Abstract
Interactions between modular domains and short linear motifs (3–10 amino acids peptide stretches) are crucial for cell signaling. The motifs typically reside in the disordered regions of the proteome and the interactions are often transient, allowing for rapid changes in response to changing stimuli. The properties that make domain-motif interactions suitable for cell signaling also make them difficult to capture experimentally and they are therefore largely underrepresented in the known protein-protein interaction networks. Most of the knowledge on domain-motif interactions is derived from low-throughput studies, although there exist dedicated high-throughput methods for the identification of domain-motif interactions. The methods include arrays of peptides or proteins, display of peptides on phage or yeast, and yeast-two-hybrid experiments. We here provide a survey of scalable methods for domain-motif interaction profiling. These methods have frequently been applied to a limited number of ubiquitous domain families. It is now time to apply them to a broader set of peptide binding proteins, to provide a comprehensive picture of the linear motifs in the human proteome and to link them to their potential binding partners. Despite the plethora of methods, it is still a challenge for most approaches to identify interactions that rely on post-translational modification or context dependent or conditional interactions, suggesting directions for further method development.
Similar content being viewed by others
Introduction
The size of the human interactome has been estimated to 650,000 interactions [1]. The known interactome is rapidly growing through the efforts of various high-through put studies such as affinity-purification coupled to mass spectrometry (AP-MS) [2] and yeast-two-hybrid (Y2H) [3]. However, less than 20 % of potential pairwise human protein-protein interactions have been explored through high-throughput studies [4]. About 15–40 % of the protein-protein interactions involve the recognition of a peptide motif (3–10 amino acid stretches) by a globular protein [5]. These interactions have crucial roles in defining cellular functions, being involved in processes such as protein scaffolding, cell signaling, targeting to subcellular compartments and post-translational modifications (PTMs) [6]. In parity with the large number of proposed interactions, a recent estimate suggested that the human proteome holds over 100,000 binding motifs [7]. The motifs are typically found in disordered regions or in exposed flexible loops and bind their target proteins through transient interactions with affinities in the low- to mid-micromolar range [8, 9]. A recent analysis revealed that 22 % of human disease mutations occur in the unstructured regions, and suggested that disease mutations in motifs are neglected players in cancer [10]. It is thus of crucial importance to systematically identify linear motifs in the proteome and link the motifs to the domains that recognize them.
A growing number of domains have been found to engage in peptide-mediated interactions. Today, there are about 200 known peptide binding domain families [11] with well studied examples being the PDZ (postsynaptic density protein 95/discs large/zona occludens 1) domains that typically bind to C-terminal peptides of target proteins [12–14], the poly proline binding WW domains [15] and SH3 (Src Homology 3) domains [16, 17], and the phosphotyrosine binding SH2 (Src Homology 2) domains [18–22] (Table 1). Manually curated databases such as the eukaryotic linear motif (ELM) resource [23] and the Linear Motif mediated Protein interaction Database (LMPID) [24] contain over 2,000 annotated instances of domain-motif interactions, most of which have been discovered by low-throughput experiments such as pulldowns, co-immunoprecipitation (co-IPs), mutational analysis and detailed structural studies of domain-peptide complexes. There is thus a striking discrepancy between the estimated number of motif-based interactions and the experimentally validated cases, suggesting that a vast number of motifs and binding domains are to be discovered. However, domain-motif interactions are difficult to capture due to their limited binding interfaces [8]. They have therefore commonly been overlooked in the methods such high-throughput AP-MS or Y2H. Indeed, an analysis of Y2H data revealed that only 1 % of the interactions rely on interactions with linear motifs [5]. The interactions can however be captured through AP-MS by the use of cross-linking [25] or by a recently developed proximity biotinylation approach [26, 27]. Although these methods may capture transient interactions, they will not necessarily report on binary interactions and they provide no direct information on the motifs that are involved in the interactions.
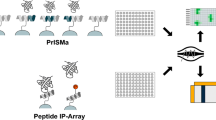
There is a variety of experimental methods dedicated to the characterization of peptide binding modules and the identification of peptide binding motifs [28]. The methods essentially fall into three main categories: arrays, display methods and protein-fragment complementation assays. Here, we summarize these methods for the identification of motif-based interactions (Fig. 1, Table 2); we introduce the basic principle of the methods and highlight recent advances in high-throughput analysis of domain-motif interactions.
Schematic representation of discussed techniques for the identification of motif-based interactions. Orange represents target protein; blue hexagon represents a binding motif; yellow, green and purple represent non-binding sequences peptides. Pink star represent a detection signal e.g. fluorescence. a Peptide microarray: Peptides with known sequences are synthesized on a solid support, incubated with the target protein and interactions are detected with specific antibodies or labeled target protein. b Protein array: A selection of different purified proteins are spotted on a solid support and incubated with a labeled peptide. c Peptide phage-display: Bait protein is immobilized and used in selections against a peptide phage library. Unbound phage particles are washed away, bound phage eluted and amplified, and used for repeated rounds of selections. Enriched binding clones are sequenced. d Yeast surface display: A library of peptides are displayed on the surface of yeast cells and incubated with a target protein. The target protein is labeled with a fluorescent tag and the cells are sorted based on peptide binding using FACS. Sorted pools are sequenced. e Yeast-two-hybrid: The binding domain (BD) of a transcription factor is linked to the target protein and the activation domain (AD) of the same transcription factor is linked to a peptide. If the protein and peptide interact BD and AD are brought together and the transcription factor reconstituted. This activates the transcription of a reporter gene
Microarrays
Peptide arrays
Peptide arrays rely on the chemically synthesis of peptides with known sequences on a solid support such as a cellulose membrane or a glass slide [29–32]. The microarray is thereafter incubated with the target protein and the bound protein is detected using for example specific antibodies or fluorescent or radioactive labeled proteins (Fig. 1a). Peptide arrays are typically semi-quantitative and allow the comparison of affinities between ligands immobilized on the same slide. An advantage of peptide array over display methods is that the peptide sequences are known and that the sequences can be systematically varied to map binding motifs. The method also provides information on non-binding peptides. A drawback of the method is a high number of false positive and false negative read-outs. This is partly due to the fact that the yield and purity of the peptides are difficult to evaluate and can vary between peptides on the same chip.
Peptide arrays were first introduced in the early nineteen’s when two groups reported techniques for parallel chemical synthesis of peptides on solid support. Fodor and co-workers described a light-directed, spatially addressable parallel chemical synthesis [33] and Frank introduced the SPOT-synthesis [34]. The majority of peptide arrays reported to date have relied on the SPOT-synthesis, which is commercially available and can be performed fully automated. Peptides are typically synthesized with a free N-terminal sequence. However, SPOT arrays have been further adapted for the synthesis of peptides with free C-terminal sequences, which was crucial for probing the binding specificities of, for example, PDZ domains [35].
A main advantage of peptide arrays is the possibility to incorporate modified and non-natural amino acids. This allow for direct and controlled mapping of interactions regulated by PTMs, such as phosphorylation [21] and acetylation [36]. For example, the tyrosine phosphopeptide binding of SH2 domains have been elucidated using a quantitative peptide microarray based approach [18] and by the use of a high-density peptide chip technology [21]. Similarly, Filippakopolous and co-workers created SPOT arrays that covered all possible sites for ε-N-acetylation of lysine residues of human histones [36]. These arrays were screened against 43 members of the bromodomain family. Affinities were determined by isothermal titration calorimetry (ITC) and a comprehensive structural characterization was performed. The study suggested that bromodomains recognize a combination of PTMs rather than single acetylates sequences.
Traditionally the throughput of peptide microarrays has been up to a few thousand peptides per chip. Ultra-dense peptide arrays now allow array sizes of 105–106 peptides [37–39]. These ultra-dense peptide arrays have been used for epitope mapping of antibodies. For example, Uhlen and co-workers developed a proteome wide peptide array, which was used for epitope mapping and cross-reactivity analysis of antibodies [38]. Using a photolithic technique they were able to in situ synthesize a total of 2.1 million overlapping peptides. This approach should be applicable for the general purpose of identifying motif-based interactions.
Apart from characterizing binding specificities of purified proteins, peptide microarrays can be used to identify targets from cell lysate. Taking such a motif centric approach, Okada and co-workers identified domains binding to proline rich peptides by synthesizing a peptide array, exposing it to cell lysate, cross-linking and identification of binding proteins through mass spectrometry. Thus, given a set of motifs, it is possible to identify proteins recognizing the given sequences [40].
Taken together, peptide arrays are useful tools for the identification and characterization of motifs-based interactions and are suitable for addressing interactions that rely on PTMs.
Protein arrays
In protein microarrays (Fig. 1b), proteins of interest are immobilized on a surface and then probed for binding to a labeled protein or peptide [41]. Proteins can be prepared by over-expression and high-throughput purification followed by spotting on the surface, or be obtained by cell-free protein expression systems [42, 43]. Proteomic microarrays allow the investigation of protein-protein interactions on a global scale [44, 45]. Protein microarrays have for example been used to elucidate the peptide binding specificities of the WW domain family [15]. Potential WW binding sites in the human proteome were identified by scanning the proteome using previously known motifs. Representative peptides were synthesized and their binding towards the WW domains tested through a quantitative ELISA-like binding assay. In another study, protein microarrays of SH2 domains and phosphotyrosine-binding (PTB) domains were used to explore their phosphorylation dependent interactions with 61 peptides representing tyrosine phosphorylation sites on the ErbB receptors [20]. Additionally, the specificities of PDZ domains were analyzed through protein microarrays paired with quantitative fluorescence polarization [13]. Protein arrays are thus useful tools for the comparative analysis of binding specificities of peptide binding modules. Among the advantages are the low sample consumption and the possibility to study interactions relying on PTMs. The method can further be used to obtain quantitative information on binding affinities. Among the disadvantages are the labor intense set-up and the requirement for rather high affinity interactions (KD <50 μM) [46].
Display methods
Peptide phage display
Peptide phage display is a powerful tool for the analysis of binding specificities of peptide binding domains [47]. Phages are viruses that infect bacteria. A link between the genotype and phenotype of the phage is provided by inserting DNA inside of the phage that encode for peptides which are displayed on the phage surface. Binding clones are enriched through selections against immobilized bait proteins and are then subjected to sequence analysis (Fig. 1c). There are various phage display systems, with the most commonly used being the p3 or p8 protein of the filamentous M13 phage or the minor coat protein 10B of the lytic T7 phage, as reviewed elsewhere [47]. The display can be either monovalent or multivalent, the former being preferred for capturing stronger interactions and the latter more suited for the identification of weaker interactions due to the avidity of the displayed peptides. The main strength of the method is that it allows the construction of highly diverse peptide libraries (1010) at a rather low cost. In a typical combinatorial peptide phage display experiment, libraries display randomized peptide sequences. The bottleneck has traditionally been the sequencing of binding clones. Today, next-generation sequencing brings down the cost of sequencing and the labor, which has opened new possibilities to exploit the potential of phage display and to gain control over the phage library compositions [48].
Peptide phage display has been used to characterize the binding specificities of various domain families. For example, the binding specificities of the yeast SH3 domains were elucidated in 2002, and the results were paired with computational predictions and with a Y2H derived protein-protein interaction network [17]. More than 10 years later, Xin et al. profiled the binding preferences of 36 SH3 domains of Caenorhabditis elegans [16], which revealed that the binding preferences were largely conserved between yeast and worm. Also the PDZ domains have been profiled through phage display. Tonikian et al. performed a large-scale characterization of PDZ binding specificities for 54 human and 28 worm PDZ domains [14], which allowed for an extended classification of their binding specificities. This information was later used to identify subspecificities among PDZ domains [49] and was paired with peptide array data [13] to construct a human PDZ domain-ligand interaction network [50].
Combinatorial phage display selections are useful for the identification of high affinity binders and the generation of consensus motifs. However, the displayed peptides may have little to do with biologically relevant targets. A study by Luck et al. highlighted that several of the consensus motifs for PDZ domains derived from combinatorial phage display are overly hydrophobic (i.e. tryptophan rich), which compromises the predictions [51]. Different attempts have been made to create phage libraries that display peptides representing parts of the human proteome, among them cDNA display and open reading frame display [47, 52]. These experiments have typically suffered from low library quality. A recent addition is the proteomic peptide phage display (ProP-PD) where phage libraries are designed to display regions of a target proteome [53, 54]. This method combines microarray synthesis of highly defined oligonucleotide libraries and next-generation sequencing. In 2011, Larman and co-workers created a T7 phage library that displays 36-mer peptides covering the human proteome [54]. More recently, this was followed by a study where M13 phage libraries were created to display the C-terminal peptides of human or viral proteins [53]. The C-terminal ProP-PD libraries were validated against a set of PDZ domains and it efficiently identified binders of potential biological relevance. ProP-PD directly identifies the binding motifs and the host proteins, thus obviating the need for predictions.
Phage display is an efficient approach for the determination of peptide binding specificities, which in case of ProP-PD provides direct information on binding sites in target proteins. Among the main benefits is the possibility to create highly diverse phage libraries and the fact that once a library has been created, it can be used over and over again. The method is suited for unbiased discovery of binding motifs, as no information is required beforehand for designing the phage display libraries. Phage display can be performed in high throughput. In such experiments, protein expression, purification and phage selections are performed in 96-well plates and the retained phage pools are analyzed by next-generation sequencing [55]. The limiting factors for these experiments are the availability of expression constructs, data analysis and the downstream validations. The main limitation of the technique is that it is not suited for capturing interactions that rely on PTMs.
Yeast surface display
Yeast surface display was developed nearly 20 years ago as a tool for in vitro evolution of proteins [56]. However, the technique can also be used for identification of protein-protein interactions and epitope mapping of antibodies. Similar to phage display, there is a direct link between the genotype and the phenotype [57–60]. Each yeast cell carries plasmid DNA that codes for a peptide that is displayed on the yeast cell surface. Typically, the Saccharomyces cerevisiae–Aga2p system is used, where peptides are displayed as fusions with the Aga2p subunit of the mating protein a-agglutinin (Fig. 1d). Aga2p is linked to the Aga1p subunit, via two disulfide bonds, which is anchored to the cell surface. Up to 50,000 copies of the peptide are displayed on a single cell. The cells are incubated with labeled protein and sorted based on binding to the protein using fluorescence-activated cell sorting (FACS) or magnetic-activated cell sorting (MACS). The sorted pools are thereafter sequenced. Signal intensities resulting from binding can be normalized against expression levels of the displayed peptide by concurrently tagging the peptide with a fluorescent tag.
Similar to phage display, next-generation sequencing has opened new possibilities to obtain comprehensive information on binding clones. The combination was for example used to identify unique major histocompability complex peptides that are recognized by T cell receptors [61]. It has also been used to identify peptides that bind to either Mcl-1 or Bcl-xL selectively, or to both with high affinity, by screening a library of randomized BH3 peptides [62]. An advantage of yeast surface display is the possibility to obtain information on non-binding clones. Another significant advantage is that yeast is eukaryotic and the system has some levels of PTMs. The main limitation with yeast surface display is the throughput, which is 100–1000 magnitudes lower than that of phage display.
Y2H
Y2H was first reported in 1989 [63]. It relies on the splitting of a DNA binding domain and an activation domain of a transcription factor that are linked to a prey or a bait protein. If the bait and prey proteins interact, the two domains of the transcription factor are brought together and the reconstituted transcription factor activate the transcription of reporter genes (Fig. 1e). The assay can be carried out against one prey at a time, or against libraries of prey proteins/peptides. Y2H is currently providing a massive amount of data on protein-protein interactions through the systematic efforts of Vidal and co-workers [3]. The method is in theory capable of capturing interactions relying on motif based interactions, but is in practice largely failing to identify these kinds of interactions [64]. Furthermore, Y2H does typically not provide information on the motifs involved in the identified binary interactions. For example, a large scale Y2H analysis of PDZ domains suggested that many PDZ domains do not rely on a free C-terminal region for binding, however the study did not identify the internal binding motifs [65]. Despite these issues, there are several successful cases of motif profiling through Y2H, such as the successful identification of SUMO interacting motifs for SUMO1 and SUMO2 [66]. In case of PDZ domains, Belotti and co-workers constructed an array for Y2H screening that contain 96 % of the human PDZ domains, and validated it against a select set of C-terminal preys, such as the E6 oncoviral protein and a set of protein kinases [67]. The interactions were further confirmed through mass spectrometry.
Y2H can also be used for characterization of peptide binding motifs by screening random peptide libraries [68]. For example, specificities of five PDZ domains were analyzed by screening of a candidate ligand library using a Y2H mating array [69]. Furthermore, the PDZ proteins PDZK1 and LNX were analyzed through Y2H screening against random peptide libraries [70, 71]. Similarly, the binding preferences for internal PDZ binding motifs was profiled by screening of 24 PDZ domains against a nearly random octapeptide Y2H library [72]. Thus, Y2H can be adopted for domain-motif interaction screening. The main issues with the method are a high percentage of false positives and false negatives read-outs. A particular issue is that the assay requires that proteins can be translocated to the nucleus. Although not reviewed here, there are other split-protein systems that may identify motif-based interactions [73, 74].
Validations of domain-motif interactions
With the development of high-throughput methods for identification of domain-motif interactions there is a need for high-throughput methods for affinity determination. In addition, if the aim is to identify biologically relevant domain-motif interactions, cell based validations are crucial. Both of these downstream validations may create bottlenecks. Typical methods for affinity determinations such as surface plasmon resonance and ITC provide high quality information, but have limited throughputs. To tackle the issue, various studies have reported methods for high-throughput measurements of protein-peptide interactions. A protocol for high-throughput affinity determinations using a protein microarray and fluorescently labeled synthetic peptides was published by Kaushansky et al. [46]. Moreover, a large-scale fluorescence polarization (FP) methodology using synthetic phosphopeptides was reported for affinity determinations of interactions involving the ErbB receptor phosphosites [19] and Reich et al. described SORTCERY, which is a method for ranking hundreds of yeast-displayed peptides according to their affinities for a target interaction partner [75]. The procedure involves fluorescence-activated cell sorting of a library, next-generation sequencing of sorted pools and computational analysis.
A recent addition is the high-throughput holdup assay [76]. The method is developed for affinity determinations of domain-motif interactions and can measure up to 1,000 binding affinities per day. Essentially, extracts of overexpressed proteins are incubated with resin saturated with ligands. This is followed by filtration where bound protein stays on the resin, while unbound protein will pass through the filter. The amount of protein in the flow-through are analyzed by microfluidic capillary electrophoresis and is inversely correlated to the affinity of the interactions. In the proof-of-principle experiments, the authors benchmarked the method against 210 PDZ-peptide interactions of known affinities.
If aiming for the identification of interactions of potential biological relevance, it is crucial to confirm interactions in the context of the full-length proteins. Such validations can, for example, be made through the high-throughput luminescence-based mammalian interactome mapping (LUMIER) assays [77, 78], the mammalian protein-protein interaction trap (MAPPIT) [79], or yellow fluorescence protein-fragment complementation assay [80]. As reviewed recently, there is a growing number of approaches for studying and validating protein-protein interactions in cell signaling networks [81].
Computational approaches
Complementing the experimental approaches, different computational approaches have been developed for the identification of motifs, such as SLiMFinder [82], DoReMi [83], and MotifHound [84]. To identify motifs in a given sequence, a combination of sequence properties is typically used such as i) a disorder propensity as motifs are enriched in disordered regions [85], ii) sequence conservation [86] and iii) a tendency to occur in functionally related proteins [82]. For example, a recent study on mitosis related proteins identified a new motif (Fx[ILV][FHY]x[DE]) termed the ABBA motif in the A type cyclins BUBR1, BUB1 and Acm1 [87].
While most approaches focus on the disorder property, Stein et al. took a structure based approach focusing on the fact that most motifs that are found in disordered regions will take defined structure(s) upon binding [88]. By scanning through the available protein complexes in the PDB, they discovered unnoticed peptide-based interactions and reported a list of novel peptide binding domains together with their recognition motifs. Following a structure- and data-based approach, De Bartolo and co-workers performed a genome-wide prediction of peptides binding to the human prosurvival Bcl-2 proteins. Predicted interactions were tested through SPOT arrays and in solution affinity measurements revealed affinities in the 1–500 nM KD range [89].
Recently, Chen et al. performed a genome-wide prediction of motif-mediated interactions by taking advantage of the known motifs in the ELM database, analyzing structures of domain-motif complexes and using non-structural information such as the gene ontology similarities and phylogenetic profile similarities [90]. They provided a list of 79,000 new predicted domain-motif interactions, although without experimental validation. In the future, it will be interesting to follow how computational analysis and experiments together map out motifs in various proteomes.
Conclusions
There is a plethora of experimental methods for the identification and characterization of domain-motif interactions (Table 2). Each method has its pros and cons, but together they provide complementary data. From our literature review it is clear that most of these methods have been developed for, and applied to, a limit set of ubiquitous domain families such as PDZ, WW, SH2 and SH3 domains, leaving many of the peptide binding domain families largely uncharted.
Interactions that rely on PTMs such as phosphorylation or acetylation are a challenge for most methods and there is a need for method development to allow for efficient identification of such interactions. Other challenges relate to fact that scaffold proteins often are composed of arrays of domains. Although information on the binding specificities of individual domains may be available, it does not necessary reflect the specificity of the domains in the context of the full-length proteins. In addition, connected domains of a bait protein might bind to linked motifs in a target protein, which may increase the apparent affinity and enhance the specificity of the interactions [91, 92]. Thus, dedicated approaches should be developed to account for such scenarios.
Nevertheless, by taking advantage of methods such as high-density peptide microarrays and proteomic display methods, and focusing the efforts on less explored peptide binding domain families it should be feasible to largely expand the knowledge on the binding motifs in the proteomes within the next ten years. By combining the finding from such efforts with the results of high-throughput Y2H and AP-MS we will obtain detailed maps of protein-protein interaction networks with assigned binding sites.
Abbreviations
- AP-MS:
-
Affinity-purification coupled to mass spectrometry
- ELISA:
-
Enzyme-linked immunosorbent assay
- ELM:
-
Eukaryotic linear motif
- ITC:
-
Isothermal titration calorimetry
- PDZ:
-
Postsynaptic density protein 95/discs large/zona occludens 1
- ProP-PD:
-
Proteomic peptide phage display
- PTB:
-
Phosphotyrosine-binding
- PTM:
-
Post-translational modification
- SH2:
-
Src Homology 2
- SH3:
-
Src Homology 3
- Y2H:
-
Yeast-two-hybrid
References
Stumpf MP, Thorne T, de Silva E, Stewart R, An HJ, Lappe M, et al. Estimating the size of the human interactome. Proc Natl Acad Sci U S A. 2008;105(19):6959–64.
Gavin AC, Maeda K, Kuhner S. Recent advances in charting protein-protein interaction: mass spectrometry-based approaches. Curr Opin Biotechnol. 2011;22(1):42–9.
Rolland T, Tasan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, et al. A proteome-scale map of the human interactome network. Cell. 2014;159(5):1212–26.
Menche J, Sharma A, Kitsak M, Ghiassian SD, Vidal M, Loscalzo J, et al. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347(6224):1257601.
Neduva V, Russell RB. Peptides mediating interaction networks: new leads at last. Curr Opin Biotechnol. 2006;17(5):465–71.
Van Roey K, Gibson TJ, Davey NE. Motif switches: decision-making in cell regulation. Curr Opin Struct Biol. 2012;22(3):378–85.
Tompa P, Davey NE, Gibson TJ, Babu MM. A million peptide motifs for the molecular biologist. Mol Cell. 2014;55(2):161–9.
Perkins JR, Diboun I, Dessailly BH, Lees JG, Orengo C. Transient protein-protein interactions: structural, functional, and network properties. Structure. 2010;18(10):1233–43.
Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, et al. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front Biosci. 2008;13:6580–603.
Uyar B, Weatheritt RJ, Dinkel H, Davey NE, Gibson TJ. Proteome-wide analysis of human disease mutations in short linear motifs: neglected players in cancer? Mol Biosyst. 2014;10(10):2626–42.
Stein A, Mosca R, Aloy P. Three-dimensional modeling of protein interactions and complexes is going ‘omics. Curr Opin Struct Biol. 2011;21(2):200–8.
Ivarsson Y. Plasticity of PDZ domains in ligand recognition and signaling. FEBS Lett. 2012;586(17):2638–47.
Stiffler MA, Chen JR, Grantcharova VP, Lei Y, Fuchs D, Allen JE, et al. PDZ domain binding selectivity is optimized across the mouse proteome. Science. 2007;317(5836):364–9.
Tonikian R, Zhang Y, Sazinsky SL, Currell B, Yeh JH, Reva B, et al. A specificity map for the PDZ domain family. PLoS Biol. 2008;6(9), e239.
Hu H, Columbus J, Zhang Y, Wu D, Lian L, Yang S, et al. A map of WW domain family interactions. Proteomics. 2004;4(3):643–55.
Xin X, Gfeller D, Cheng J, Tonikian R, Sun L, Guo A, et al. SH3 interactome conserves general function over specific form. Mol Syst Biol. 2013;9:652.
Tong AH, Drees B, Nardelli G, Bader GD, Brannetti B, Castagnoli L, et al. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science. 2002;295(5553):321–4.
Engelmann BW, Kim Y, Wang M, Peters B, Rock RS, Nash PD. The development and application of a quantitative peptide microarray based approach to protein interaction domain specificity space. Mol Cell Proteomics. 2014;13(12):3647–62.
Hause Jr RJ, Leung KK, Barkinge JL, Ciaccio MF, Chuu CP, Jones RB. Comprehensive binary interaction mapping of SH2 domains via fluorescence polarization reveals novel functional diversification of ErbB receptors. PLoS One. 2012;7(9), e44471.
Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439(7073):168–74.
Tinti M, Kiemer L, Costa S, Miller ML, Sacco F, Olsen JV, et al. The SH2 domain interaction landscape. Cell reports. 2013;3(4):1293–305.
Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72(5):767–78.
Dinkel H, Michael S, Weatheritt RJ, Davey NE, Van Roey K, Altenberg B, et al. ELM--the database of eukaryotic linear motifs. Nucleic Acids Res. 2012;40(Database issue):D242–51.
Sarkar D, Jana T, Saha S. LMPID: A manually curated database of linear motifs mediating protein-protein interactions. Database (Oxford). 2015;2015.
Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8(8):645–54.
Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196(6):801–10.
Roux KJ. Marked by association: techniques for proximity-dependent labeling of proteins in eukaryotic cells. Cell Mol Life Sci. 2013;70(19):3657–64.
Liu BA, Engelmann BW, Nash PD. High-throughput analysis of peptide-binding modules. Proteomics. 2012;12(10):1527–46.
Katz C, Levy-Beladev L, Rotem-Bamberger S, Rito T, Rudiger SG, Friedler A. Studying protein-protein interactions using peptide arrays. Chem Soc Rev. 2011;40(5):2131–45.
Volkmer R, Tapia V, Landgraf C. Synthetic peptide arrays for investigating protein interaction domains. FEBS Lett. 2012;586(17):2780–6.
Briant DJ, Murphy JM, Leung GC, Sicheri F. Rapid identification of linear protein domain binding motifs using peptide SPOT arrays. Methods Mol Biol. 2009;570:175–85.
Foong YM, Fu J, Yao SQ, Uttamchandani M. Current advances in peptide and small molecule microarray technologies. Curr Opin Chem Biol. 2012;16(1–2):234–42.
Fodor SP, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251(4995):767–73.
Frank R. Spot-synthesis: An easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron. 1992;48(42):9217–32.
Boisguerin P, Leben R, Ay B, Radziwill G, Moelling K, Dong L, et al. An improved method for the synthesis of cellulose membrane-bound peptides with free C termini is useful for PDZ domain binding studies. Chem Biol. 2004;11(4):449–59.
Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149(1):214–31.
Buus S, Rockberg J, Forsstrom B, Nilsson P, Uhlen M, Schafer-Nielsen C. High-resolution mapping of linear antibody epitopes using ultrahigh-density peptide microarrays. Mol Cell Proteomics. 2012;11(12):1790–800.
Forsstrom B, Axnas BB, Stengele KP, Buhler J, Albert TJ, Richmond TA, et al. Proteome-wide epitope mapping of antibodies using ultra-dense peptide arrays. Mol Cell Proteomics. 2014;13(6):1585–97.
Carmona SJ, Nielsen M, Schafer-Nielsen C, Mucci J, Altcheh J, Balouz V, et al. Towards high-throughput immunomics for infectious diseases: use of next-generation peptide microarrays for rapid discovery and mapping of antigenic determinants. Mol Cell Proteomics. 2015;4(7):1871–84.
Okada H, Uezu A, Soderblom EJ, Moseley 3rd MA, Gertler FB, Soderling SH. Peptide array X-linking (PAX): a new peptide-protein identification approach. PLoS One. 2012;7(5), e37035.
MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289(5485):1760–3.
Fasolo J, Snyder M. Protein microarrays. Methods Mol Biol. 2009;548:209–22.
Zarate X, Galbraith DW. A cell-free expression platform for production of protein microarrays. Methods Mol Biol. 2014;1118:297–307.
Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, et al. Global analysis of protein activities using proteome chips. Science. 2001;293(5537):2101–5.
Uzoma I, Zhu H. Interactome mapping: using protein microarray technology to reconstruct diverse protein networks. Genomics Proteomics Bioinformatics. 2013;11(1):18–28.
Kaushansky A, Allen JE, Gordus A, Stiffler MA, Karp ES, Chang BH, et al. Quantifying protein-protein interactions in high throughput using protein domain microarrays. Nat Protoc. 2010;5(4):773–90.
Sundell GN, Ivarsson Y. Interaction Analysis through Proteomic Phage Display. BioMed Res Int. 2014;2014:176172.
Rentero Rebollo I, Sabisz M, Baeriswyl V, Heinis C. Identification of target-binding peptide motifs by high-throughput sequencing of phage-selected peptides. Nucleic Acids Res. 2014;42(22), e169.
Gfeller D, Butty F, Wierzbicka M, Verschueren E, Vanhee P, Huang H, et al. The multiple-specificity landscape of modular peptide recognition domains. Mol Syst Biol. 2011;7:484.
Kim J, Kim I, Yang JS, Shin YE, Hwang J, Park S, et al. Rewiring of PDZ domain-ligand interaction network contributed to eukaryotic evolution. PLoS Genet. 2012;8(2), e1002510.
Luck K, Trave G. Phage display can select over-hydrophobic sequences that may impair prediction of natural domain-peptide interactions. Bioinformatics. 2011;27(7):899–902.
Di Niro R, Sulic AM, Mignone F, D’Angelo S, Bordoni R, Iacono M, et al. Rapid interactome profiling by massive sequencing. Nucleic Acids Res. 2010;38(9), e110.
Ivarsson Y, Arnold R, McLaughlin M, Nim S, Joshi R, Ray D, et al. Large-scale interaction profiling of PDZ domains through proteomic peptide-phage display using human and viral phage peptidomes. Proc Natl Acad Sci U S A. 2014;111(7):2542–7.
Larman HB, Zhao Z, Laserson U, Li MZ, Ciccia A, Gakidis MA, et al. Autoantigen discovery with a synthetic human peptidome. Nat Biotechnol. 2011;29(6):535–41.
Huang H, Sidhu SS. Studying binding specificities of peptide recognition modules by high-throughput phage display selections. Methods Mol Biol. 2011;781:87–97.
Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15(6):553–7.
Gai SA, Wittrup KD. Yeast surface display for protein engineering and characterization. Curr Opin Struct Biol. 2007;17(4):467–73.
Gera N, Hussain M, Rao BM. Protein selection using yeast surface display. Methods. 2013;60(1):15–26.
Pepper LR, Cho YK, Boder ET, Shusta EV. A decade of yeast surface display technology: where are we now? Comb Chem High Throughput Screen. 2008;11(2):127–34.
Cherf GM, Cochran JR. Applications of Yeast Surface Display for Protein Engineering. Methods Mol Biol. 2015;1319:155–75.
Birnbaum ME, Mendoza JL, Sethi DK, Dong S, Glanville J, Dobbins J, et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 2014;157(5):1073–87.
Dutta S, Gulla S, Chen TS, Fire E, Grant RA, Keating AE. Determinants of BH3 binding specificity for Mcl-1 versus Bcl-xL. J Mol Biol. 2010;398(5):747–62.
Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340(6230):245–6.
Shoemaker BA, Panchenko AR. Deciphering protein-protein interactions. Part I. Experimental techniques and databases. PLoS Comput Biol. 2007;3(3), e42.
Lenfant N, Polanowska J, Bamps S, Omi S, Borg JP, Reboul J. A genome-wide study of PDZ-domain interactions in C. elegans reveals a high frequency of non-canonical binding. BMC Genomics. 2010;11:671.
Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem. 2006;281(23):16117–27.
Belotti E, Polanowska J, Daulat AM, Audebert S, Thome V, Lissitzky JC, et al. The human PDZome: a gateway to PSD95-Disc large-zonula occludens (PDZ)-mediated functions. Mol Cell Proteomics. 2013;12(9):2587–603.
Yang M, Wu Z, Fields S. Protein-peptide interactions analyzed with the yeast two-hybrid system. Nucleic Acids Res. 1995;23(7):1152–6.
Song E, Gao S, Tian R, Ma S, Huang H, Guo J, et al. A high efficiency strategy for binding property characterization of peptide-binding domains. Mol Cell Proteomics. 2006;5(8):1368–81.
Hu S, Song E, Tian R, Ma S, Yang T, Mu Y, et al. Systematic analysis of a simple adaptor protein PDZK1: ligand identification, interaction and functional prediction of complex. Cell Physiol Biochem. 2009;24(3–4):231–42.
Guo Z, Song E, Ma S, Wang X, Gao S, Shao C, et al. Proteomics strategy to identify substrates of LNX, a PDZ domain-containing E3 ubiquitin ligase. J Proteome Res. 2012;11(10):4847–62.
Mu Y, Cai P, Hu S, Ma S, Gao Y. Characterization of diverse internal binding specificities of PDZ domains by yeast two-hybrid screening of a special peptide library. PLoS One. 2014;9(2), e88286.
Shekhawat SS, Ghosh I. Split-protein systems: beyond binary protein-protein interactions. Curr Opin Chem Biol. 2011;15(6):789–97.
Lam MH, Stagljar I. Strategies for membrane interaction proteomics: no mass spectrometry required. Proteomics. 2012;12(10):1519–26.
Reich LL, Dutta S, Keating AE. SORTCERY-A High-Throughput Method to Affinity Rank Peptide Ligands. J Mol Biol. 2015;427(11):2135–50.
Vincentelli R, Luck K, Poirson J, Polanowska J, Abdat J, Blemont M, et al. Quantifying domain-ligand affinities and specificities by high-throughput holdup assay. Nat Methods. 2015.
Blasche S, Koegl M. Analysis of protein-protein interactions using LUMIER assays. Methods Mol Biol. 2013;1064:17–27.
Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307(5715):1621–5.
Eyckerman S, Verhee A, der Heyden JV, Lemmens I, Ostade XV, Vandekerckhove J, et al. Design and application of a cytokine-receptor-based interaction trap. Nat Cell Biol. 2001;3(12):1114–9.
Nyfeler B, Michnick SW, Hauri HP. Capturing protein interactions in the secretory pathway of living cells. Proc Natl Acad Sci U S A. 2005;102(18):6350–5.
Yao Z, Petschnigg J, Ketteler R, Stagljar I. Application guide for omics approaches to cell signaling. Nat Chem Biol. 2015;11(6):387–97.
Edwards RJ, Davey NE, Shields DC. SLiMFinder: a probabilistic method for identifying over-represented, convergently evolved, short linear motifs in proteins. PLoS One. 2007;2(10), e967.
Horn H, Haslam N, Jensen LJ. DoReMi: context-based prioritization of linear motif matches. PeerJ. 2014;2, e315.
Kelil A, Dubreuil B, Levy ED, Michnick SW. Fast and accurate discovery of degenerate linear motifs in protein sequences. PLoS One. 2014;9(9), e106081.
Davey NE, Haslam NJ, Shields DC, Edwards RJ. SLiMSearch 2.0: biological context for short linear motifs in proteins. Nucleic Acids Res. 2011;39(Web Server issue):W56–60.
Davey NE, Cowan JL, Shields DC, Gibson TJ, Coldwell MJ, Edwards RJ. SLiMPrints: conservation-based discovery of functional motif fingerprints in intrinsically disordered protein regions. Nucleic Acids Res. 2012;40(21):10628–41.
Di Fiore B, Davey NE, Hagting A, Izawa D, Mansfeld J, Gibson TJ, et al. The ABBA motif binds APC/C activators and is shared by APC/C substrates and regulators. Dev Cell. 2015;32(3):358–72.
Stein A, Aloy P. Novel peptide-mediated interactions derived from high-resolution 3-dimensional structures. PLoS Comput Biol. 2010;6(5), e1000789.
DeBartolo J, Taipale M, Keating AE. Genome-wide prediction and validation of peptides that bind human prosurvival Bcl-2 proteins. PLoS Comput Biol. 2014;10(6), e1003693.
Chen TS, Petrey D, Garzon JI, Honig B. Predicting Peptide-mediated interactions on a genome-wide scale. PLoS Comput Biol. 2015;11(5), e1004248.
Ye F, Zhang M. Structures and target recognition modes of PDZ domains: recurring themes and emerging pictures. Biochem J. 2013;455(1):1–14.
Dodson EJ, Fishbain-Yoskovitz V, Rotem-Bamberger S, Schueler-Furman O. Versatile communication strategies among tandem WW domain repeats. Exp Biol Med (Maywood). 2015;240(3):351–60.
Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol. 2005;1:2005 0008.
Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, et al. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature. 1993;363(6424):85–8.
Grootjans JJ, Zimmermann P, Reekmans G, Smets A, Degeest G, Durr J, et al. Syntenin, a PDZ protein that binds syndecan cytoplasmic domains. Proc Natl Acad Sci U S A. 1997;94(25):13683–8.
Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, et al. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem. 2001;276(18):15164–73.
Acknowledgements
This work was supported by grants from the Swedish Research Council (YI: C0509201), the Ake Wiberg foundation (YI: 3773397), the Magnus Bergvall’s foundation (YI), Lennanders foundation (CB) and Ingegerd Bergh’s foundation (CB). The authors apologize to all authors whose work has not been cited due to the limited space of the review.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CB made the figure. Both authors wrote the paper. Both authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Blikstad, C., Ivarsson, Y. High-throughput methods for identification of protein-protein interactions involving short linear motifs. Cell Commun Signal 13, 38 (2015). https://doi.org/10.1186/s12964-015-0116-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12964-015-0116-8