Abstract
This study presents a framework linkage map based on microsatellite markers for Muscadinia rotundifolia (1n = 20). The mapping population consisted of 206 progeny generated from a cross of two M. rotundifolia varieties, ‘Fry’ and ‘Trayshed’. A total of 884 primers were tested for their ability to amplify markers: 686 amplified and 312 simple sequence repeat (SSR) primer pairs generated 322 polymorphic markers for either one or both parents. The map for the female parent ‘Fry’ consisted of 212 markers and covered 879 cM on 18 chromosomes. The average distance between the markers was 4.1 cM and chromosome 6 was not represented due to a lack of polymorphic markers. The map for the male parent ‘Trayshed’ consisted of 191 markers and covered 841 cM on 19 chromosomes. The consensus map consisted of 314 markers on 19 chromosomes with a total distance of 1,088 cM, which represented 66 % of the distance covered by the Vitis vinifera reference linkage map. Marker density varied greatly among chromosomes from 5 to 35 mapped markers. Relatively good synteny was observed across 19 chromosomes based on markers in common with the V. vinifera reference map. Extreme segregation distortion was observed for chromosome 8 and 14 on the female parent map, and 4 on the male parent map. The lack of mapping coverage for the 20th M. rotundifolia chromosome is discussed in relation to possible evolutionary events that led to the reduction in chromosome number from 21 to 19 in the ancestral genome.




Similar content being viewed by others
References
Adam-Blondon AF, Roux C, Claux D, Butterlin G, Merdinoglu D, This P (2004) Mapping 245 SSR markers on the Vitis vinifera genome: a tool for grape genetics. Theor Appl Genet 109:1017–1027
Barker CL, Donald T, Pauquet J, Ratnaparkhe MB, Bouquet A, Adam-Blondon A-F, Thomas MR, Dry I (2005) Genetic and physical mapping of the grape powdery mildew resistant gene, Run1, using a bacterial artificial chromosome library. Theor Appl Genet 111:370–377
Bellin D, Peressotti E, Merdinoglu D, Wiedemann-Merdinoglu S, Adam-Blondon A-F, Cipriani G, Morgante M, Testolin R, Di Gaspero G (2009) Resistance to Plasmopara viticola in grapevine ‘Bianca’ is controlled by a major dominant gene causing localized necrosis at the infection site. Theor Appl Genet 120:163–176
Blasi P, Blanc B, Wiedemann-Merdinoglu S, Prado E, Rühl EH, Mestre P, Merdinoglu D (2011) Construction of a reference linkage map of Vitis amurensis and genetic mapping of Rpv8, a locus conferring resistance to grapevine downy mildew. Theor Appl Genet 123:43–53
Bloodworth PJ, Nesbitt WB, Barker KR (1980) Resistance to root-knot nematodes in Euvitis × Muscadinia hybrids. In: Proceedings of third international symposium on grape breeding, University of California, Davis, 15–18 June 1980, pp 275–292
Bouquet A (1980) Vitis × Muscadinia hybridization: a new way in grape breeding for disease resistance in France (Acta Hort). In: Proceedings of third international symposium on grape breeding, University of California, Davis, 15–18 June 1980, pp 42–61
Bowers JE, Dangl GS, Vignani R, Meredith CP (1996) Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome 39:628–633
Bowers JE, Dangl GS, Meredith CP (1999) Development and characterization of additional microsatellite DNA markers for grape. Am J Enol Vitic 50:243–246
Brizicky GK (1965) The genera of Vitaceae in the southeastern United States. J. Arnold Arboretum 46:48–67
Cipriani G, Marrazzo MT, Di Gaspero G, Pfeiffer A, Morgante M, Testolin R (2008) A set of microsatellite markers with long core repeat optimized for grape (Vitis spp.) genotyping. BMC Plant Biol 8:127
Clayton CN (1975) Diseases of muscadine and bunch grapes in North Carolina and their control. NC Agr Expt Stn Bull 451:31
Comeaux BL, Nesbitt WB, Fantz PR (1987) Taxonomy of the native grapes of North Carolina. Castanea 52:197–215
Costantini L, Grando MS, Feingold S, Ulanovsky S, Mejía N, Hinrichsen P, Doligez A, This P, Cabezas JA, Martínez-Zapater JM (2007) Generation of a common set of mapping markers to assist table grape breeding. Am J Enol Vitic 58:102–111
Dalbó MA, Ye GN, Weeden NF, Steinkellner H, Sefc KM, Reisch BI (2000) A gene controlling sex in grapevines placed on a molecular marker-based genetic map. Genome 43:333–340
Davidis UX, Olmo HP (1964) The Vitis vinifera × V. rotundifolia hybrids as phylloxera resistant rootstocks. Vitis 4:129–143
Decroocq V, Favé MG, Hagen L, Bordenave L, Decroocq S (2003) Development and transferability of apricot and grape EST microsatellite markers across taxa. Theor Appl Genet 106:912–922
Detjen LR (1919) The limits in hybridization of Vitis rotundifolia with related species and genera. NC Agric Expt Sta Bull 17:42
Devos KM (2005) Updating the “crop circle”. Curr Opin Plant Biol 8:155–162
Di Gaspero G, Cipriani G, Marrazzo MT, Andreetta D, Prado Castro MJ, Peterlunger E, Testolin R (2005) Isolation of (AC)n—microsatellites in Vitis vinifera L. and analysis of genetic background in grapevines under marker assisted selection. Mol Breed 15:11–20
Di Gaspero G, Cipriani G, Adam-Blondon AF, Testolin R (2007) Linkage maps of grapevine displaying the chromosomal locations of 420 microsatellite markers and 82 markers for R-gene candidates. Theor Appl Genet 114:1249–1263
Doligez A, Adam-Blondon A-F, Cipriani G, Di Gaspero G, Laucou V, Merdinoglu D, Meredith CP, Riaz S, Roux C, This P (2006) An integrated SSR map of grapevine based on five different populations. Theor Appl Genet 113:369–382
Dunstan RT (1962) Some fertile hybrids of bunch and muscadine grapes. J Hered 53:299–303
Esmenjaud D, Van Ghelder C, Roger V, Bordenave L, Decroocq S, Bouquet A, Ollat N (2010) Host suitability of Vitis and Vitis–Muscadinia material to the nematode Xiphinema index over one to four years. Am J Enol Vitic 61:96–101
Farré A, Benito L, Cistué L, de Jong JH, Romagosa I, Jansen J (2011) Linkage map construction involving a reciprocal translocation. Theor Appl Genet 122:1029–1037
Firoozabady E, Olmo HP (1982) Resistance to grape Phylloxera in Vitis vinifera × Vitis rotundifolia grape hybrids. Vitis 21:1–4
Fischer BM, Salakhutdinov I, Akkurt M, Eibach R, Edwards KJ, Töpfer R, Zyprian EM (2004) Quantitative trait locus analysis of fungal disease resistance factors on a molecular map of grapevine. Theor Appl Genet 108:501–515
Fry B (1967) Value of certain varieties and selections in the breeding of high quality, large-fruited muscadine grapes. Proc Am Soc Hort Sci 91:213–216
Galet P (1988) Cépages et Vignobles de France. Tome 1. Lesvignes Américaines, 2nd edn, Pierre Galet, Imprimerie Dehan, Montpellier, p 286
Goldy RG (1992) Breeding muscadine grapes. Hort Rev 14:357–405
Grando MS, Bellin D, Edwards KJ, Pozzi C, Stefanini M, Velasco R (2003) Molecular linkage maps of Vitis vinifera L. and Vitis riparia Mchx. Theor Appl Genet 106:1213–1224
Grzegorczyk W, Walker MA (1998) Evaluating resistance to grape phylloxera in Vitis species with an in vitro dual culture assay. Am J Enol Vitic 49:17–22
Hoffmann S, Di Gaspero G, Kovacs L, Howard S, Kiss E, Galbacs Z, Testolin R, Kozma P (2008) Resistance to Erysiphe necator in the grapevine ‘Kishmish vatkana’ is controlled by a single locus through restriction of hyphal growth. Theor Appl Genet 116:427–438
Huang H, Lu J, Ren Z, Hunter W, Dowd SE, Dang P (2010) Mining and validating grape (Vitis vinifera L.) ESTs to develop EST-SSR markers for genotyping and mapping. Mol Breed 28(2):241–254. doi:10.1007/s11032-010-9477-2
Husmann GC, Dearing C (1913) Muscadine grapes. US Dept Agric Farmers Bull, p 273
Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyere C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, Del Fabbro C, Alaux M, Di Gaspero G, Dumas V, Felice N, Paillard S, Juman I, Moroldo M, Scalabrin S, Canaguier A, Le Clainche I, Malacrida G, Durand E, Pesole G, Laucou V, Chatelet P, Merdinoglu D, Delledonne M, Pezzotti M, Lecharny A, Scarpelli C, Artiguenave F, Pe ME, Valle G, Morgante M, Caboche M, Adam-Blondon AF, Weissenbach J, Quetier F, Wincker P (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467
Jáuregui B, de Vicente MC, Messeguer R, Felipe A, Bonnet A, Salesses G, Arús P (2001) A reciprocal translocation between “Garfi” almond and “Nemared” peach. Theor Appl Genet 102:1169–1176
Karkamkar SP, Patil SG, Misra SC (2010) Cyto-morphological studies and their significance in evolution of family Vitaceae. Nucleus 53:37–43
Kirchheimer F (1939) Rhamnales/Vitaceae in Jongmanns W. fossilium, Catalogus Plantae 24. Feller Neubrandburg 2:153
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Lodhi MA, Reisch BI (1995) Nuclear DNA content of Vitis species, cultivars, and other genera of the Vitaceae. Theor Appl Genet 90:11–16
Lodhi MA, Reisch BI, Weeden NF (1994) A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Mol Bio Rep 12:6–13
Lowe KM, Walker MA (2006) Genetic linkage map of the interspecific grape rootstock cross Ramsey (Vitis champinii) × Riparia Gloire (Vitis riparia). Theor Appl Genet 112:1582–1592
Lysak M, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I (2006) Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci USA 103:5224–5229
Marguerit E, Boury C, Manicki A, Donnart M, Butterlin G, Némorin A, Wiedemann-Merdinoglu S, Merdinoglu D, Ollat N, Decroocq S (2009) Genetic dissection of sex determinism, inflorescence morphology and downy mildew resistance in grapevine. Theor Appl Genet 118:1261–1278
Merdinoglu D, Wiedemann-Merdinoglu S, Coste P, Dumas V, Haetty S, Butterlin G, Greif C (2003) Genetic analysis of downy mildew resistance derived from Muscadinia rotundifolia. Acta Hort 603:451–456
Merdinoglu D, Butterlin G, Bevilacqua L, Chiquet V, Adam-Blondon AF, Decroocq S (2005) Development and characterization of a large set of microsatellite markers in grapevine (Vitis vinifera L.) suitable for multiplex PCR. Mol Breed 15:349–366
Moisy C, Garrison K, Meredith CP, Pelsy F (2008) Characterization of ten novel Ty1 copia-like retrotransposon families of the grapevine genome. BMC Genomics 9:469
Moroldo M, Paillard S, Marconi R, Fabrice L, Canaguier A, Cruaud C, De Berardinis V, Guichard C, Brunaud V, Le Clainche I, Scalabrin S, Testolin R, Di Gaspero G, Morgante M, Adam-Blondon AF (2008) A physical map of the heterozygous grapevine ‘Cabernet Sauvignon’ allows mapping candidate genes for disease resistance. BMC Plant Bio 8:38
Mortensen JA (1981) Sources and inheritance of resistance to anthracnose in Vitis. J Hered 72:423–426
Munson TV (1909) Foundations of American grape culture. Munson TV and Son, Texas, p 252
Murphy WJ, Pevzner PA, O’Brien SJ (2004) Mammalian phylogenomics comes of age. Trends Genet 20:631–639
Olien WC (1990a) Muscadine—a classic southeastern fruit. HortScience 25:726–831
Olien WC (1990b) The muscadine grape: botany, viticulture, history, and current industry. HortScience 25:732–739
Olmo HP (1971) Vinifera × Rotundifolia hybrids as wine grapes. Am J Enol Vitic 22:87–91
Olmo HP (1986) The potential role of (vinifera × rotundifolia) hybrids in grape variety improvement. Experientia 42:921–926
Patel GI, Olmo HP (1955) Cytogenetics of Vitis: 1. The hybrid V. vinifera × V. rotundifolia. Am J Bot 42:141–159
Pauquet J, Bouquet A, This P, Adam-Blondon A-F (2001) Establishment of a local map of AFLP markers around the powdery mildew resistance gene Run1 in grapevine and assessment of their usefulness for marker aided selection. Theor Appl Genet 103:1201–1210
Péros JP, Berger G, Portemont A, Boursiquot JM, Lacombe T (2011) Genetic variation and biogeography of the disjunct Vitis subgenus Vitis (Vitaceae). J Biogeogr 38:471–486
Reimer FC, Detjen RL (1914) Breeding rotundifolia grapes: a study of transmission of character. NC Agr Expt Sta Tech Bul 10:1–47
Ren H, Wen J (2007) Vitis. In: Wu ZY, Raven PH, Hong DY (eds) Flora of China, vol 12. Science Press, Beijing, pp 210–222 (Missouri Botanical Garden Press, St. Louis)
Riaz S, Dangl GS, Edwards KJ, Meredith CP (2004) A microsatellite marker based framework linkage map of Vitis vinifera L. Theor Appl Genet 108:864–872
Riaz S, Krivanek AF, Xu K, Walker MA (2006) Refined mapping of the Pierce’s disease resistance locus, PdR1, and sex on an extended genetic map of Vitis rupestris × V. arizonica. Theor Appl Genet 113:1317–1329
Riaz S, Tenscher AC, Ramming DW, Walker MA (2011) Using a limited mapping strategy to identify major QTLs for resistance to grapevine powdery mildew (Erysiphe necator) and their use in marker-assisted breeding. Theor Appl Genet 122:1059–1073
Rozen S, Skaletsky H (2000) Primer 3 on the WWW for general users and for biologist programmers. Methods Mol Bio 132:365–386
Ruel JJ, Walker MA (2006) Resistance to Pierce’s disease in Muscadinia rotundifolia and other native grape species. Am J Enol Vitic 57:158–165
Scalabrin S, Troggio M, Moroldo M, Pindo M, Felice N, Coppola G, Prete G, Malacarne G, Marconi R, Faes G, Jurman I, Grando S, Jesse T, Segala C, Valle G, Policriti A, Fontana P, Morgante M, Velasco R (2010) Physical mapping in highly heterozygous genomes: a physical contig map of the Pinot Noir grapevine cultivar. BMC Genomics 11:204
Schranz ME, Lysak MA, Mitchell-Olds T (2006) The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci 11:535–542
Schranz ME, Song BH, Windsor AJ, Mitchell-Olds T (2007a) Comparative genomics in the Brassicaceae: a family wide perspective. Curr Opin Plant Biol 10:168–175
Schranz ME, Windsor AJ, Song BH, Lawton-Rauh A, Mitchell-Olds T (2007b) Comparative genetic mapping in Boechera stricta, a close relative of Arabidopsis. Plant Physiol 144:286–298
Scott KD, Eggler P, Seaton G, Rossetto M, Ablett EM, Lee LS, Henry RJ (2000) Analysis of SSRs derived from grape ESTs. Theor Appl Genet 100:723–726
Sefc KM, Regner F, Turetschek E, Glossl J, Steinkellner H (1999) Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome 42:367–373
Small J (1913) Flora of the Southeastern United States, 2nd edn. The Author, New York
Smith BP (2010) Genetic and molecular mapping studies on a population derived from Vitis vinifera × Muscadinia rotundifolia and genetic diversity of wild Muscadinia rotundifolia. PhD dissertation, University of California, Davis
This P, Lacombe T, Thomas MR (2006) Historical origins and genetic diversity of wine grapes. Trends Genet 22:511–519
Thomas MR, Scott NS (1993) Microsatellite repeats in grapevine reveal DNA polymorphism when analysed as sequence-tagged sites (STSs). Theor Appl Genet 86:985–990
Troggio M, Malacarne G, Coppola G, Segala C, Cartwright DA, Pindo M, Stefanini M, Mank R, Moroldo M, Morgante M, Grando MS, Velasco R (2007) A dense single-nucleotide polymorphism-based genetic linkage map of grapevine (Vitis vinifera L.) anchoring Pinot Noir bacterial artificial chromosome contigs. Genetics 176:2637–2650
Tröndle D, Schröder S, Kassemeyer H-H, Kiefer C, Koch MA, Nick P (2010) Molecular phylogeny of the genus Vitis (Vitaceae) based on plastid markers. Am J Bot 97:1168–1178
van Ooijen JW (2006) JoinMap®4.0 software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen
Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, Pindo M, Fitzgerald LM, Vezzulli S, Reid J, Malacarne G, Iliev D, Coppola G, Wardell B, Micheletti D, Macalma T, Facci M, Mitchell JT, Perazzolli M, Eldredge G, Gatto P, Oyzerski R, Moretto M, Gutin N, Stefanini M, Chen Y, Segala C, Davenport C, Demattè L, Mraz A, Battilana J, Stormo K, Costa F, Tao Q, Si-Ammour A, Harkins T, Lackey A, Perbost C, Taillon B, Stella A, Solovyev V, Fawcett JA, Sterck L, Vandepoele K, Grando MS, Toppo S, Moser C, Lanchbury J, Bogden R, Skolnick M, Sgaramella V, Bhatnagar SK, Fontana P, Gutin A, Van de Peer Y, Salamini F, Viola R (2007) A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2:e1326
Voorips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Walker MA, Lider LA, Goheen AC, Olmo HP (1991) VR 039–16. HortScience 26:1224–1225
Walker MA, Ferris H, Eyre M (1994a) Resistance in Vitis and Muscadinia species to Meloidogyne incognita. Plant Dis 78:1055–1058
Walker MA, Wolpert JA, Weber E (1994b) Viticultural characteristics of VR hybrid rootstocks in a vineyard site infected with grapevine fanleaf virus. Vitis 33:19–23
Wan Y, Schwaninger H, Li D, Simon CJ, Wang Y, Zhang C (2008) A review of the taxonomic research on Chinese wild grapes. Vitis 47:81–88
Wang J, Roe B, Macmil S, Yu Q, Murray JE, Tang H, Chen C, Najar F, Wiley G, Bowers J, Van Sluys MA, Rokhsar DS, Hudson ME, Moose SP, Paterson AH, Ming R (2010) Microcollinearity between autopolyploid sugarcane and diploid sorghum genomes. BMC Genomics 11:261
Wen J, Nie Z-L, Soejima A, Meng Y (2007) Phylogeny of Vitaceae based on the nuclear GAI1 gene sequences. Can J Bot 85:731–745
Zecca G, Richard Abbott J, Sun W-B, Spada A, Sala F, Grassi F (2012) The timing and the mode of evolution of wild grapes (Vitis). Mol Phylogenet Evol 62:736–747
Zhang J, Hausmann L, Eibach R, Welter LJ, Töpfer R, Zyprian EM (2009) A framework map from grapevine V3125 (Vitis vinifera ‘Schiava grossa’ × ‘Riesling’) × rootstock cultivar ‘Börner’ (Vitis riparia × Vitis cinerea) to localize genetic determinants of phylloxera root resistance. Theor Appl Genet 119:1039–1051
Zyprian E, Welter LJ, Akkurt M, Ebert S, Salakhutdinov I, Gokurk-Baydar N, Eibach R, Topfer R (2009) Genetic analysis of fungal disease resistance in grapevine. Acta Hort 827:535–538
Acknowledgments
Research funding from the California Grape Rootstock Improvement Commission, the California Grapevine Rootstock Research Foundation, the California Department of Food and Agriculture PD/GWSS Board and the Louis P. Martini Endowed Chair funds is gratefully acknowledged. The authors also thank Dr. John E. Bowers, Institute of Plant Breeding Genetics and Genomics, University of Georgia, and Didier Merdinoglu, Institut National de la Recherche Agronomique—Colmar, for helpful suggestions on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Charcosset.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2012_1906_MOESM2_ESM.ppt
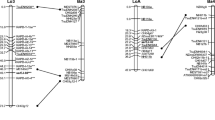
Supplementary Figure 2. Comparison of the consensus genetic map of M. rotundifolia (center) to two different maps of V. vinifera. On the left (A) is the integrated map (Doligez et al. 2006) of five different grape populations: 1) Syrah × Grenache; 2) Selfed Riesling; 3) Chardonnay × Bianca (Bouvier × Villard Blanc); 4) MTP2223-27 (Dattier de Beyrouth × 75 Pirovano) × MTP2121-30 (Alphonse Lavallée × Sultanine); and 5) Riesling × Cabernet Sauvignon. The map on the right (B) is a consensus of two populations (Chardonnay × Bianca (Bouvier × Villard Blanc), and Cabernet Sauvignon × Vitis ‘breeding line 20/3’. Lines join markers common among the three maps. Marker names in red indicate those SSR primer pairs that amplified V. vinifera ‘Thompson Seedless’ but failed to generate any product for the Muscadinia samples (PPT 4242 kb)
Rights and permissions
About this article
Cite this article
Riaz, S., Hu, R. & Walker, M.A. A framework genetic map of Muscadinia rotundifolia . Theor Appl Genet 125, 1195–1210 (2012). https://doi.org/10.1007/s00122-012-1906-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-012-1906-7