Abstract
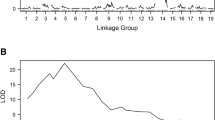
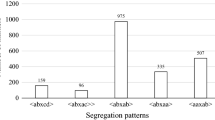
Grapevine rootstock cultivar ‘Börner’ is a hybrid of Vitis riparia and Vitis cinerea Arnold that shows high resistance to phylloxera (Daktulosphaira vitifoliae Fitch). To localize the determinants of phylloxera root resistance, the susceptible grapevine V3125 (Vitis vinifera ‘Schiava grossa’ × ‘Riesling’) was crossed to ‘Börner’. Genetic framework maps were built from the progeny. 235 microsatellite markers were placed on the integrated parental map. They cover 1,155.98 cM on 19 linkage groups with an average marker distance of 4.8 cM. Phylloxera resistance was scored by counting nodosities after inoculation of the root system. Progeny plants were triplicated and experimentally infected in 2 years. A scan of the genetic maps indicated a quantitative trait locus on linkage group 13. This region was targeted by six microsatellite-type markers newly developed from the V. vinifera model genome sequence. Two of these appear closely linked to the trait, and can be useful for marker-assisted breeding.




Similar content being viewed by others
References
Ablett E, Seaton G, Scott K, Shelton D, Graham MW, Baverstock P, Lee SL, Henry R (2000) Analysis of grape ESTs: global gene expression patterns in leaf and berry. Plant Sci 159:87–95
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bowers JE, Dangl GS, Vignani R, Meredith C (1996) Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome 39:628–633
Bowers JE, Dangl GS, Meredith C (1999) Development and characterization of additional microsatellite DNA markers for grape. Am J Enol Vitic 50:243–246
Cabezas JA, Cervera MT, Ruiz-Garcia L, Carreno J, Martinez-Zapater JM (2006) A genetic analysis of seed and berry weight in grapevine. Genome 49:1572–1585
Dalbó MA, Ye GN, Weeden NF, Steinkellner H, Sefc KM, Reisch BI (2000) A gene controlling sex in grapevine placed on a molecular marker-based genetic map. Genome 43:333–340
Di Gaspero G, Cipriani G, Marrazzo MT, Andretta D, Prado, Castro MJ, Peterlunger E, Testolin R (2005) Isolation of (AC)n -microsatellites in Vitis vinifera L. and analysis of genetic background in grapevines under marker assisted selection. Mol Breed 15:11–20
Di Gaspero G, Cipriani G, Adam-Blondon A-F, Testolin R (2007) Linkage maps of grapevine displaying the chromosomal locations of 420 microsatellite markers and 82 markers for R-gene candidates. Theor Appl Genet 114:1249–1263
Doligez A, Bouquet A, Danglot Y, Lahogue F, Riaz S, Meredith CP, Edwards KJ, This P (2002) Genetic mapping of grapevine (Vitis vinifera L.) applied to the detection of QTLs for seedlessness and berry weight. Theor Appl Genet 105:780–795
Doligez A, Adam-Blondon A-F, Cipriani G, Di Gaspero G, Laucou V, Merdinoglu D, Meredith CP, Riaz S, Roux C, This P (2006) An integrated SSR map of grapevine based on five mapping populations. Theor Appl Genet 113:369–382
Doucleff M, Jin Y, Gao F, Riaz S, Krivanek AF, Walker MA (2004) A genetic linkage map of grape, utilizing Vitis rupestris and Vitis arizonica. Theor Appl Genet 109:1178–1187
El-Nady MF (2001) Untersuchungen zum Mechanismus der Reblausresistenz der Unterlagsrebsorte ‘Börner’. Dissertation, Johannes Gutenberg-Universität Mainz
Fischer B, Salakhutdinov I, Akkurt M, Eibach R, Edwards KJ, Toepfer R, Zyprian EM (2004) Quantitative trait locus analysis of fungal disease resistance factor on a molecular map of grapevine. Theor Appl Genet 108:505–515
Grando MS, Bellin D, Edwards KJ, Pozzi C, Stefanini M, Velasco R (2003) Molecular linkage maps of Vitis vinifera L. and Vitis riparia Mchx. Theor Appl Genet 106:1213–1224
Grando MS, Sevini F, Moser S, Marino R, Dalla Serra A, Versini G (2004) Genetic mapping of aroma compounds in grape. Bull de l′OIV 2004:881–882
Grattapaglia D, Sederoff R (1994) Genetic linkage maps of Eucalyptus grandis and Eucalytus urophylla using a pseudo-testcross mapping strategy and RAPD markers. Genetics 137:1121–1137
Hulbert SH, Ilott TW, Legg EJ, Lincoln SE, Lander ES, Michelmore RW (1988) Genetic analysis of the fungus Bremia lactucae, using restriction fragment length polymorphisms. Genetics 120:947–958
Jaillon O, Aury J-M, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyere C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, DelFabbro C, Alaux M, DiGaspero G, Dumas V, Felice N, Paillard S, Juman I, Moroldo M, Scalabrin S, Canaguier A, LeClainche I, Malacrida G, Durand E, Pesole G, Laucou V, Chatelet P, Merdinoglu D, Delledonne M, Pezotti M, Lecharny A, Scarpelli C, Artiguenave F, Pe ME, Valle G, Morgante M, Caboche M, Adam-Blondon A-F, Weissenbach J, Quetier F, Wincker O (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–468
Kellow AV, Sedgley M, Van Heeswijck R (2004) Interaction between Vitis vinifera and grape phylloxera: changes in root tissue during nodosity formation. Ann Bot 93:581–590
Kosambi DD (1944) The estimation of distances from recombination values. Ann Eugenet 12:172–175
Lamoureux D, Bernole A, LeClainche I, Tual S, Thareau V, Paillard S, Legeai F, Dossat C, Wincker P, Oswald M, Merdinoglu D, Vignault C, Delrot S, Caboche M, Chalhoub B, Adam-Blondon A-F (2006) Anchoring of a large set of markers onto a BAC library for the development of a draft physical map of the grapevine genome. Theor Appl Genet 113:344–356
Lodhi MA, Daly MJ, Ye GN, Weeden NF, Reisch BI (1995) A molecular marker based linkage map of Vitis. Genome 38:786–794
Lowe KM, Walker MA (2006) Genetic linkage map of the interspecific grape rootstock cross Ramsey (Vitis champinii) × Riparia Gloire (Vitis riparia). Theor Appl Genet 112:1582–1592
Mandl K, Santiago J-L, Hack R, Fardossi A, Regner F (2006) A genetic map of Welschriesling × Sirius for the identification of magnesium-deficiency by QTL analysis. Euphytica 149:133–144
Merdinoglu D, Butterlin G, Bevilacqua L, Chiquet V, Adam-Blondon AF, Decroocq S (2005) Development and characterization of a large set of microsatellite markers in grapevine (Vitis vinifera L.) suitable for multiplex PCR. Mol Breed 15:349–366
Moser C, Segala C, Fontana P, Salakhutdinov I, Gatto P, Pindo M, Zyprian E, Toepfer R, Grando MS, Velasco R (2005) Comparative analysis of expressed sequence tags from different organs of Vitis vinifera L. Funct Integr Genomics 5:208–217
Niklowitz X (1954) Histologische Studien an Reblausgallen und Reblausabwehrnekrosen (Viteus vulpinae CB. auf Vitis vinifera und Vitis riparia). Phytopathologische Z 24:299–340
Peng FY, Reid K, Liao N, Schlosser J, Lijavetzky D, Holt R, Martinez Zapater JM, Jones S, Marra M, Bohlmann J, Lund ST (2007) Generation of ESTs in Vitis vinifera wine grape (Cabernet Sauvignon) and table grape (Muscat Hamburg) and discovery of new candidate genes with potential roles in berry development. Gene 402:40–50
Riaz S, Dangl GS, Edwards KJ, Meredith CP (2004) A Microsatellite based framework linkage map of Vitis vinifera L. Theor Appl Genet 108:864–872
Riaz S, Krivanek AF, Xu K, Walker MA (2006) Refined mapping of the Pierce’s disease resistance locus, PdR1, and sex on the extended genetic map of Vitis rupestris × V. arizonica. Theor Appl Genet 113:1317–1329
Riaz S, Tenscher AC, Rubin J, Graziani R, Pao SS, Walker MA (2008) Fine-scale genetic mapping of two Pierce’s disease resistance loci and a major segregation distortion region on chromosome 14 of grape. Theor Appl Genet 117:671–681
Roush TL, Granett J, Walker MA (2007) Inheritance of gall formation relative to phylloxera resistance levels in hybrid grapevines. Am J Enol Vitic 58:234–241
Salmaso M, Malacarne G, Troggio M, Faes G, Stefanini M, Grando MS, Velasco R (2008) A grapevine (Vitis vinifera L.) genetic map integrating the position of 139 expressed genes. Theor Appl Genet 116:1129–1143
Schäller G (1965) Ergebnisse der Rebenunterlagenzüchtung mit Vitis cinerea Arnold. Der Züchter 35:250–255
Schmid J, Rühl EH (2003) Performance of Vitis cinerea hybrids in motherblock and nursery—preliminary results. Acta Hort 617, ISHS 2003:141–145
Schmid J, Manty F, Rühl EH (2003) Experience with phylloxera tolerant and resistant rootstocks at different vineyard sites. Acta Hort 617, ISHS 2003:85–93
Scott KD, Eggler P, Seaton G, Rossetto M, Ablett EM, Lee LS, Henry RJ (2000) Analysis of SSRs derived from grape ESTs. Theor Appl Genet 100:723–726
Sefc KM, Regner F, Turetschek E, Glössl J, Steinkellner H (1999) Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome 42:367–373
Thomas MR, Scott N (1993) Microsatellite repeats in grapevine reveal DNA polymorphisms when analyzed as sequence-tagged sites (STSs). Theor Appl Genet 6:985–990
Troggio M, Malacarne G, Coppola G, Segala C, Cartwright DA, Pindo M, Stefanini M, Mank R, Moroldo M, Morgante M, Grando MS, Velasco R (2007) A dense single-nucleotide polymorphism based genetic linkage map of grapevine (Vitis vinifera L.) anchoring Pinot noir bacterial artificial chromosome contigs. Genetics 176:2637–2650
Van Heeswijck R, Bondar A, Croser L, Franks T, Kellow A, Powell K (2003) Molecular and cellular events during the interaction of phylloxera with grapevine roots. In: Rühl EH and Schmid J (eds) Proceedings of phylloxera infested vineyards. Acta Hort 617, ISHS 2003, pp 13–15
Van Oijen JW (2004) MapQTL®5, software for the mapping of quantitative trait loci in experimental populations. Kyazma B.V, Wageningen
Van Ooijen JW (2006) JoinMap®4.0, software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V, Wageningen
Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss A, Pruss D, Pindo M, FitzGerald LM, Vezulli S, Reid J, Malacarne G, Iliev D, Coppola G, Wardell B, Micheletti D, Macalma T, Facci M, Mitchell JT, Perrazzolli M, Eldredge G, Gatto P, Oyzerski R, Moretto M, Gutin N, Stefanini M, Chen Y, Segala C, Davenport C, Dematte L, Mraz A, Battilana J, Stormo K, Costa F, Tao Q, Si-Ammour A, Harkins T, Lackey A, Perbost C, Taillon B, Stella A, Solovyev V, Fawcett JA, Sterck L, Vandepoele K, Grando SM, Toppo S, Moser C, Lanchbury J, Bogden R, Skolnick M, Sgaramella V, Bhatnagar SK, Fontana P, Gutin A, Van de Peer Y, Salamini F, Viola R (2007) A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS One 2(12):e1326. doi:10.1371/journal.pone.0001326
Welter LJ, Göktürk-Baydar N, Akkurt M, Maul E, Eibach R, Töpfer R, Zyprian EM (2007) Genetic mapping and localization of quantitative trait loci affecting fungal disease resistance and leaf morphology in grapevine (Vitis vinifera L.). Mol Breed 20:359–374
Xu K, Riaz S, Roncoroni NC, Jin Y, Hu R, Zhou R, Walker MA (2008) Genetic and QTL analysis of resistance to Xiphinema index in a grapevine cross. Theor Appl Genet 116:305–311
Acknowledgments
The expert technical assistance of Andreas Preiss, Dagmar Andrä and Margrit Daum is highly acknowledged. Junke Zhang was funded as a visiting scientist under the frame of a Sino-Germany Cooperation by the Federal Ministry of Food, Agriculture and Consumer Protection and the Foundation of Friends and Promoters of the Institute for Grapevine Breeding Geilweilerhof.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Graner.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2009_1107_MOESM1_ESM.xls
Primer list of the STMS markers specifically developed for linkage group 13 based on the genome sequence of PN40024 (sc = scaffold) (XLS 19 kb)
Rights and permissions
About this article
Cite this article
Zhang, J., Hausmann, L., Eibach, R. et al. A framework map from grapevine V3125 (Vitis vinifera ‘Schiava grossa’ × ‘Riesling’) × rootstock cultivar ‘Börner’ (Vitis riparia × Vitis cinerea) to localize genetic determinants of phylloxera root resistance. Theor Appl Genet 119, 1039–1051 (2009). https://doi.org/10.1007/s00122-009-1107-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-009-1107-1