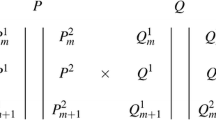Abstract
Sugarcane-breeding programs take at least 12 years to develop new commercial cultivars. Molecular markers offer a possibility to study the genetic architecture of quantitative traits in sugarcane, and they may be used in marker-assisted selection to speed up artificial selection. Although the performance of sugarcane progenies in breeding programs are commonly evaluated across a range of locations and harvest years, many of the QTL detection methods ignore two- and three-way interactions between QTL, harvest, and location. In this work, a strategy for QTL detection in multi-harvest-location trial data, based on interval mapping and mixed models, is proposed and applied to map QTL effects on a segregating progeny from a biparental cross of pre-commercial Brazilian cultivars, evaluated at two locations and three consecutive harvest years for cane yield (tonnes per hectare), sugar yield (tonnes per hectare), fiber percent, and sucrose content. In the mixed model, we have included appropriate (co)variance structures for modeling heterogeneity and correlation of genetic effects and non-genetic residual effects. Forty-six QTLs were found: 13 QTLs for cane yield, 14 for sugar yield, 11 for fiber percent, and 8 for sucrose content. In addition, QTL by harvest, QTL by location, and QTL by harvest by location interaction effects were significant for all evaluated traits (30 QTLs showed some interaction, and 16 none). Our results contribute to a better understanding of the genetic architecture of complex traits related to biomass production and sucrose content in sugarcane.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sugarcane (Saccharum spp.) is a clonally propagated outcrossing polyploid crop of great importance in tropical agriculture as a source of sugar and bioethanol. Modern commercial sugarcane cultivars are derived from interspecific crosses between Saccharum officinarum (basic chromosome number: x = 10; 2n = 8x = 80) and its wild relative S. spontaneum (x = 8; 5x ≤ 2n ≤ 16x), followed by few cycles of intercrossing and selection. Due to the intercrossings, these modern cultivars have chromosome number in somatic cells (2n) ranging from 100 to 130 (D’Hont et al. 1998; Irvine 1999; Grivet and Arruda 2001; D’Hont 2005; Piperidis et al. 2010).
Quantitative trait loci (QTL) mapping is a useful tool to dissect and to understand the genetic architecture of complex traits. However, two main complicating factors make QTL mapping more challenging in sugarcane than other species. (1) Ploidy level: the polyploidy and aneuploidy nature of sugarcane cultivars cause a complex pattern of chromosomal segregation in meiosis (Heinz and Tew 1987); (2) Outbred parents: since sugarcane inbred lines are not available, linkage map construction and QTL mapping rely on segregating progenies derived from biparental cross of highly heterozygous outbred parents. These two factors combined enable the appearance of different allele dosages (copy number variation) in each locus (marker or QTL), therefore, a mixture of segregating patterns can be observed in the segregating progenies (Ripol et al. 1999; Wu et al. 2002a, b; Lin et al. 2003). Moreover, due to the usage of outbred parents, linkage phases between markers are unknown.
The estimation of genetic linkage maps in sugarcane started after the development of single-dose markers (SDMs) (Wu et al. 1992). In a biparental cross, an SDM has either a single copy of an allele in one parent only or a single copy of the same allele in both parents, thus segregating in 1:1 (presence : absence) or 3:1 (presence:absence) ratio, respectively. The double pseudo-testcross strategy uses SDMs segregating in 1:1 ratio for each parent separately to build two independent genetic maps (one for each parent) for any cross between heterozygous parents with bivalent pairing in meiosis (Grattapaglia and Sederoff 1994; Porceddu et al. 2002; Shepherd et al. 2003; Carlier et al. 2004; Chen et al. 2008; Cavalcanti and Wilkinson 2007). In spite of the relative success of the double pseudo-testcross strategy in sugarcane (for example, Al-janabi et al. 1993; Ming et al. 1998; Hoarau et al. 2001; McIntyre et al. 2005a), an integrated map combining SDMs segregating in 1:1 and 3:1 ratio (Garcia et al. 2006; Oliveira et al. 2007) permits better genome saturation and characterization of the polymorphic variation in the biparental cross, therefore, being a more realistic framework for QTL mapping.
Although many statistical methods have been specifically developed to map QTLs in outcrossing species (Knott and Haley 1992; Haley et al. 1994; Schäfer-Pregl et al. 1996; Knott et al. 1997; Sillanpää and Arjas 1999; Lin et al. 2003; Wu et al. 2007; Hu and Xu 2009), the general double pseudo-testcross method has been widely used to study QTL in sugarcane through single marker analysis (SM), interval mapping (IM) and composite interval mapping (CIM) (Sills et al. 1995; Daugrois et al. 1996; Ming et al. 2001; 2002a, b; Hoarau et al. 2002; Jordan et al. 2004; da Silva and Bressiani 2005; McIntyre et al. 2005a, b, 2006; Reffay et al. 2005; Aitken et al. 2006, 2008; Raboin et al. 2006; Al-Janabi et al. 2007; Piperidis et al. 2008; Pinto et al. 2010; Pastina et al. 2010). In this approach, statistical analyses are carried out with the well-established backcross model using softwares developed for inbred-based populations. However, for the reasons stated previously, an integrated-map-based model might be a better choice for outcrossing species, such as sugarcane.
In addition to its genetic complexity, sugarcane is a perennial crop, in which individuals are usually harvested in multiple years. Thus, traits are often repeatedly measured not only across different locations but also along successive years (harvests), adding a time dimension to the phenotypic data. Quantitative-trait-based sugarcane varietal selection is commonly based on information from a series of field trials, considering different harvests and locations, here called multi-harvest-location trials (MHLT). QTL studies in sugarcane usually are carried out for each harvest-location trial separately, ignoring QTL-by-harvest (QTL × H), QTL-by-location (QTL × L) and QTL-by-harvest-by-location (QTL × H × L) interactions (Hoarau et al. 2002; Jordan et al. 2004; McIntyre et al. 2005b; Reffay et al. 2005; Pinto et al. 2010; Pastina et al. 2010). The use of statistical models that allow the identification of stable QTL across different environments (an environment is any combination of location and harvest) can provide powerful and useful information for breeding purposes, such as breeding values in marker-assisted selection (MAS).
Mixed models have been successfully employed to study genotype-by-environment (G × E) interaction (Denis et al. 1997; Piepho 1997; Cullis et al. 1998; Chapman 2008; Smith et al. 2001, 2007; van Eeuwijk et al. 2007), as well as QTL-by-environment (QTL × E) interaction (Piepho 2000, 2005; Verbyla et al. 2003; Malosetti et al. 2004, 2008; van Eeuwijk et al. 2005; Boer et al. 2007; Mathews et al. 2008). They provide great flexibility to represent the complex variance-covariance structures that follow from the patterns of genetic correlations between harvests and locations. In this article, we propose a mixed model QTL mapping strategy for sugarcane, paying special attention to model dependencies (correlations) between harvests and locations, which allows us to find stable QTLs that can be distinguished from environment-sensitive QTLs.
Materials and methods
Plant material
Phenotypic and molecular data were collected in a segregating population of 100 individuals derived from a cross between two pre-commercial Brazilian cultivars, SP80-180 (B3337 × polycross) and SP80-4966 (SP71-1406 × polycross). SP80-180 was the female parent and had lower sucrose content and high stalk production, whereas SP80-4966 (male parent) had higher sucrose and lower stalk production. Both parents and population were developed at the Experimental Station of the Centro de Tecnologia Canavieira (CTC), Camamu county, State of Bahia, Brazil.
Molecular data
Restriction fragment length polymorphism (RFLP), RFLP and simple sequence repeat (SSR) markers derived from expressed sequence tag (EST-RFLP and EST-SSR) were used to genotype parents and progeny. All these markers had already been generated and coded, as detailed in Garcia et al. (2006) and Oliveira et al. (2007). Each segregating allele was scored as a dominant marker, based on its presence or absence in the progeny. Only SDMs were considered. The observed segregation pattern of each marker was tested against its expected ratio using chi-square tests (χ2): 1:1 if it is a SDM present in only one parent or 3:1 if it is a SDM present in both parents. All loci with strong deviations from expected proportions were discarded after Bonferroni correction.
Phenotypic data
The mapping population was planted in 2003 at two locations (Piracicaba and Jaú, both in the State of São Paulo, Brazil), and evaluated in the first, second and third harvest years for cane yield (tonnes of cane per hectare, TCH), sugar yield (tonnes of sugar per hectare, TSH), fiber percent and sucrose content (Pol). In each location, the experimental design consisted of an augmented randomized complete block design with two replicates. However, genotypes were not fully randomized within blocks, instead they were randomly split into three groups with 36, 38, and 26 individuals each. Then, individuals were randomized within each group, but groups were not randomized within blocks. In the experiments, each group of individuals was augmented by four checks (commercial cultivars SP80-1842, SP81-3250, SP80-1816 and RB72454). Both parents were also included in one of the groups, but not considered in the statistical analysis.
Linkage map
Based on a multipoint approach (Wu et al. 2002a, b), map construction was carried out using the OneMap package (Margarido et al. 2007). For this purpose, 741 molecular markers were used, including 459 loci displaying an 1:1 segregation ratio (100 RFLP, 27 EST-RFLP, 332 EST-SSR) and 282 loci segregating in a 3:1 ratio (88 RFLP, 10 EST-RFLP, 184 EST-SSR). Following the notation in Wu et al. (2002a), markers segregating for the parent SP80-180 (P 1) were denoted by D 1, corresponding to the configuration ‘ao × oo’, in which the a allele is dominant over the o (null) allele. Informative loci for the parent SP80-4966 (P 2) were denoted by D 2, with the configuration ‘oo × ao’, and markers segregating for both parents were denoted by C, with configuration ‘ao × ao’. Markers were assigned to linkage groups (LGs) based on two point analysis, considering a minimum LOD threshold of 6. LGs with a maximum of five loci were ordered through the comparison of all possible orders, in a procedure analogous to the compare command in the MAPMAKER/EXP software (Lander et al. 1987). For LGs with more than 5 markers, the order algorithm started with the five most informative markers, which were ordered through the comparison of all possible orders, and then the other markers were sequentially placed on the LG at the position with largest likelihood, in a similar way to that performed by the try command in the MAPMAKER/EXP software. Afterward, the ripple command was applied to verify if local inversions had occurred. Map distances were expressed in centiMorgans (cM) based on the Kosambi function (Kosambi 1944). LGs were assembled into putative homology groups (HGs) when at least two loci (from the same or different marker type: RFLP, EST-RFLP or EST-SSR) were shared (Jannoo et al. 2004; Okada et al. 2010).
Genetic predictors
For notation purposes, in a similar way to that proposed by Lin et al. (2003), consider a full-sib progeny obtained from a cross between two outbred diploid parents, denoted as P and Q (Fig. 1). The illustration in Fig. 1 could be seen as a general case when compared with loci configuration observed in sugarcane, where only SDMs were considered. The genotypes of two adjacent markers m and m + 1 can be represented by P {1,2} m , Q {1,2} m , P {1,2} m+1 and Q {1,2} m+1 , in which {1, 2} indicates the allelic possibilities for each locus. However, since we are using dominant markers, we let allele 2 in parents P and Q representing possibly a series of alleles in polyploid species. Allele 2 could be thought as “all but allele 1”. Suppose that there is a QTL between these two markers, with alleles P 1 and P 2 for parent P, Q 1 and Q 2 for parent Q. Thus, QTL segregation in the progeny will fit into four genotypic classes (P 1 Q 1, P 1 Q 2, P 2 Q 1 and P 2 Q 2), with an 1:1:1:1 ratio. Therefore, it is possible to define three orthogonal contrasts involving these four genotypic classes (Lin et al. 2003; Gazaffi 2009):
The first and second contrasts relate to additive QTL effects in parents P and Q respectively, while the third refers to dominance effect (intra-locus interaction) between the additive effects in each parent. Genetic predictors were constructed for a discrete grid of evaluation points (w) along the genome (w = 1, …, W). These genetic predictors were used as explanatory variables in the mixed models. For individual i and evaluation point w, the genetic predictors are:
where \({x_p}_{iw}, {x_q}_{iw}\) and \({x_{pq}}_{iw}\) are the expected values of α p , α q and δ pq respectively, conditional on all marker information M i in a particular LG (Haley and Knott 1992; Martínez and Curnow 1992; Lynch and Walsh 1998). The conditional multipoint probabilities \(p(P^1Q^1|{\bf M}_{\rm i}), p(P^1Q^2|{\bf M}_{\rm i}),p(P^2Q^1|{\bf M}_{\rm i})\) and \(p(P^2Q^2|{\bf M}_{\rm i})\) were calculated via hidden Markov chain model (OneMap package, Margarido et al. 2007) for all marker positions and discrete grid of evaluation points with step size of 1 cM along the genome.
Due to the lack of information of SDMs (i.e. only 1:1 and 3:1 segregation patterns could be obtained), some genetic predictors could be linear combinations of others at some genomic positions, therefore, the matrix of genetic predictors could be singular. Since collinearity could cause serious problems with estimation and interpretation of parameters, its presence was investigated by examining the singular values and the condition number of the matrix of genetic predictors at all genomic positions. Only informative contrasts (without collinearity) were then considered. For example, LGs with only marker type D 1 have enough information solely for the estimation of one contrast for the additive effect in parent P. The same principle was applied to all LGs and genomic positions.
Multi-harvest-location phenotypic analysis
Prior to QTL detection, the identification of an appropriate mixed model for the phenotypic data was done by comparing different structures of variance-covariance (VCOV) matrix for the genetic effects (Table 1). For mathematical description of the model, a notation similar to that presented by Eckermann et al. (2001), Verbyla et al. (2003) and Boer et al. (2007) was used. The statistical model, in which the underlining indicates a random variable, is:
\(\underline {y}_{isjkr}\) is the phenotype of the rth replicate (r = 1, 2) of the ith individual (\(i = 1, 2, \ldots, n\)) of group s (s = 1, 2, 3) in location j (j = 1, J = 2) and harvest k (k = 1, 2, K = 3); μ is the overall mean; L j is the location effect; H k is the harvest effect; LH jk is the location by harvest interaction effect; \(\underline {G}_{ijk}\) is the effect of individual i at location j and harvest k; and \(\underline {\varepsilon}_{isjkr}\) is a non-genetic effect. The individuals can be separated into two groups, n = n g + n c, where n g is the number of genotypes in the progeny (i = 1, …, n g ), and n c is the number of checks (i = n g + 1, …, n g + n c). The model for \(\underline {G}_{ijk}\) is:
where \(\underline {g}_{ijk}\) is a random genetic effect of genotype i at location j and harvest k, and c ijk represents a fixed effect for check i at location j and harvest k. Although checks (c ijk ) are not relevant to the detection of QTL, adding them to the model helps to account for non-genetic variation that may be present (Verbyla et al. 2003; Boer et al. 2007). It was assumed that the vector \(\underline {{\bf g}} = (\underline {g}_{111}, \ldots, \underline {g}_{IJK})\) has a multivariate normal distribution with zero mean and VCOV matrix G M ⊗ \({\bf I}_{n_{\rm g}},\) in which M = JK, ⊗ is the Kronecker direct product of matrices, and \({\bf I}_{n_{\rm g}}\) is an identity (co)variance matrix of genotypes. Seven different models for the G M matrix (Table 1) were examined and compared via AIC (Akaike Information Criterion) (Akaike 1974) and BIC (Bayesian Information Criterion) (Schwarz 1978). Models (a–e) do not structure the G M matrix on the basis of harvests and locations, whereas models (f–g) do so via direct products of (co)variance matrices for locations and harvests separately (Smith et al. 2007; Malosetti et al. 2008). Model (a) considers homogeneous variation (ID), i.e. there are no genetic correlations between environments, and genetic variances are homogeneous across environments. Model (b) allows for heterogeneous genetic variances but assumes no genetic correlations between environments. Model (c) considers heterogeneous genetic variance and common genetic covariance between environments. Model (d) uses a multiplicative model called factor analytic model of order 1 to approximate a fully unstructured (co)variance matrix (Oman 1991; Gogel et al. 1995). Model (e) allows for the G M matrix to contain specific genetic variances or covariances for each environment. Model (f) combines a heterogeneous autoregressive model (of order 1) for harvests and an unstructured model for the locations. In the heterogeneous autoregressive model (of order 1), the correlations between harvests decay with time and each harvest has its own genetic variance. Model (g) combines unstructured matrices for both harvests and locations.
For the non-genetic term (\(\underline {\varepsilon}_{isjkr}\)), the model was:
where t s is the group effect, t sjk is the effect of group s at location j and harvest k; b sjkr is the effect of block r within group s, location j and harvest k; \(\underline {\eta}_{isjkr}\) represents a non-genetic residual error term. In a similar way to what was done for matrix G M , several VCOV structures were compared for the matrix of non-genetic residual effects (R M ) to allow for residual heterocedasticity as well as correlation between repeated measures (same individual plots were observed in different harvests).
QTL analysis
Based on the IM approach (Lander and Botstein 1989), the presence of a putative QTL was tested along the genome. In this context, the phenotypic model (Eq. 1) was expanded to include marker information:
where \(\alpha_{p_{jkw}}, \alpha_{q_{jkw}}\) and \(\delta_{pq_{jkw}}\) are the harvest-location-specific effects of the additive genetic predictor for parent P and Q, and dominance genetic predictor, respectively, at evaluation point \(w;\,{\underline {G}^{*}}_{ijk}\) now indicates the genetic residual effect of individual i at location j and harvest k not explained by the QTL already in the model (genetic residual effect). The VCOV matrix used for \(\underline {g}_{ijk}\) was selected in the previous multi-harvest-location phenotypic analyses. The null hypothesis of a putative QTL without effect across locations and harvests can be stated as:
Search for QTL main effects were also performed along the genome using a simpler model in which QTL effects were equal across harvests and locations:
Genomic positions with p ≤ 0.01 (Wald test, Verbeke and Molenberghs 2000) in the QTL profile produced by models (4) and (5) were selected to build a multi-QTL model.
One at a time, each unlinked marker (424 total) was fitted in the phenotypic model (Eq. 1) and tested with the Wald test to further identify putative QTL effects associated with individual markers. Unlinked markers were coded as either −1 (allele o) or 1 (allele a).
A multi-QTL model was built through a five-steps procedure. At step I, significant effects were searched using genome-wide interval mapping with models (4) and (5) separately. For each model, three genome-wide searches were carried out: a search for additive effect of parent P, in which the interval model had only the α p effect; a search for additive effect of parent Q, in which the interval model had only the effect α q ; and, a search for dominance effect, in which the interval model had all three effects, but only the δ pq was tested. At step II, unlinked markers were tested for association with putative QTL via single marker (SM) analyses. At step III, genomic positions (from step I) and unlinked markers (from step II) with significant effects were put together in a multi-QTL model. Subsequently, the statistical significance of each effect in this multi-QTL model was assessed via the Wald test. Non-significant QTL effects, with p-value greater than 0.05 were excluded from the model. At step IV, each remaining effect in the multi-QTL model was tested for QTL-effect × E: first a test was performed on QTL-effect × Harvest × Location (six degrees of freedom), and when this interaction was not significant, we tested the significances of both QTL-effect × Location (one degree of freedom) and QTL-effect × Harvest (two degrees of freedom). A last step consisted of estimating parameters in the final multi-QTL model. Throughout all steps, if a dominance effect was found to be significant, its respective additive effects from both parents were also added to the model even if they were not marginally significant. All the statistical analysis involving mixed models were performed in Genstat 12th edition (Payne et al. 2009) using Residual Maximum Likelihood (REML).
To show the advantages of the mixed-model approach, its results were compared with those from the IM analyses of each harvest-location combination (the R/qtl software was used for the IM analyses, Broman et al. 2003). As the majority of published QTL studies of sugarcane (Pastina et al. 2010) used the IM strategy without considering integrated linkage maps (markers D 1, D 2 and C combined), we then disregarded in our R/qtl analyses all LGs in which markers D 1, D 2 and C were linked.
Results
Linkage map
From a total of 741 molecular markers, 317 (42.8%) were mapped to 96 LGs with a total map length of 2468.14 cM and average distance between markers (marker density) of 7.5 cM. Forty-two LGs (43.7%) had only two linked markers; 27 had 3 markers; 9 had 4; 9 had 5; and, 6 had 6 markers. The three largest LGs had 10, 11 and 13 markers. Markers were mostly clustered along the LGs, while other parts of the genome were sparsely covered. In the linkage map, 11.8% of the adjacent markers showed gaps larger than 20 cM. While 91 LGs were assembled into 11 putative HGs, the remaining five did not contain enough loci in common with any HG to allow them to be assigned with a certain degree of confidence. The number of LGs assembled in each HG ranged from 2 to 23 (Online Supplementary Material).
Multi-harvest-location phenotypic analysis
While the selected VCOV models for the G matrix based on AIC and BIC criteria (Table 2) coincided for TSH (both criteria selected model e) and so for Pol (both criteria selected model e), different VCOVs were selected for the G matrix for TCH (models e and g) as well as for Fiber (models e and d). However, the differences of AIC values from first and second best models were greater than the respective BIC differences for both TCH and Fiber. Therefore, we decided to use VCOVs selected via AIC criterion, which in this study led to model (e) for all traits. Although model (e) requires estimation of a larger number of parameters, it had the smallest AIC throughout traits. For the non-genetic residual effects, the model combining an unstructured matrix \({\bf R}_{K \times K}^{\hbox{H}}\) for harvests and a diagonal matrix \({\bf R}_{J \times J}^{\hbox{L}}\) for locations had the smallest AIC when compared with simpler models, such as the model assuming heterogeneity of non-genetic residual variances and absence of correlation between harvest-location combinations (DIAG). The selected R M matrix was included in the final phenotypic model, taking into account the existence of non-genetic residual correlations and heterogeneity of non-genetic residual variances across harvests and locations.
QTL analysis
The search for QTL via IM (see step I in QTL analysis of "Materials and methods") led to the identification of 29 putative QTLs, 9 for TCH, 9 for TSH, 5 for Pol, and 6 for Fiber (Fig. 2). Each QTL was located on a different LG. Twenty-six marker-QTL associations were found in the SM analyses (see step II in QTL analysis "Materials and methods"): five for TCH, eight for TSH, eight for Pol, and five for Fiber (Online Supplementary Material). Genomic positions and single markers significantly associated with putative QTL in the IM and SM analyses were included in the multi-QTL model for the estimation of QTL main effects and QTL harvest-location-specific effects.
Interval mapping search of putative QTL (red- and yellow-inverted triangles) associated with cane yield (tonnes of cane per hectare, TCH), sugar yield (tonnes of sugar per hectare, TSH), fiber content, and sucrose content (Pol) using a mixed model with unstructured G M matrix (model e; Table 2). Two different situations were considered: (1) using only main effects, model 5 (α p and α q : additive effects on parents P and Q, respectively; and δ pq : dominance); (2) using genetic effects specifically for each harvest-location combination, through model 4 (\(\alpha_{p_{jk}}, \alpha_{q_{jk}}\) and \(\delta_{pq_{jk}}\)). Not all effects were estimated for all genomic positions due to lack of information conveyed by SDMs (see "Materials and methods"); black triangles marker positions, dot-dashed line −log10(0.001) and dotted line −log10(0.01). Marker types D 1 and D 2 segregate for parent P and Q, respectively, while marker C segregates for both parents
IM and SM analyses identified 14 QTLs for TCH, 13 of which after been included in a multi-QTL model and been tested (see steps III and IV of QTL analysis "Materials and methods") remained in the final multi-QTL model (Table 3). The QTLs identified on LG9 and LG19 had significant additive main effect. QTLs detected on LG25, LG32, LG72, LG92, and unlinked markers EST3EC and ESTC81m3C had significant QTL × H interaction, indicating that these QTLs showed the same behavior along the two locations, but not along harvests. The QTL identified on LG66 and the QTL associated with marker ESTB64m3C had QTL × L interaction, i.e. the effects of each QTL are significantly different across locations, but equal across harvests. The QTLs on LG8, LG28, and marker SG61BD1 had QTL × H × L interactions, which means that the effects of each QTL are significantly different for at least one harvest-location combination.
For TSH, 17 QTLs were identified via IM and SM analyses, 14 of them remained in the final multi-QTL model (Table 4). QTLs detected on LG19, LG21, and unlinked markers SG61BD1, SG105AD1 and EST9BD2 had significant additive main effects. QTLs associated with unlinked markers SG140CC and ESTC03m2D2 had QTL × H × L interaction, which means that each QTL had a different effect for at least one harvest-location combination. Other QTLs had interaction with location (LG9 and LG92) and with harvest (LG25, LG32, LG42, and unlinked markers EST3EC and ESTC02m1D2).
From a total of 13 QTLs identified via IM and SM analyses of Pol, 8 QTLs remained significant in the multi-QTL model (Table 4). QTLs detected on LG6, LG35, LG55, and unlinked markers SG06AD1 and ESTA03m4C showed significant additive main effects but no interaction was found. QTL × H × L interaction was significant for those QTLs detected on LG64, and associated with unlinked marker ESTB122m8D2. QTL detected on LG81 showed a QTL × L interaction.
In the IM and SM analyses of Fiber, 11 QTLs were identified, all of them remained in the final multi-QTL model (Table 4). A QTL with dominance effect was identified on LG3. QTLs detected on LG35, and unlinked markers SG25BC and ESTC110m2C showed significant main effects but no interaction was found. QTL × L interaction was detected for unlinked marker SG99DC, which means that each QTL had different effects across locations, but not across harvests. Other QTLs identified on LG37, LG44, LG55, LG83, and unlinked markers ESTA32m1C and ESTB153m1D2 showed QTL × H × L interaction.
Discussion
The number of LGs found in this study (96) is close to the expected chromosome number (2n = 100 − 130) for modern sugarcane cultivars (Grivet and Arruda 2001; Hoarau et al. 2001). However, the large number of unlinked markers (424), the small length of most LGs, and the reduced number of markers (loci) per LG, indicate that the map is still not well-saturated. Probably, most of the small LGs represent unconnected groups.
On one hand, usually only SDMs are used for linkage map estimation (Ming et al. 1998), thus gaps in sugarcane maps are commonly expected due to the exclusion of multiple dose markers, such as, duplex of monoparental origin, triplex or higher multiplex markers. Therefore, linkage maps based solely on SDMs are not optimal for QTL mapping and lower statistical power is possibly expected. On the other hand, we estimated an integrated map via multipoint likelihood (OneMap, Margarido et al. 2007). Our integrated map had higher likelihood than other single-dose-based maps estimated from our population (Garcia et al. 2006; Oliveira et al. 2007). Since multipoint likelihood can put together in the same LG markers with 3:1 and 1:1 segregation patterns, the resulting integrated map is more saturated and conveys higher representation of the biparental genetic polymorphism than its counter part double pseudo-testcross maps, hence, higher statistical power is expected in the QTL analysis. Moreover, the use of an integrated map allowed us to estimate additive effects in each parent (α p and α q ) and dominance effect (δ pq ), which to the best of our knowledge is being proposed for the first time to map QTL in sugarcane.
In spite of the interspecific origin of modern commercial sugarcane cultivars with genome composition of about 70–80% of Saccharum officinarum, 10–20% of S. spontaneum and 5–17% of recombinant chromosomes (D’Hont et al. 1996; Grivet and Arruda 2001; Jannoo et al. 2004; D’Hont 2005; Piperidis et al. 2010), and the high level of polyploidy and aneuploidy, the number of putative HGs identified (11) are in close agreement with the expected number for sugarcane, as the basic number of chromosomes (x) of the genus Saccharum can range from x = 8 to x = 10 (D’Hont et al. 1998; Irvine 1999; Grivet and Arruda 2001; Piperidis et al. 2010).
Despite varietal selection of sugarcane based on quantitative traits is usually done with measurements taken from series of field trials in multiple locations and multiple harvests, fitting alternative VCOV structures for modeling genetic effect across locations and harvests is seldom pursued (Smith et al. 2007). Mixed models were used in this study due to their flexibility to model VCOV structures that appears when repeated measures are taken across locations and harvests. In the mixed model analyses, genotypes in the progeny were assumed to be random because the main interest is in the genetic variation of genotypes in the progeny rather than the genotypes themselves. The effects of location (L) and harvest (H) were taken as fixed. Models that exploit the direct product of (co)variance matrices (models f–g) have fewer parameters, and therefore, we would expect them to show smaller AIC values. However, the unstructured VCOV matrix (model e), modeling specific genetic variances or covariances for each environment, showed smaller AIC values throughout all traits, despite its larger number of parameters. Although it is well-documented in the literature that AIC tends to select models with more parameters as compared with BIC, the choice of unstructured VCOV model shows some evidence for the presence of heterogeneity of variances and covariances across different harvest-location combinations (Table 5).
For QTL mapping, a VCOV model for the genetic effects selected in the phenotypic analysis was combined with fixed QTL main effects and harvest-location-specific QTL effects. Thus, QTL effects were tested taking into account the background genetic correlation in the data. Piepho (2005), via simulation, showed that ignoring genetic correlations in multi-environment data leads to substantial increase in type I error rate when testing for QTL effects. Therefore, it is expected that our multi-harvest-location mixed model approach will reduce the risk of over-optimistic conclusions, since an unstructured genetic (co)variance matrix was considered. Another important feature of our approach is that all analyses are undertaken within the same modeling framework, avoiding the combination of results from different analyses, as in the so-called two-stage analyses: one analysis to obtain genotypic means for individual trials (BLUEs) and another analysis on the means for QTL detection (Welham et al. 2010).
Amongst all traits, many QTLs (65%) showed significant interaction: QTL × H (24%), QTL × L (13%), QTL × H × L (28%) interaction; and 17 QTLs (35%) had stable effect across harvests and locations. The number of detected interactions was greater for QTL × H than for QTL × L, possibly because genotype by harvest (G × H) interaction accounted for great part of the genotype by environment interaction for each trait, and, moreover, there was no significant genotype by location (G × L) interaction for Pol and Fiber.
On one hand, QTL whose effects are not statistically different across harvests and locations are important for studies that seek to identify major genes controlling agronomic traits, as the expression of these genes would not be expected to change drastically across harvest-location combinations. For example: QTLs identified on LG9 and LG19 (TCH), LG19, LG21, unlinked markers SG61BD1, SG105AD1 and EST9BD2 (TSH), LG6, LG35, LG55, unlinked markers SG06AD1 and ESTA03m4C (Pol), LG3, LG35, unlinked markers SG25BC and ESTC110m2C (Fiber). It is worth mentioning that 62.5% of QTLs identified for Pol were stable across all harvest-location combinations, corroborating the speculated fact raised by many breeders that Pol has reached the plateau of adaptability and stability. On the other hand, QTLs with stable effects across harvests within locations (likewise, stable effects across locations within harvests) are also important to identify genes with similar expression across harvests (likewise, across locations). For instance: QTLs located on LG66 and unlinked marker ESTB64m3C (TCH), LG66 and LG92 (TSH), LG81 (Pol), and unlinked marker SG99DC (Fiber). Not only QTL effect stability is important to applications, but also its sign and magnitude, as for example in MAS. To exemplify, QTLs on LG8 and LG28 of TCH changed signs across some harvest-location combinations, and QTL on LG25 had negative effect with increasing magnitude across harvests, which is particularly interesting in sugarcane, since yield decreases across harvests.
Assignment of LGs to HGs may help us to infer whether QTLs mapped at distinct LGs, but in genomic regions that share at least a common locus, are the same or not. For example, while QTLs detected on LG8 and LG50 (TSH) were assigned to HGIV, they were positioned far apart at 4.7 cM and 17.1 cM from their common locus ESTA47, respectively. Therefore, we cannot infer that these genomic regions share the same QTL. Likewise, although QTLs detected on LG25 and LG28 (TCH), LG3 and LG35 (Fiber) belong to HGI and HGV, respectively, they are far apart from their common locus ESTA15 (TCH), ESTB65 and ESTB69 (Fiber), hence, they represent different QTLs. It is important to notice that the linkage map estimated in this study is not well-saturated. Adding more markers to the data may change the number, length and marker ordering of LGs, therefore, possibly conveying more information about whether QTLs mapped on LGs belonging to an HG are the same or not.
Some QTLs of different traits were identified in common linkage groups or associated with common markers. For example, both TCH and TSH had one QTL mapped on each of the following LGs and unlinked markers: LG19, LG25, LG32, LG66 and LG92, and markers SG61BD1, EST3EC and ESTC03m2D2. As all the common QTLs were close by, it is possible that they are pleiotropic QTLs. It was expected that these traits would have some QTLs in common, since they are strongly correlated. Both Pol and Fiber had a QTL on LG35, possibly they are just one pleiotropic QTL. In breeding programs, special attention should be given to these two QTLs when simultaneous improvement is aimed for Pol and Fiber, since the QTLs had opposite signs on these traits. Moreover, the negative correlation between Pol and Fiber is interesting to the modern trend of industrial production of second-generation (cellulosic) ethanol, which seeks for sugarcane varieties specialized in biomass production with higher fiber content.
We aimed to compare our multi-harvest-location modeling strategy (mixed model) with other strategies of modeling QTL × H × L interaction in sugarcane, but no other study of this nature has been pursued, to the best of our knowledge (Pastina et al. 2010). However, some attempts to study QTL × H × L interaction have been made via SM analyses of each harvest or harvest-location combination (when available) separately, for each parent through the pseudo-testcross strategy. Stability of QTLs across environments were inferred based on their effect sizes (Hoarau et al. 2002; Jordan et al. 2004; McIntyre et al. 2005a, b; Reffay et al. 2005; Aitken et al. 2006, 2008; Al-Janabi et al. 2007; Piperidis et al. 2008). Nevertheless, none of these studies could be compared to ours due to differences in the data. Therefore, IM was carried out through R/qtl for each trait and harvest-location combination separately (univariate QTL analyses, Online Supplementary Material) to be compared with our mixed-model approach. Through this separate analyses it was possible to identify only one putative QTL for TCH and two for Pol. For TCH, the QTL was positioned on LG32, which had a significant effect for harvests 1 and 2 in location 1. For location 2 there were some evidences of QTLs on LG32 with different effects across harvests, however, the LOD values were smaller than the threshold considered (LOD = 3). These QTL may correspond to the QTL identified in the same LG using mixed model, which showed unstable effect across harvests. For Pol, two different QTLs were identified, one on LG6 and other on LG55, which were also identified in the mixed model analysis. The QTLs identified on LG6 and LG55 had significant effects for harvest 2 in location 1 and harvest 2 in location 2, respectively, which do not agree with the mixed model analysis, since stable effects were found across all harvest-location combinations. Overall, while separate analyses found only three QTLs, the mixed model analyses found forty-six, clearly showing the overwhelming advantage of the mixed model approach.
QTL mapping in sugarcane still presents several difficulties, such as the use of only SDMs, low saturated linkage maps, small sample size (n g), the occurrence of collinearity between the additive genetic predictors estimated for parents (as a consequence of the lack of information conveyed by SDMs). The latter difficulty restricted the estimation of dominance genetic predictor for only a limited number of linkage groups (LG2, LG3, LG14, LG18, LG37 and LG41). Thus, the fact that only one QTL with dominance effect was found for Fiber is not necessarily related to the genetic basis of this trait, but due to the fact that we simply often could not estimate and test for it. We are also aware that the small sample size used (n g = 100) has reduced statistical power, but the focus of our work was on the illustration of how to use a mixed-model framework that takes into account heterogeneity of genetic and non-genetic residual variances and covariances. Despite these limitations, the present study provides many contributions, such as, the identification of a considerable number of QTLs for the evaluated traits, with information about effect sizes, positions, stability of QTLs, and presence of QTL × H, QTL × L, and QTL × H × L interactions. Therefore, unveiling the genetic architecture of sugarcane production and sucrose content, which are complex traits. In addition, the statistical models used here can be used in future QTL studies involving multiplex markers in addition to SDMs.
References
Aitken KS, Jackson PA, McIntyre CL (2006) Quantitative trait loci identified for sugar related traits in a sugarcane (Saccharum spp.) cultivar x Saccharum officinarum population. Theor Appl Genet 112:1306–1317
Aitken KS, Hermann S, Karno K, Bonnett GD, McIntyre LC, Jackson PA (2008) Genetic control of yield related stalk traits in sugarcane. Theor Appl Genet 117:1191–1203
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Automat Contr AC 19:716–723
Al-janabi SM, Honeycutt RJ, Mcclelland M, Sobral BWS (1993) A genetic linkage map of Saccharum spontaneum L. ’SES 208’. Genetics 134:1249–1260
Al-Janabi SM, Parmessur Y, Kross H, Dhayan S, Saumtally S, Ramdoyal K, Autrey LJC, Dookun-Saumtally A (2007) Identification of a major quantitative trait locus (QTL) for yellow spot (Mycovellosiella koepkei) disease resistance in sugarcane. Mol Breed 19:1–14
Boer MP, Wright D, Feng L, Podlich DW, Luo L, Cooper M, van Eeuwijk FA (2007) A mixed-model quantitative trait loci (QTL) analysis for multiple-environment trial data using environmental covariables for QTL-by-environment interactions, with an example in maize. Genetics 177:1801–1813
Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890
Carlier JD, Reis A, Duval MF, Coppens D’Eeckenbrugge G, Leitao JM (2004) Genetic maps of RAPD, AFLP and ISSR markers in Ananas bracteatus and A. comosus using the pseudo-testcross strategy. Plant Breed 123:186–192
Cavalcanti JJV, Wilkinson MJ (2007) The first genetic maps of cashew (Anacardium occidentale L.). Euphytica 157:131–143
Chapman SC (2008) Use of crop models to understand genotype by environment interactions for drought in real-world and simulated plant breeding trials. Euphytica 161:195–208
Chen C, Bowman KD, Choi YA, Dang PM, Rao MN, Huang S, Soneji JR, McCollum TG, Gmitter FG (2008) EST-SSR genetic maps for Citrus sinensis and Poncirus trifoliata. Tree Genetics Genomes 4:1–10
Cullis B, Gogel B, Verbyla A, Thompson R (1998) Spatial analysis of multi-environment early generation variety trials. Biometrics 54(1):1–18
Daugrois JH, Grivet L, Roques D, Hoarau JY, Lombard H, Glaszmann JC, D’Hont A (1996) A putative major gene for rust resistance linked with a RFLP marker in sugarcane cultivar ’R570’. Theor Appl Genet 92:1059–1064
da Silva JA, Bressiani JA (2005) Sucrose synthase molecular marker associated with sugar content in elite sugarcane progeny. Genet Mol Biol 28(2):294–298
Denis JB, Piepho HP, van Eeuwijk FA (1997) Modelling expectation and variance for genotype by environment data. Heredity 79:162–171
D’Hont A (2005) Unravelling the genome structure of polyploids using FISH and GISH; examples in sugarcane and banana. Cytogenet Genome Res 109:27–33
D’Hont A, Grivet L, Feldman P, Rao S, Berding N, Glaszmann JC (1996) Characterisation of the double genome structure of modern sugarcane cultivares (Saccharum spp.) by molecular cytogenetics. Mol Gen Genet 250:405–413
D’Hont A, Ison D, Alix K, Roux C, Glaszmann JC (1998) Determination of basic chromosome numbers in the genus Saccharum by physical mapping of ribosomal RNA genes. Genome 41:221–225
Eckermann PJ, Verbyla AP, Cullis BR, Thompson R (2001) The abalysis of quantitative traits in wheat mapping populations. Aust J Agric Res 52:1195–1206
Garcia AAF, Kido EA, Meza AN, Silva JAGD, Souza AP, Pinto LR, Pastina MM, Leite CS, da Silva JAG, Ulian EC, Figueira A, Souza HMB (2006) Development of an integrated genetic map of a sugarcane (Saccharum spp.) commercial cross, based on a maximum-likelihood approach for estimation of linkage and linkage phases. Theor Appl Genet 112:298–314
Gazaffi R (2009) Desenvolvimento de modelo genético estatístico para mapeamento de QTLs em progênie de irmãos completos, com aplicação em cana-de-açúcar. PhD Thesis, Escola Superior de Agricultura Luiz de Queiroz, Universidade de São Paulo, Brazil
Gogel BJ, Cullis BR, Verbyla AP (1995) REML estimation of multiplicative effects in multi-environment variety trials. Biometrics 51(2):744–749
Grattapaglia D, Sederoff R (1994) Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137:1121–1137
Grivet L, Arruda P (2001) Sugarcane genomics: depicting the complex genome of an important tropical crop. Curr Opin Plant Biol 5:122–127
Haley CS, Knott SA (1992) A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69:315–324
Haley CS, Knott SA, Elsen JM (1994) Mapping quantitative trait loci in crosses between outbred lines using least squares. Genetics 136:1195–1207
Heinz DJ, Tew TL (1987) Hybridization procedures. In: Heinz DJ (eds) Sugarcane Improvement through Breeding, Elsevier, Amsterdam, pp 313–342
Hoarau JY, Offmann B, D’Hont A, Risterucci AM, Roques D, Glaszmann JC, Grivet L (2001) Genetic dissection of a modern sugarcane cultivar (Saccharum spp.). I. Genome mapping with AFLP markers. Theor Appl Genet 103:84–97
Hoarau JY, Grivet L, Offmann B, Raboin LM, Diorflar JP, Payet J, Hellmann M, D’Hont A, Glaszmann JC (2002) Genetic dissection of a modern sugarcane cultivar (Saccharum spp.). II. Detection of QTLs for yield components. Theor Appl Genet 105:1027–1037
Hu Z, Xu S (2009) Proc qtl—a sas procedure for mapping quantitative trait loci. Int J Plant Genomics. doi:10.1155/2009/141234
Irvine JE (1999) Saccharum species as horticultural classes. Theor Appl Genet 98:186–194
Jannoo N, Grivet L, David J, D’Hont A, Glaszmann JC (2004) Differential chromosome pairing affinities at meiosis in polyploid sugarcane revealed by molecular markers. Heredity 93:460–467
Jordan DR, Casu RE, Besse P, Carroll BC, Berding N, McIntyre CL (2004) Markers associated with stalk number and suckering in sugarcane colocate with tillering and rhizomatousness QTLs in sorghum. Genome 47:988–993
Knott SA, Haley CS (1992) Maximum likelihood mapping of quantitative trait loci using full-sib families. Genetics 132:1211–1222
Knott SA, Neale DB, Sewell MM, Haley CS (1997) Multiple marker mapping of quantitative trait loci in an outbred pedigree of loblolly pine. Theor Appl Genet 94:810–820
Kosambi DD (1944) The estimation of map distances from recombination values. Annu Eugene 12:172–175
Lander E, Botstein D (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–199
Lander E, Green P, Abrahamson J, Barlow A, Daley M, Lincoln S, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lin M, Lou X, Chang M, Wu R (2003) A general statistical framework for mapping quantitative trait loci in nonmodel systems: issue for characterizing linkage phases. Genetics 165:901–913
Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits. Sinauer Associates, Sunderland
Malosetti M, Voltas J, Romagosa I, Ullrich SE, van Eeuwijk FA (2004) Mixed models including environmental covariables for studying QTL by environment interaction. Euphytica 137:139–145
Malosetti M, Ribaut JM, Vargas M, Crossa J, van Eeuwijk FA (2008) A multi-trait multi-environment QTL mixed model with an application to drought and nitrogen stress trials in maize (Zea mays L.). Euphytica 161:241–257
Margarido GRA, Souza AP, Garcia AAF (2007) OneMap: software for genetic mapping in outcrossing species. Hereditas 144:78–79
Martínez O, Curnow RN (1992) Estimating the locations and the sizes of the effects of quantitative trait loci using flanking markers. Theor Appl Genet 85:480–488
Mathews KL, Malosetti M, Chapman S, McIntyre L, Reynolds M, Shorter R, van Eeuwijk F (2008) Multi-environment QTL mixed models for drought stress adaptation in wheat. Theor Appl Genet 117:1077–1091
McIntyre C, Jackson M, Cordeiro G, Amouyal O, Hermann S, Aitken K, Eliott F, Henry R, Casu R, Bonnett G (2006) The identification and characterisation of alleles of sucrose phosphate synthase gene family III in sugarcane. Mol Breed 18:39–50
McIntyre CL, Whan VA, Croft B, Magarey R, Smith GR (2005) Identification and validation of molecular markers associated with Pachymetra root rot and brown rust resistance in sugarcane using map- and association-based approaches. Mol Breed 16:151–161
McIntyre CL, Casu RE, Drenth J, Knight D, Whan VA, Croft BJ, Jordan DR, Manners JM (2005) Resistance gene analogues in sugarcane and sorghum and their association with quantitative trait loci for rust resistance. Genome 48:391–400
Ming R, Liu SC, Lin YR, da Silva J, Wilson W, Braga D, van Deynze A, Wenslaff TF, Wu KK, Moore PH, Burnquist W, Sorrells ME, Irvine JE, Paterson AH (1998) Detailed alignment of saccharum and sorghum chromosomes: comparative organization of closely related diploid and polyploid genomes. Genetics 150:1663–1682
Ming R, Liu SC, Moore PH, Irvine JE, Paterson AH (2001) QTL analysis in a complex autopolyploid: genetic control of sugar content in sugarcane. Genome Res 11:2075–2084
Ming R, Wang W, Draye X, Moore H, Irvine E, Paterson H (2002) Molecular dissection of complex traits in autopolyploids: mapping QTLs affecting sugar yield and related traits in sugarcane. Theor Appl Genet 105:332–345
Ming R, Del Monte TA, Hernandez E, Moore PH, Irvine JE, Paterson AH (2002) Comparative analysis of QTLs affecting plant height and flowering among closely-related diploid and polyploid genomes. Genome 45:794–803
Okada M, Lanzatella C, Saha MC, Bouton J, Wu R, Tobias CM (2010) Complete switchgrass genetic maps reveal subgenome collinearity, preferential pairing and multilocus interactions. Genetics 185:745–760
Oliveira KM, Pinto LR, Marconi TG, Margarido GRA, Pastina MM, Teixeira LHM, Figueira AV, Ulian EC, Garcia AAF, Souza AP (2007) Functional integrated genetic linkage map based on EST-markers for a sugarcane (Saccharum spp.) commercial cross. Mol Breed 20:189–208
Oman SD (1991) Multiplicative effects in mixed model analysis of variance. Biometrika 78(4):729–739
Pastina MM, Pinto LR, Oliveira KM, Souza AP, Garcia AAF (2010) Molecular mapping of complex traits. In: Henry R, Kole C (eds) Genetics, genomics and breeding of sugarcane, Science Publishers, Enfield, pp 117–148
Payne RW, Murray DA, Harding SA, Baird DB, Soutar DM (2009) GenStat for Windows (12th Edition) Introduction. VSN International, Hemel Hempstead
Piepho HP (1997) Analyzing genotype-environment data by mixed models with multiplicative terms. Biometrics 53(2):761–766
Piepho HP (2000) A mixed-model approach to mapping quantitative trait loci in barley on the basis of multiple environment data. Genetics 156:2043–2050
Piepho HP (2005) Statistical tests for QTL and QTL-by-environment effects in segregating populations derived from line crosses. Theor Appl Genet 110:561–566
Pinto LR, Garcia AAF, Pastina MM, Teixeira LHM, Bressiani JA, Ulian EC, Bidoia MAP, Souza AP (2010) Analysis of genomic and functional RFLP derived markers associated with sucrose content, fiber and yield QTLs in a sugarcane (Saccharum spp.) commercial cross. Euphytica 172:313–327
Piperidis N, Jackson PA, D’Hont A, Besse P, Hoarau JY, Courtois B, Aitken KS, McIntyre CL (2008) Comparative genetics in sugarcane enables structured map enhancement and validation of marker-trait associations. Mol Breed 21:233–247
Piperidis N, Piperidis G, D’Hont A (2010) Molecular cytogenetics. In: Henry R, Kole C (eds) Genetics, genomics and breeding of sugarcane, Science Publishers, Enfield, pp 9–18
Porceddu A, Albertini E, Barcaccia G, Falistocco E, Falcinelli M (2002) Linkage mapping in apomictic and sexual Kentucky bluegrass (Poa pratensis L.) genotypes using a two way pseudo-testcross strategy based on AFLP and SAMPL markers. Theor Appl Genet 104:273–280
Raboin LM, Oliveira KM, Raboin LM, Lecunff L, Telismart H, Roques D, Butterfield M, Hoarau JY, D’Hont A (2006) Genetic mapping in sugarcane, a high polyploid, using bi-parental progeny: identification of a gene controlling stalk colour and a new rust resistance gene. Theor Appl Genet 112:1382–1391
Reffay N, Jackson PA, Aitken KS, Hoarau JY, D’Hont A, Besse P, McIntyre CL (2005) Characterisation of genome regions incorporated from an important wild relative into Australian sugarcane. Mol Breed 15:367–381
Ripol MI, Churchill GA, da Silva JAG, Sorrells M (1999) Statistical aspects of genetic mapping in autopolyploids. Gene 235:31–41
Schäfer-Pregl R, Salamini F, Gebhardt C (1996) Models for mapping quantitative trait loci (QTL) in progeny of non-inbred parents and their behaviour in presence of distorted segregation rations. Genet Res 67:43–54
Schwarz G (1978) Estimating the dimension of a model. Ann Stat 6:461–464
Shepherd M, Cross M, Dieters MJ, Henry R (2003) Genetic maps for Pinus elliottii var hondurensis using AFLP and microsatellite markers. Theor Appl Genet 106:1409–1419
Sillanpää M, Arjas E (1999) Bayesian mapping of multiple quantitative trait loci from incomplete outbred offspring data. Genetics 151:1605–1619
Sills GR, Bridges W, Al-Janabi SM, Sobral BWS (1995) Genetic analysis of agronomic traits in a cross between sugarcane (Saccharum officinarum L.) and its presumed progenitor (S. robustum Brandes & Jesw. ex Grassl). Mol Breed 1:355–363
Smith A, Cullis B, Thompson R (2001) Analyzing variety by environment data using multiplicative mixed models and adjustments for spatial field trend. Biometrics 57(4):1138–1147
Smith AB, Stringer JK, Wei X, Cullis BR (2007) Varietal selection for perennial crops where data relate to multiple harvests from a series of field trials. Euphytica 157:253–266
Verbeke G, Molenberghs G (2000) Linear mixed models for longitudinal data. Spinger, New York
van Eeuwijk FA, Malosetti M, Yin X, Struik PC, Stam P (2005) Statistical models for genotype by environment data: from conventional ANOVA models to eco-physiological QTL models. Aust J Agric Res 56:883–894
van Eeuwijk FA, Malosetti M, Boer MP (2007) Modelling the genetic basis of response curves underlying genotype × environment interaction. In: Spiertz JHJ, Struik PC, van Laar HH (ed) Scale and complexity in plant systems research: gene–plant–crop relations. Springer, Dordrecht, pp 115–126
Verbyla A, Eckerman PJ, Thompson R, Cullis B (2003) The analysis of quantitative trait loci in multi-environment trials using a multiplicative mixed model. Aust J Agric Res 54:1395–1408
Welham S, Gogel B, Smith A, Thompson R, Cullis B (2010) A comparison of analysis methods for late-stage variety evaluation trials. Aust N Z J Stats 52:125–149
Wu K, Burnquist W, Sorrels M, Tew T, Moore P, Tanksley S (1992) The detection and estimation of linkage in polyploids using single-dose restriction fragments. Theor Appl Genet 83:294–300
Wu R, Ma CX, Painter I, Zeng ZB (2002) Simultaneous maximum likelihood estimation of linkage and linkage phases in outcrossing species. Theor Popul Biol 61:349–363
Wu R, Ma CX, Wu SS, Zeng ZB (2002) Linkage mapping of sex-specific differences. Genet Res 79:85–96
Wu R, Ma CX, Casella G (2007) Statistical genetics of quantitative traits: linkage, maps, and QTL. Springer, New York
Acknowledgments
The authors want to thank the anonymous reviewers for their suggestions, and Luciano da Costa e Silva to carefully read and give suggestions for the final English version. This research was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, grant 140680/2005-5 and 201409/2008-9) and Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP, grant 2008/52197-4 and 2010/00083-5), both from Brazil. It was also part of the researches of the Instituto Nacional de Ciência e Tecnologia do Bioetanol (granted by CNPq, 574002/2008-1, and FAPESP, 2008/57908-6) and of the PhD Thesis of M.M. Pastina, presented at Escola Superior de Agricultura “Luiz de Queiroz”, Universidade de São Paulo. M.M. Pastina was working under the supervision of F.A. van Eeuwijk at Biometris Department, Plant Sciences Group, Wageningen University, from February/2009 to September/2009. A.A.F. Garcia and A.P. Souza are recipients of research fellowship from CNPq.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Charcosset.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Pastina, M.M., Malosetti, M., Gazaffi, R. et al. A mixed model QTL analysis for sugarcane multiple-harvest-location trial data. Theor Appl Genet 124, 835–849 (2012). https://doi.org/10.1007/s00122-011-1748-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1748-8