoA carboxylase (Acc-2) and nine other full-length (FL) cDNAs together with tandem repeats as multicolor FISH probes to map Acc-2 sequences, identify chromosomes and reveal chromosome rearrangements in wheat and its closest diploid and tetraploid relatives T. urartu Tumanian ex Gandilyan, T. monococcum L., Ae. speltoides Tausch., Ae. tauschii Coss., T. turgidum subsp. dicoccoides (Körn. Ex Asch. & Graebn.) Thell., T. turgidum subsp. dicoccum (Schrank) Thell., T. turgidum subsp. durum (Desf.) Husn. and T. timopheevii (Zhuck.) Zhuck. (Danilova et al. 2012). Unlike RFLPs, compensation analysis and chromosome pairing tests, FISH analysis does not require the development of wheat-alien substitution lines or segregating mapping populations; it visualizes homoeologous regions directly on alien chromosomes in a simple and fast experiment.
The purpose of this work was to develop and map a set of FISH cDNA probes specific to wheat homoeologous groups to mark all chromosome arms at distal and proximal positions. The markers were employed in detecting chromosome rearrangements and analyzing homoeologous relationships within wheat genome and genomes of three wild Triticeae species.
Materials and methods
Plant material
The material used in this study included T. aestivum cv. Chinese Spring TA3008, Aegilops umbellulata Zhuk. TA1851, Ae. caudata L. TA1908 and the wheat-Thinopyrum intermedium (Host) Barkworth and D.R. Dewey translocation line TA5634 homozygous for the translocation chromosome T7BS·7S#3L (Liu et al. 2011) from the collection of the Wheat Genetics Resource Center, Kansas State University. Accession TA1851 was used for developing wheat-Ae. umbellulata chromosome addition lines (Friebe et al. 1995; Zhang et al. 1998). Accession TA1908 was used for developing wheat-Ae. caudata addition lines (Friebe et al. 1992).
Slide preparation and FISH procedure
Somatic chromosome preparations using the drop technique, direct probe labeling by nick translation and the FISH procedure were as described previously (Kato et al. 2004, 2006) with minor modifications as described in (Danilova et al. 2012). To make FISH probes, cDNAs were amplified with standard primers T3 and T7; PCR products were purified with Invitrogen PCR purification kit (Life Technologies, Grand Island, NY, USA Cat. # K310001) and labeled with Texas red-5-dCTP (PerkinElmer, Waltham, MA, USA, cat # NEL426001EA). GAA- and pAs1- oligonucleotide probes (Danilova et al. 2012) were synthesized by Integrated DNA Technologies with a flourochrome attached to the 5′ end. For nucleolus organizing region (NOR) probe, clone pTa71 was used (Gerlach and Bedbrook 1979). Genomic in situ hybridization (GISH) was performed according to Zhang et al. (2001) with modifications described in Liu et al. (2011). Chromosome preparations were mounted and counterstained with 4′,6-diamidino-2-phenylindole solution (DAPI) or propidium iodide (PI) in Vectashield (Vector Laboratories, Burlingham, CA, USA, cat # H-1200, H-1300). Images were captured with a Zeiss Axioplan 2 microscope using a cooled charge-coupled device camera CoolSNAP HQ2 (Photometrics, Tucson, AZ, USA) and AxioVision 4.8 software (Zeiss). Images were processed using the Adobe Photoshop software (Adobe Systems Incorporated, San Jose, CA, USA).
FLcDNA map development and chromosome measurements
To develop detectable FISH probes, long (>2.5 kb) wheat FLcDNA sequences (Kawaura et al. 2009) were selected from the Triticeae FLcDNA database (Mochida et al. 2009, http://www.shigen.nig.ac.jp/wheat/komugi/ests/tissueBrowse.jsp). The chromosomal position of cDNAs was detected by BLAST (Altschul et al. 1997) against expressed sequence tags (ESTs) mapped in the deletion bins of Chinese Spring (Qi et al. 2004; GrainGenes Database http://wheat.pw.usda.gov/cgi-bin/westsql/map_locus.cgi) and verified by BLAST against the barley (Deng et al. 2007), rice and Brachypodium genome sequences. The cDNAs that showed similarity to ESTs, mapped on more than one homoeologous group were rejected. The presence of repetitive sequences was checked using Repeatmasker software (Smit et al. 1996–2004). The cDNA clones were supplied by the National BioResource Project-Wheat, Japan. Acc2 cDNA pooled probe was produced from wheat RNA by Reverse Transcriptase-PCR (Danilova et al. 2012). Ten out of 61 FLcDNAs were used in our previous work (Table 1).
Selected cDNAs were used as FISH probes in a mixture with probes to tandem repeats and microsatellites, which allowed chromosome identification. The relative distance of a cDNA FISH site from the centromere was measured and calculated using the MicroMeasure 3.3 software (Reeves and Tear 2000). For measurements, the centromeres were marked on DAPI images of chromosomes as the position of the primary constriction. For most cDNA probes, the signal position was measured on five chromosomes from different metaphase spreads. Average relative distance from the centromere, standard deviation and confidence intervals with a significance level of 0.05 were calculated using Microsoft Office Excel functions. The idiogram was constructed using the wheat standard karyotype (Gill et al. 1991). The heterochromatic N-bands that corresponded to GAA- FISH bands were left on the idiogram and pAs1- FISH bands were added.
Results
Mapping FLcDNAs on wheat cv Chinese Spring chromosomes using FISH
For each wheat chromosome arm, we selected at least three cDNA clones to develop a distal, interstitial and proximal FISH marker. In total, 61 cDNA sequences were selected to develop FISH probes (Table 1). Fifty-one cDNA probes produced distinguishable FISH signals, no unspecific background and hybridized to all three homoeologous chromosome pairs (Fig. 1). Three cDNA probes hybridized to several homoeologous chromosome sets or on nonhomoeologous chromosomes, 4S-3 (chromosome arms 4BS, 4DS, 4AL and 5AL, 5BL, 5DL and 7AL), 4S-4 (2AS, 2BS, 2DS and 4BS, 4DS, 4AL) and Acc-2 (3AL, 3BL, 3DL, 4AS, 5DL). One probe tplb0006e16 (1,110 bp) was too short and produced a very weak signal on the expected chromosome arm and labeled NOR sites. Six probes showed staining of repeat clusters or unspecific staining on all chromosomes (Table 1). Three cDNAs tplb0006a16, tplb0018i03 and tplb0017c03 contain simple-sequence repeats (CCT)11, (CCG)5 and (GGA)10, respectively, and produced FISH patterns in accordance with the SSRs distribution reported by Cuadrado et al. (2008a). Repeats (GGA) n /(CCT) n are distributed as clusters on specific chromosome sites and repeats (CCG) n are dispersed throughout all chromosomes. Three other probes, tplb0028l15, AK331504 and tplb0007d03 label unknown repeats (Supplemental Fig. 1).
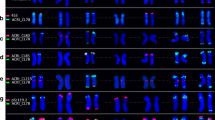
Mapping 5L-3 probe (AK334748) on wheat chromosomes of cv Chinese Spring. a Merged image. The cDNA probe is red, GAA and pAs1 oligonucleotide probes are green, chromosomes, counterstained with DAPI are blue. b Red channel image. The probe hybridizes to three pairs of homoeologous chromosomes. Bar corresponds to 10 μm
FISH mapping of group-1 chromosomes is shown in Fig. 2. Some probes showed a difference in the sequence copy number between homoeologs; for example probes 1S-3 and 1S-1 produced double signals on chromosome 1D and probe 1L-3 on chromosome 1A (Fig. 2). Probe 7S-4 hybridizes as a cluster of at least three copies on all group-7 chromosomes and showed a very bright FISH signal (Supplemental Fig. 1).
The FISH positions of 49 cDNAs out of 54 mapped by BLAST against ESTs correlate well with the corresponding EST bin positions (Table 1). The sequence tplb0028l15 matches an EST mapped on the short arms of group-1 chromosomes but is similar to a sequence on chromosome 4HS of barley and was mapped by FISH on chromosome arms 4BS, 4DS and 4AL. The probe 1L-3 (tplb0014k07) was mapped by BLAST against EST sequences on the long arms of group-3 chromosomes, but hybridized to group-1 chromosomes. The cDNA sequence has matches on barley chromosomes 1H (92 % similarity) and 3H (76 %). Probe 6L-4 (AK333670) was mapped by EST similarity on the short arms of group-6 chromosomes, but matches (92 %) to a sequence on barley chromosome arm 6HL and hybridized to the long arms of group-6 chromosomes of wheat. The probe 5S-5 (tplb0027f03) was mapped by EST similarity on the long arms of group-3 chromosomes but hybridized to group 5. This cDNA clone may contain a wrong insert because instead of the expected 2,416 bp, its length is about 3,100 bp. Probe 4S-1 (AK330261) showed FISH signals on the short arms of chromosomes 4A, 4B and 4D. This sequence is 98 % similar to the EST mapped on the proximal part of 4BL and 4DL arms (Table 1), but has 95 % similarity to a sequence mapped on barley chromosome 4HS. For Acc-2 cDNA no matching bin mapped ESTs were found.
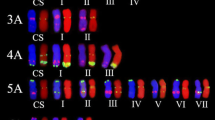
Most of the cDNA probes hybridized to all three homoeologous chromosomes in the same order and at a similar relative position, except for chromosome arms 2AS, 4A, 4BL, 6AS and 7BS, where the positions of several probes are significantly different from those of the other wheat homoeologs (Table 1; Fig. 3). On chromosome 4A, known to be rearranged (Naranjo et al. 1987; Devos et al. 1995; Mickelson-Young et al. 1995; Miftahudin et al. 2004), all three group-4 long arm specific cDNA probes hybridized to the short arm of 4A and five cDNAs mapped on short arm of group-4 chromosomes hybridized to the long arm of 4A. One exception is probe 4S-1 (AK330261), which showed FISH signals on the short arms of all group-4 chromosomes.
Idiogram of wheat chromosomes. Heterochromatic bands, which can be visualized with GAA oligonucleotide probes, are shown in black, pAs1 bands are shown in green, cDNA probe positions are shown as red dots. The names of the probes, which hybridize to more than one homoeologous group are highlighted in red
The positions of the cDNA FISH probes, GAA and pAs1 bands are shown on the idiogram (Fig. 3). There are three or more FISH markers on the distal, interstitial and proximal positions on the long arms of all chromosomes except those of group 4. The cDNA FISH markers are distributed unequally along the short arms of most chromosomes and those of group 4, leaving some parts uncovered. The short arm of group -6 chromosomes has markers only at interstitial positions. Thus, we did not find markers at desirable positions for some chromosome arms, because there were either no long cDNAs with similarity to any physically mapped EST found in the Triticeae FLcDNA database or the ESTs were mapped to a very large bin, but corresponding cDNAs hybridized at adjacent positions, as on short arms of chromosome groups 5, 6 and 7.
FISH on chromosomes of Triticeae species
To test whether wheat probes produce detectable signals on chromosomes of other Triticeae species and reveal chromosome rearrangements, we selected Ae. umbellulata as a model. Its genetic map was constructed using RFLP markers and homoeologous relationships with wheat were studied by Zhang et al. (1998). The genetic map provided evidence for multiple translocations in Ae. umbellulata chromosomes relative to those of hexaploid wheat. At first, we developed a FISH karyotype of Ae. umbellulata by labeling GAA microsatellites and NOR sites. All chromosomes were identified (Fig. 4a) by comparing their FISH pattern, size and arm ratio with the C-banding karyotype (Friebe et al. 1995). Then, each of six wheat probes, specific for group-1 chromosomes, was applied to Ae. umbellulata preparations together with the GAA oligonucleotide probe. All wheat cDNA probes produced distinct FISH signals on Ae. umbellulata chromosomes; five were detected on chromosome 1U at similar positions and order as on wheat group-1 chromosomes (Fig. 4b). Probe 1L-3, located on the distal end of the long arms of wheat group-1 chromosomes, was detected on the distal end of chromosome arm 6UL. According to Zhang et al. (1998), chromosome 1U is colinear with most of wheat chromosome 1D except the distal segment, which was translocated to the distal end of 6U. Thus, our results correspond with those of the RFLP analysis.
Application of wheat cDNA FISH probes to three Triticeae species: Ae. umbellulata (a, b), Ae. caudata (c, d) and Th. intermedium (e). a, c Chromosomes of Ae. umbellulata and Ae. caudata were identified by FISH pattern of tandem repeats and chromosome morphology; a GAA is green, NOR is red, c GAA is white, NOR is green, pAs1 is red. Karyotypes were constructed with chromosomes from a single metaphase. b, d Group-1 chromosome-specific probes are red, the GAA repeat is green. e The translocation chromosome T7BS 7S#3L with 7BL substituted by 7S#3L of Th. intermedium was identified by GISH (Th. intermedium gDNA is green, wheat DNA, counterstained with PI, is red) and by GAA pattern (green). Group-7 chromosome-specific probes are red. Bar corresponds to 5 μm
We applied the same approach and the same set of probes to Ae. caudata, where the chromosome homoeology to wheat is not known. All chromosomes of Ae. caudata were identified (Fig. 4c) by comparing their GAA-FISH pattern, positions of NOR sites and morphology with C-banding karyotype developed by Friebe et al. (1992). The six wheat cDNA probes produced clear signals on chromosome A of Ae. caudata in the same order and similar positions as those on wheat chromosomes of group 1 (Fig. 4d), conferring its homoeology and suggesting that this chromosome is not rearranged.
Thinopyrum intermedium was another Triticeae species with an unknown chromosome structure selected for the FISH analysis. We have developed a line with the translocation chromosome T7BS·7S#3L, which confers resistance to wheat streak mosaic and Triticum mosaic virus (Liu et al. 2011). We employed induced homoeologous recombination (reviewed in Qi et al. 2007) to shorten the 7S#3L segment, however, after screening 400 progenies, no recombinants were recovered. This result prompted us to verify the colinearity of chromosome arm 7S#3L. The translocation chromosome was identified by its GAA pattern in which the 7BS arm has specific bands and the 7S#3L arm has no GAA bands. The four wheat 7L-specific cDNA probes produced clear signals on the Th. intermedium chromosome arm in the same order and positions as those on the long arms of wheat group-7 chromosomes (Fig. 4d), indicating no rearrangements.
Discussion
FISH map of wheat chromosomes
The coding sequences of wheat genomes are highly conserved; mean synteny estimates between the A-, B- and D-genome loci, based on Southern blot analysis of ESTs collection, ranged from 0.91 to 0.96 (Akhunov et al. 2003). Although it is not known from which genome of T. aestivum originate the FLcDNAs used in our experiment, 51 out of 61 cDNAs hybridized to all three homoeologous chromosomes and can be used as homoeologous group specific wheat markers. Seven probes contain repetitive sequences and label dispersed or tandem repeats and three other probes hybridize to more than one homoeologous or nonhomeologous chromosome (Table 1); consequently, some chromosome arms are missing FISH markers at desirable positions.
For 91 % cDNAs hybridized to all three homoeoloci, the positions of FISH site correspond to the matching EST positions on bin deletion map. The disagreement between EST and FISH maps may be caused by presence of paralogous genes. The small size probes used in Southern analysis to map ESTs (Qi et al. 2004) may detect sequences shared by several genes. An example is sequence tplb0014k07 (probe 1L-3) which matches to sequences on barley chromosomes 1H (92 % similarity) and 3H (76 %) and to wheat EST mapped on group-3 chromosomes, but produced FISH signal on group-1 chromosomes (Table 1). Some wheat deletion chromosomes may be rearranged or telosomics may be actually acrocentrics which may cause errors in mapping of pericentromeric regions (probe 4S-1). For all cDNAs mapped, their position on wheat chromosomes corresponds to positions of homoeoloci on barley sequence physical map (The International Barley Genome Sequencing Consortium 2012). Thus, our FISH map can be integrated with the wheat deletion bin map and together with the barley sequence physical map can help to verify mismapped wheat loci.
The homoeologous genomes of T. aestivum were found to be largely collinear, i.e. composed of homoeoloci at equivalent positions along the chromosomes, except chromosomes 2B, 4A, 5A, 6B and 7B involved in translocations (Devos et al. 1993b, 1995; Mickelson-Young et al. 1995; Dvorak 2009). The order and location of the cDNA FISH probes on the homoeologous genomes were colinenar as well (Table 1; Fig. 3), except chromosome arms 2AS, 4A, 4BL, 6BS, and 7BS. The proximal displacement of two probes mapped on the 6BS arm may be explained by overestimation of the arm length because of the large size of the secondary constriction at the NOR site (Dvorak et al. 1984). Chromosome 4A is known to be rearranged as a result of 4AL/5AL translocation followed by a 7BS translocation and one paracentric and two pericentric inversions (Naranjo et al. 1987; Devos et al. 1995; Mickelson-Young et al. 1995; Miftahudin et al. 2004), and all but one group-4 cDNA probe hybridized accordingly. The difference in the position of probe 4S-1 on the short arm of chromosomes 4B and 4D and the proximal displacement of three cDNA probes on 4BL (Table 1; Fig. 3) can be explained by a pericentric inversion, specific for chromosome 4B of Chinese Spring and some other cultivars (Endo and Gill 1984; Friebe and Gill 1994). Probe 4S-1 hybridized to the short arm of chromosome 4A which may result from an additional pericentric inversion. The displacement of all four 7BS FISH probes to the distal end of 7BS reflects the translocation of a large part of chromosome arm 7BS to 4AL. Devos et al. (1993b) found that the distal end of chromosome arm 2BS was deleted as a result of 2BS/6BS translocation. We found that on chromosome arm 2AS the positions of all five cDNA probes differ significantly from 2BS and 2DS, they all displaced distally. We found the same displacement on Chinese Spring deletion bin map: among 100 EST probes physically mapped to 2AS5-0.78-1.00 bin, 20 are mapped to more proximal bins of 2BS1-0.53-0.75, and 4 mapped to 2DS1-0.33-0.47 bin (http://wheat.pw.usda.gov/cgi-bin/westsql/map_locus.cgi). For mapping chromosome arm 2DS a stock with very large distal deletion was used (1–0.47), which makes it less informative. Based on our measurements and EST mapping, we assume that 2AS has either a distal deletion or a translocation in cv Chinese Spring. Thus, our data confirm previously known wheat chromosomal rearrangements and can be used to detect the new rearrangements.
Application of wheat FISH markers in studying chromosome homoeology of Triticeae
The success of gene transfer by induced homoeologous recombination (Riley et al. 1968; Sears 1977) depends on genome affinity and chromosome colinearity. A significant difference in meiotic pairing of alien chromosomes or chromosome arms with wheat homoeologs correspond to the level of structural rearrangements (Ceoloni et al. 1988; Cuadrado et al. 1997; Devos et al. 1993a; Lukaszewski et al. 2004). The lack of knowledge on the evolutionary and cytogenetic relationship among wheat and its relatives hampers the alien gene transfer or causes non-compensating translocations, which is the reason why only few alien genes were intensively being used in cultivar improvement (Friebe et al. 1996; Ceoloni and Jauhar 2006). RFLP analysis was so far the most informative approach to study homoeology of wheat and alien chromosomes and to reveal rearrangements. Cytogenetic studies of Triticeae employ a large collection of tandem repeats, which allow to identify individual chromosomes. But because the abundance and distribution of the repeats is highly variable among species, they are not suitable for detecting chromosome homoeology, unless applied in chromosome pairing analysis. We developed wheat cytogenetic map using FISH with probes specific to coding regions of homoeologous chromosomes. Earlier we showed that wheat FLcDNA probes can be used to detect homoeologous chromosomes and reveal rearrangements in genomes of closest wheat diploid and tetraploid relatives T. urartu and T. monococcum (genome AA), Ae. speltoides (SS), Ae. tauschii (DD), T. turgidum subsp. dicoccoides, T. turgidum subsp. dicoccum, T. turgidum subsp. durum (AABB) and T. timopheevii (AAGG) (Danilova et al. 2012). In the current study we extended this approach to more distant wheat relatives Ae. umbellulata, Ae. caudata and Th. intermedium.
Aegilops umbellulata and Ae. caudata are annual, diploid, self-pollinated grasses. Aegilops umbellulata (2n = 14, UU) belongs to Aegilops L. section which includes nine species of different ploidy level; all of them have the U-genome combined with one of C, M, N or S genomes. Aegilops caudata (2n = 14, CC) belongs to section Cylindropyron (Jaub. et Spach) Zhuk., which has only one other species, Ae. cylindrica Host (2n = 28, DC) (Kilian et al. 2011). Thinopyrum intermedium is a perennial, cross-pollinating, allohexaploid species (2n = 6x = 42, genome JJJsJsSS); the J genome is related to Th. bessarabicum (Savul. and Rayss) A. Löve, Js is related to Dasypyrum villosum L. Candargy and S is related to a Pseudoroegneria-like progenitors (Chen et al. 1998; Liu et al. 2011; Mahelka et al. 2011). Phylogenetic analyses of several genic regions showed that sequences of Aegilops and Triticum species, including the C and U genomes, are similar and form a tight cluster, separate from Pseudoroegneria species and D. villosum. Within the Aegilops-Triticum clade of diploid species, the U and C genomes form a tighter subcluster, which shows their high similarity (Kellogg et al. 1996; Mason-Gamer 2005; Petersen et al. 2006; Escobar et al. 2011; Mahelka et al. 2011). Based on the phylogenetic data, we can assume that the genic sequences of Ae. umbellulata, Ae. caudata and other Aegilops and Triticum species are similar enough to be detected by wheat FISH probes. Indeed, the hybridization of wheat chromosome 1 -specific cDNA probes to their chromosomes was successful. FISH confirmed the rearrangement of Ae. umbellulata chromosome 1U, detected earlier by RFLP analysis (Zhang et al. 1998). The homoeologous relationships of Ae. caudata chromosomes to wheat have not been studied previously, although this species is an important source of many agronomic traits (Friebe et al. 1992; Makkouk et al. 1994; Riar et al. 2012). Our data revealed that chromosome A of Ae. caudata is homoeologous to wheat group-1 chromosomes and is not involved in any major rearrangements.
In spite of the large phylogenetic distance and dissimilarity between wheat and S-genome of Th. intermedium, wheat cDNA probes were detected on the 7S#3L chromosome arm. FISH mapping did not reveal any structural rearrangements, hence, our failure in recovering any recombinant chromosomes can be explained by low affinities of evolutionary distant genomes, which was confirmed by chromosome pairing analysis. The frequency of meiotic pairing between chromosome arms 7S#3L and 7BL in plants, homozygous for mutant gene ph1b (Sears 1977), was very low −0.3 %, (B. Friebe, T.V. Danilova, unpublished data), indicating, that for recovering recombinants with shortened Th. intermedium segments, a larger population needs to be screened.
The successful detection of wheat cDNA probes on all three wheat homoeologs and on chromosomes of eight Triticeae species and the high sequence similarity between wheat and barley homoeoloci (90 % or more) allows us to expect, that FISH with probes specific to genic regions can be used to detect chromosome homoeology and rearrangements in Triticeae. We plan to map additional FISH markers on wheat chromosomes to increase the resolution of this approach.
References
Akhunov ED, Akhunova AR, Linkiewicz AM, Dubcovsky Hummel JD et al (2003) Synteny perturbations between wheat homoeologous chromosomes caused by locus duplications and deletions correlate with recombination rates. Proc Natl Acad Sci USA 100:10836–10841. doi:10.1073/pnas.1934431100
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25:3389–3402. doi:10.1093/nar/25.17.3389
Ceoloni C, Jauhar PP (2006) Chromosome engineering of the durum wheat genome: strategies and applications of potential breeding value. In: Singh RJ, Jauhar PP (eds) Genetic resources, chromosome engineering, and crop improvement: Cereals. CRC Press, Boca Raton, pp 27–59
Ceoloni C, Del Signore G, Pasquini M, Testa A (1988) Transfer of mildew resistance from Triticum longissimum into wheat by ph1 induced homoeologous recombination. In: Miller TE, Koebner RMD (eds) 7th International wheat genetics symposium. Institute of Plant Science Research, Cambridge (UK), pp 221–226
Chen Q, Conner RL, Laroche A, Thomas JB (1998) Genome analysis of Thinopyrum intermedium and Thinopyrum ponticum using genomic in situ hybridization. Genome 41:580–586. doi:10.1139/g98-055
Cuadrado A, Schwarzacher T (1998) The chromosomal organization of simple sequence repeats in wheat and rye genomes. Chromosoma 107:587–594. doi:10.1007/s004120050345
Cuadrado A, Vitellozzi F, Jouve N, Ceoloni C (1997) Fluorescence in situ hybridization with multiple repeated DNA probes applied to the analysis of wheat-rye chromosome pairing. Theor Appl Genet 94:347–355
Cuadrado A, Cardoso M, Jouve N (2008a) Increasing the physical markers of wheat chromosomes using SSRs as FISH probes. Genome 51:809–815. doi:10.1139/G08-065
Cuadrado A, Cardoso M, Jouve N (2008b) Physical organization of simple sequence repeats (SSRs) in Triticeae: structural, functional and evolutionary implications. Cytogenet Genome Res 120:210–219. doi:10.1159/000121069
Danilova TV, Birchler JA (2008) Integrated cytogenetic map of mitotic metaphase chromosome 9 of maize: resolution, sensitivity, and banding paint development. Chromosoma 117:345–356
Danilova TV, Friebe B, Gill BS (2012) Single-copy gene fluorescence in situ hybridization and genome analysis: Acc-2 loci mark evolutionary chromosomal rearrangements in wheat. Chromosoma 121:597–611. doi:10.1007/s00412-012-0384-7
Deng W, Nickle DC, Learn GH, Maust B, Mullins JI (2007) ViroBLAST: a stand-alone BLAST web server for flexible queries of multiple databases and user’s datasets. Bioinformatics 23:2334–2336. doi:10.1093/bioinformatics/btm331
Devos KM, Atkinson MD, Chinoy CN, Francis HA, Harcourt RL, Koebner RMD, Liu CJ, Masojc P, Xie DX, Gale MD (1993a) Chromosomal rearrangements in the rye genome relative to that of wheat. Theor Appl Genet 85:673–680. doi:10.1007/Bf00225004
Devos KM, Millan T, Gale MD (1993b) Comparative RFLP maps of the homoeologous group-2 chromosomes of wheat, rye and barley. Theor Appl Genet 85:784–792. doi:10.1007/BF00225020
Devos KM, Dubcovsky J, Dvorak J, Chinoy CN, Gale MD (1995) Structural evolution of wheat chromosomes 4A, 5A, and 7B and its impact on recombination. Theor Appl Genet 91:282–288. doi:10.1007/BF00220890
Dvorak J (1980) Homoeology between Agropyron elongatum chromosomes and Triticum aestivum chromosomes. Can J Genet Cytol 22:237–259. doi:10.1139/g80-029
Dvorak J (2009) Triticeae genome structure and evolution. In: Feuillet C, Muehlbauer GJ (eds) Genetics and genomics of the Triticeae. Springer, London, pp 685–711
Dvorak J, Mcguire PE, Mendlinger S (1984) Inferred chromosome morphology of the ancestral genome of Triticum. Plant Syst Evol 144:209–220. doi:10.1007/BF00984134
Endo TR, Gill BS (1984) Somatic karyotype, heterochromatin distribution, and nature of chromosome differentiation in common wheat, Triticum aestivum L. em Thell. Chromosoma 89:361–369. doi:10.1007/BF00331253
Escobar JS, Scornavacca C, Cenci A, Guilhaumon C, Santoni S et al (2011) Multigenic phylogeny and analysis of tree incongruences in Triticeae (Poaceae). BMC Evol Biol 11:181. doi:10.1186/1471-2148-11-181
Feuillet C, Keller B (2002) Comparative genomics in the grass family: molecular characterization of grass genome structure and evolution. Annals Bot 89:3–10. doi:10.1093/aob.2002.mcf008
Friebe B, Gill BS (1994) C-band polymorphism and structural rearrangements detected in common wheat (Triticum aestivum). Euphytica 78:1–5. doi:10.1007/BF00021392
Friebe B, Schubert V, Bluthner WD, Hammer K (1992) C-Banding pattern and polymorphism of Aegilops caudata and chromosomal constitutions of the amphiploid Triticum aestivum, Ae. caudata and 6 derived chromosome addition lines. Theor Appl Genet 83:589–596. doi:10.1007/BF00226902
Friebe B, Tuleen N, Jiang JM, Gill BS (1993) Standard karyotype of Triticum longissimum and its cytogenetic relationship with Triticum aestivum. Genome 36:731–742. doi:10.1139/G93-098
Friebe B, Jiang J, Tuleen N, Gill BS (1995) Standard karyotype of Triticum umbellulatum and the characterization of derived chromosome addition and translocation lines in common wheat. Theor Appl Genet 90:150–156. doi:10.1007/BF00221010
Friebe B, Jiang J, Raupp WJ, McIntosh RA, Gill BS (1996) Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica 91:59–87. doi:10.1007/BF00035277
Friebe BR, Tuleen NA, Gill BS (1999) Development and identification of a complete set of Triticum aestivum, Aegilops geniculata chromosome addition lines. Genome 42:374–380. doi:10.1139/Gen-42-3-374
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucl Acids Res 7:1869–1885. doi:10.1093/nar/7.7.1869
Gill BS, Friebe B, Endo TR (1991) Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum). Genome 34:830–839. doi:10.1139/g91-128
Karafiátová M, Bartoš J, Kopecký D, Ma L, Sato K, Houben A, Stein N, Doležel J (2013) Mapping nonrecombining regions in barley using multicolor FISH. Chrom Res. doi:10.1007/s10577-013-9380-x
Kato A, Lamb JC, Birchler JA (2004) Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci USA 101:13554–13559. doi:10.1073/pnas.0403659101
Kato A, Albert PS, Vega JM, Birchler JA (2006) Sensitive fluorescence in situ hybridization signal detection in maize using directly labeled probes produced by high concentration DNA polymerase nick translation. Biotech Histochem 81:71–78. doi:10.1080/10520290600643677
Kawaura K, Mochida K, Enju A, Totoki Y, Toyoda A et al (2009) Assessment of adaptive evolution between wheat and rice as deduced from full-length common wheat cDNA sequence data and expression patterns. BMC Genom 10:271. doi:10.1186/1471-2164-10-271
Kellogg EA, Appels R, MasonGamer RJ (1996) When genes tell different stories: the diploid genera of Triticeae (Gramineae). Syst Botany 21:321–347. doi:10.2307/2419662
Kilian B, Mammen K, Millet E, Sharma R, Graner A et al (2011) Aegilops. In: Kole C (ed) Wild crop relatives: genomic and breeding resources. Cereals, Springer, Berlin, pp 1–76
Komuro S, Endo R, Shikata K, Kato A (2013) Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56:131–137. doi:10.1139/gen-2013-0003
Lamb JC, Danilova T, Bauer MJ, Meyer JM, Holland JJ, Jensen MD, Birchler JA (2007) Single-gene detection and karyotyping using small-target fluorescence in situ hybridization on maize somatic chromosomes. Genetics 175:1047–1058
Liu W, Seifers DL, Qi LL, Friebe B, Gill BS (2011) A Compensating wheat, Thinopyrum intermedium Robertsonian translocation conferring resistance to wheat streak mosaic virus and Triticum mosaic virus. Crop Sci 51:2382–2390. doi:10.2135/cropsci2011.03.0118
Lukaszewski AJ, Rybka K, Korzun V, Malyshev SV, Lapinski B et al (2004) Genetic and physical mapping of homoeologous recombination points involving wheat chromosome 2B and rye chromosome 2R. Genome 47:36–45. doi:10.1139/G03-089
Ma L, Vu GTH, Schubert V, Watanabe K, Stein N, Houben A, Schubert I (2010) Synteny between Brachypodium distachyon and Hordeum vulgare as revealed by FISH. Chromosome Research 18(7):841–850
Mahelka V, Kopecky D, Pastova L (2011) On the genome constitution and evolution of intermediate wheatgrass (Thinopyrum intermedium: Poaceae, Triticeae). BMC Evol Biol 11:127. doi:10.1186/1471-2148-11-127
Makkouk KM, Comeau A, Ghulam W (1994) Resistance to barley yellow dwarf luteovirus in Aegilops Species. Can J Plant Sci 74:631–634. doi:10.4141/cjps94-113
Mason-Gamer RJ (2005) The beta-amylase genes of grasses and a phylogenetic analysis of the Triticeae (Poaceae). Amer J Botany 92:1045–1058. doi:10.3732/ajb.92.6.1045
McArthur RI, Zhu X, Oliver RE, Klindworth DL, Xu SS et al (2012) Homoeology of Thinopyrum junceum and Elymus rectisetus chromosomes to wheat and disease resistance conferred by the Thinopyrum and Elymus chromosomes in wheat. Chrom Res 20:699–715. doi:10.1007/s10577-012-9307-y
Mickelson-Young L, Endo TR, Gill BS (1995) A cytogenetic ladder-map of the wheat homoeologous group-4 chromosomes. Theor Appl Genet 90:1007–1011. doi:10.1007/BF00222914
Miftahudin RK, Ma XF, Mahmoud AA, Layton J, Milla MAR et al (2004) Analysis of expressed sequence tag loci on wheat chromosome group 4. Genetics 168:651–663. doi:10.1534/genetics.104.034827
Mochida K, Yoshida T, Sakurai T, Ogihara Y, Shinozaki K (2009) TriFLDB: a database of clustered full-length coding sequences from Triticeae with applications to comparative grass genomics. Plant Physiol 150:1135–1146. doi:10.1104/pp.109.138214
Mukai Y, Nakahara Y, Yamamoto M (1993) Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total genomic and highly repeated DNA probes. Genome 36:489–494. doi:10.1139/g93-067
Naranjo T, Roca A, Goicoechea PG, Giraldez R (1987) Arm homoeology of wheat and rye chromosomes. Genome 29:873–882. doi:10.1139/g87-149
Pedersen C, Langridge P (1997) Identification of the entire chromosome complement of bread wheat by two-colour FISH. Genome 40:589–593. doi:10.1139/g97-077
Pedersen C, Rasmussen SK, LindeLaursen I (1996) Genome and chromosome identification in cultivated barley and related species of the Triticeae (Poaceae) by in situ hybridization with the GAA-satellite sequence. Genome 39:93–104. doi:10.1139/g96-013
Petersen G, Seberg O, Yde M, Berthelsen K (2006) Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum). Mol Phylogenet Evol 39:70–82. doi:10.1016/j.ympev.2006.01.023
Qi L, Echalier B, Friebe B, Gill BS (2003) Molecular characterization of a set of wheat deletion stocks for use in chromosome bin mapping of ESTs. Funct Integr Genomics 3:39–55. doi:10.1007/s10142-002-0063-5
Qi LL, Echalier B, Chao S, Lazo GR, Butler GE et al (2004) A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168:701–712. doi:10.1534/genetics.104.034868
Qi L, Friebe B, Zhang P, Gill BS (2007) Homoeologous recombination, chromosome engineering and crop improvement. Chrom Res 15:3–19. doi:10.1007/s10577-006-1108-8
Reeves A, Tear J (2000) MicroMeasure for Windows, version 3.3
Riar AK, Kaur S, Dhaliwal HS, Singh K, Chhuneja P (2012) Introgression of a leaf rust resistance gene from Aegilops caudata to bread wheat. J Genetics 91:155–161. doi:10.1007/s12041-012-0161-7
Riley R, Chapman V, Johnson R (1968) Incorporation of alien disease resistance in wheat by genetic interference with regulation of meiotic chromosome synapsis. Genet Res 12:199–219. doi:10.1017/S0016672300011800
Sears ER (1952) Homoeologous chromosomes in Triticum aestivum. Genetics 37:624
Sears ER (1977) An induced mutant with homoeologous pairing in common wheat. Can J Gen Cytol 19:585–593. doi:10.1139/g77-063
Smit AFA, Hubley R, Green P (1996-2004) RepeatMasker Open-3.0. http://www.repeatmasker.org
The International Barley Genome Sequencing Consortium (2012) A physical, genetical and functional sequence assembly of the barley genome. Nature 491:711–716
Yang YC, Tuleen NA, Hart GE (1996) Isolation and identification of Triticum aestivum L. em. Thell. cv Chinese Spring, T.peregrinum Hackel disomic chromosome addition lines. Theor Appl Genet 92:591–598. doi:10.1007/s001220050168
Zhang H, Jia J, Gale MD, Devos KM (1998) Relationships between the chromosomes of Aegilops umbellulata and wheat. Theor Appl Genet 96:69–75. doi:10.1007/s001220050710
Zhang P, Friebe B, Lukaszewski AJ, Gill BS (2001) The centromere structure in Robertsonian wheat-rye translocation chromosomes indicates that centric breakage-fusion can occur at different positions within the primary constriction. Chromosoma 110:335–344. doi:10.1007/s004120100159
Acknowledgments
This research was funded by the Kansas Wheat Commission and the Kansas Crop Improvement Association. We thank W. John Raupp for critical editorial review of the manuscript. This is contribution number 14-021-J from the Kansas Agricultural Experiment Station, Kansas State University, Manhattan KS 66506 – 5502, USA. Publication of this article was funded in part by the Kansas State University Open Access Publication Fund.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Pat. Heslop-Harrison.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2013_2253_MOESM1_ESM.tif
Supplemental Fig. 1. a, b) Probe tplb0007d03 label unknown repeats on chromosomes 5B and 4B. c, d) Probe 7S-4 (tplb0015e09) labels a gene cluster on short arms of group-7 chromosomes. BLAST analysis showed 90 % similarity of tplb0015e09 to Brachypodium distachyon and Setaria italica ABC transporter C family member 10-like mRNA and to Zea mays multidrug-resistance associated protein 3 (MRP3) gene. a, c) Merged images; cDNA probe is red, GAA and pAs1 oligonucleotide probes are green, chromosomes, conterstained with DAPI are blue. b, d) Red channel image. Bar corresponds to 10 μm. (TIFF 60245 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Danilova, T.V., Friebe, B. & Gill, B.S. Development of a wheat single gene FISH map for analyzing homoeologous relationship and chromosomal rearrangements within the Triticeae. Theor Appl Genet 127, 715–730 (2014). https://doi.org/10.1007/s00122-013-2253-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-013-2253-z