Abstract
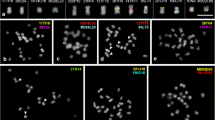
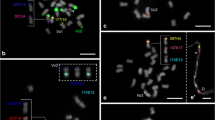
We investigated by fluorescence in situ hybridization (FISH) the synteny between Brachypodium distachyon with a small genome (1C = 320 Mb) and barley with a large genome (1C = 5,100 Mb) at the chromosome level. Reciprocal genomic in situ hybridization (GISH) between B. distachyon and barley labeled mainly 45S ribosomal DNA loci, indicating that most high copy DNA is weakly conserved between both grasses. Of 13 BAC clones with inserts from different B. distachyon chromosomes, only two belonging to chromosome 1 yielded hybridization signals on a barley metaphase chromosome (on 7HS and 7HL, respectively), confirming synteny between both chromosomes. FISH experiments to characterize the synteny of single-copy loci were performed. Two of four Brachypodium sylvaticum BACs spanning a 223-kb interval homologous to the region of barley that harbors a gibberellic-acid-insensitive semi-dwarfing gene, sdw3, hybridized specifically to a central position of B. distachyon chromosome 1 short arm but not to the homologous region of the barley genome. Repeat-free sequences PCR amplified from four non-overlapping barley BACs linked to the core of Sdw3 region yielded signals at distinct positions in the middle of barley chromosome arm 2HS. Together, these results (1) confirmed the synteny between B. distachyon chromosome 1 and barley chromosomes 2H and 7H at the cytological level, (2) indicated mid-arm position for the Sdw3 locus genetically mapped at the centromere of barley chromosome 2H, and (3) proved that the sdw3 core interval of <100 kb in B. distachyon corresponds to a megabase-sized syntenic region in barley.







Similar content being viewed by others
Abbreviations
- BAC:
-
Bacterial artificial chromosome
- CCD:
-
Charge-coupled device
- cM:
-
CentiMorgan
- dUTP:
-
2’-deoxyuridine-5’-triphosphate
- FISH:
-
Fluorescence in situ hybridization
- GISH:
-
Genomic in situ hybridization
- HCl:
-
Hydrogen chloride
- kb:
-
Kilo-base pairs
- Mb:
-
Mega-base pairs
- MDR:
-
Mathematically defined repeat
- PCR:
-
Polymerase chain reaction
- sdw:
-
Semi-dwarfing
- SSC:
-
Saline sodium citrate
- TE:
-
Tris-EDTA buffer
References
Bevan MW, Garvin DF, Vogel JP (2010) Brachypodium distachyon genomics for sustainable food and fuel production. Curr Opin Biotechnol 21:211–217
Catalan P, Shi Y, Armstrong L, Draper J, Stace CA (1995) Molecular phylogeny of the grass genus Brachypodium P. Beauv. based on RFLP and RAPD analysis. Bot J Linn Soc 117:263–280
Cuadrado A, Jouve N (2007) The nonrandom distribution of long clusters of all possible classes of trinucleotide repeats in barley chromosomes. Chromosome Res 15:711–720
Danilova TV, Birchler JA (2008) Integrated cytogenetic map of mitotic metaphase chromosome 9 of maize: resolution, sensitivity, and banding paint development. Chromosoma 117:345–356
de Jong H, Fransz P, Zabel P (1999) High resolution FISH in plants—techniques and applications. Trends Plant Sci 4:258–263
Devos KM (2010) Grass genome organization and evolution. Curr Opin Plant Biol 13:139–145
Dolezel J, Greilhuber J, Lucretti S, Meister A, Lysak MA, Nardi L, Obermayer R (1998) Plant genome size estimation by flow cytometry: inter-laboratory comparison. Ann Bot 82:17–26
Draper J, Mur LA, Jenkins G, Ghosh-Biswas GC, Bablak P, Hasterok R, Routledge AP (2001) Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol 127:1539–1555
Foote TN, Griffiths S, Allouis S, Moore G (2004) Construction and analysis of a BAC library in the grass Brachypodium sylvaticum: its use as a tool to bridge the gap between rice and wheat in elucidating gene content. Funct Integr Genomics 4:26–33
Fukui K, Kamisugi Y, Sakai F (1994) Physical mapping of 5S rDNA loci by direct-cloned biotinylated probes in barley chromosomes. Genome 37:105–111
Hasterok R, Draper J, Jenkins G (2004) Laying the cytotaxonomic foundations of a new model grass, Brachypodium distachyon (L.) Beauv. Chromosome Res 12:397–403
Hasterok R, Marasek A, Donnison IS, Armstead I, Thomas A, King IP, Wolny E, Idziak D, Draper J, Jenkins G (2006) Alignment of the genomes of Brachypodium distachyon and temperate cereals and grasses using bacterial artificial chromosome landing with fluorescence in situ hybridization. Genetics 173:349–362
International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463:763–768
Jackson SA, Cheng Z, Wang ML, Goodman HM, Jiang J (2000) Comparative fluorescence in situ hybridization mapping of a 431-kb Arabidopsis thaliana bacterial artificial chromosome contig reveals the role of chromosomal duplications in the expansion of the Brassica rapa genome. Genetics 156:833–838
Jiang J, Gill BS (2006) Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49:1057–1068
Kato A, Albert PS, Vega JM, Birchler JA (2006) Sensitive fluorescence in situ hybridization signal detection in maize using directly labeled probes produced by high concentration DNA polymerase nick translation. Biotech Histochem 81:71–78
Koumbaris GL, Bass HW (2003) A new single-locus cytogenetic mapping system for maize (Zea mays L.): overcoming FISH detection limits with marker-selected sorghum (S. propinquum L.) BAC clones. Plant J 35:647–659
Künzel G, Korzun L, Meister A (2000) Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics 154:397–412
Lamb JC, Danilova T, Bauer MJ, Meyer JM, Holland JJ, Jensen MD, Birchler JA (2007) Single-gene detection and karyotyping using small-target fluorescence in situ hybridization on maize somatic chromosomes. Genetics 175:1047–1058
Lysak MA, Koch MA, Pecinka A, Schubert I (2005) Chromosome triplication found across the tribe Brassiceae. Genome Res 15:516–525
Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I (2006) Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci U S A 103:5224–5229
Mandakova T, Lysak MA (2008) Chromosomal phylogeny and karyotype evolution in x = 7 crucifer species (Brassicaceae). Plant Cell 20:2559–2570
Moore G, Gale MD, Kurata N, Flavell RB (1993) Molecular analysis of small grain cereal genomes: current status and prospects. Nat Biotechnol 11:584–589
Moore G, Devos KM, Wang Z, Gale MD (1995) Cereal genome evolution. Grasses, line up and form a circle. Curr Biol 5:737–739
Paterson AH, Bowers JE, Bruggmann R et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Phillips D, Nibau C, Ramsay L, Waugh R, Jenkins G (2010) Development of a molecular cytogenetic recombination assay for barley. Cytogenet Genome Res 129:154–161
Schnable PS, Ware D, Fulton RS et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Schulte D, Close TJ, Graner A, Langridge P, Matsumoto T, Muehlbauer G, Sato K, Schulman AH, Waugh R, Wise RP, Stein N (2009) The international barley sequencing consortium—at the threshold of efficient access to the barley genome. Plant Physiol 149:142–147
Vu GT, Wicker T, Buchmann JP, Chandler PM, Matsumoto T, Graner A, Stein N (2010) Fine mapping and syntenic integration of the semi-dwarfing gene sdw3 of barley. Funct Integr Genomics. doi:10.1007/s10142-010-0173-4
Wicker T, Narechania A, Sabot F, Stein J, Vu GT, Graner A, Ware D, Stein N (2008) Low-pass shotgun sequencing of the barley genome facilitates rapid identification of genes, conserved non-coding sequences and novel repeats. BMC Genomics 9:518
Wicker T, Taudien S, Houben A, Keller B, Graner A, Platzer M, Stein N (2009) A whole-genome snapshot of 454 sequences exposes the composition of the barley genome and provides evidence for parallel evolution of genome size in wheat and barley. Plant J 59:712–722
Wolny E, Hasterok R (2009) Comparative cytogenetic analysis of the genomes of the model grass Brachypodium distachyon and its close relatives. Ann Bot 104:873–881
Yu Y, Tomkins JP, Waugh R, Frisch DA, Kudrna D, Kleinhofs A, Brueggeman RS, Muehlbauer GJ, Wise RP, Wing RA (2000) A bacterial artificial chromosome library for barley (Hordeum vulgare L.) and the identification of clones containing putative resistance genes. Theor Appl Genet 101:1093–1099
Yu WC, Lamb JC, Han FP, Birchler JA (2007) Cytological visualization of DNA transposons and their transposition pattern in somatic cells of maize. Genetics 175:31–39
Acknowledgments
We are grateful to Martina Kühne and Katrin Kumke (IPK, Gatersleben) for excellent technical assistance. We thank Glyn Jenkins (IBERS, Aberystwyth, UK) and Robert Hasterok (Silesian University, Katowice, Poland) for providing B. distachyon seeds and BACs. This project was supported by the Leibniz Association (WGL).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Walther Traut.
Rights and permissions
About this article
Cite this article
Ma, L., Vu, G.T.H., Schubert, V. et al. Synteny between Brachypodium distachyon and Hordeum vulgare as revealed by FISH. Chromosome Res 18, 841–850 (2010). https://doi.org/10.1007/s10577-010-9166-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-010-9166-3