Abstract
A key challenge to advance the efficiency of bioprocesses is the uncoupling of biomass from product formation, as biomass represents a by-product that is in most cases difficult to recycle efficiently. Using the example of rhamnolipid biosurfactants, a temperature-sensitive heterologous production system under translation control of a fourU RNA thermometer from Salmonella was established to allow separating phases of preferred growth from product formation. Rhamnolipids as bulk chemicals represent a model system for future processes of industrial biotechnology and are therefore tied to the efficiency requirements in competition with the chemical industry. Experimental data confirms function of the RNA thermometer and suggests a major effect of temperature on specific rhamnolipid production rates with an increase of the average production rate by a factor of 11 between 25 and 38 °C, while the major part of this increase is attributable to the regulatory effect of the RNA thermometer rather than an unspecific overall increase in bacterial metabolism. The production capacity of the developed temperature sensitive-system was evaluated in a simple batch process driven by a temperature switch. Product formation was evaluated by efficiency parameters and yields, confirming increased product formation rates and product-per-biomass yields compared to a high titer heterologous rhamnolipid production process from literature.
Similar content being viewed by others
Introduction
A major part of a future bioeconomy-driven industrial biotechnology is based on sustainable bulk products with typically low added-value. Formerly mainly focused on products that are difficult to synthesize by chemical means, such as proteins or chiral molecules etc., industrial biotechnology will include typical low-molecular weight products that were exclusively produced by chemical means. As such, the efficiency of substrate conversion is of particular importance, as future processes of industrial biotechnology are therefore tied to efficiency requirements in competition with the chemical industry. A key challenge to advance the efficiency of bioprocesses is the uncoupling of biomass from product formation, as biomass represents a by-product that is in most cases difficult to recycle efficiently.
Rhamnolipids
Microbial rhamnolipid biosurfactants (RL) as bulk chemicals represent a model system for future industrial biotechnology in the context of a bio-based economy. Biosurfactants are based on renewable substrates, whereas many conventional surfactants are derived from petrochemical sources1. Biosurfactants exhibit beneficial properties like low toxicity, biodegradability, high surfactant efficiencies and high stability at extreme pH, temperature and salinity when compared to many chemical surfactants2. The fields of application for surfactants range from the cosmetic sector (personal care products), the food sector (emulsifiers or stabilizers) and the pharmaceutical sector (antimicrobial and antifungal activities) to enhanced oil recovery and cleaning agents (household detergents)2,3,4,5,6. Unsurprisingly, an increase in research interest on microbial biosurfactants could be observed over the past decade7. However, production of biosurfactants is still cost intensive, especially the downstream processing8. Furthermore, the use of the model organism P. aeruginosa, even though it is reported to produce the highest RL titers (39 g/L)9, bares two disadvantages for a commercial production of the biosurfactant RL. First, P. aeruginosa is a human pathogen10 and second, RL production is controlled by a complex regulatory cell density-dependent mechanism called quorum sensing. To overcome these obstacles, heterologous RL production was first described by Pseudomonas putida11, 12. In 2016, the rhlAB genes were transferred from P. aeruginosa with the pBBR1MCS-3 based plasmid pSynPro8oT_rhlAB into the nonpathogenic host P. putida KT2440 resulting in heterologous constitutive expression of rhamnosyltransferase genes needed for mono-RL production13. A maximum concentration of 14.9 g/L RL could be reached in a fed batch process with a two-phase glucose feeding profile. However, this approach for heterologous RL production suffers from considerable biomass production which represents a major carbon sink (roughly 50% more biomass than product)14. This challenge is addressed with the presented study.
RNA thermometer
Temperature fluctuations can be detected by different molecular thermosensors made of DNA, RNA, proteins or lipids. The different principles of temperature sensing have been extensively reviewed15,16,17. The thermosensors of interest for this study are molecular structures mostly found in the 5'-UTR of the mRNA that enable thermosensing. Due to the physical properties of RNA, so-called hairpin structures can be formed. In these hairpin structures, base pairing blocks the Shine-Dalgarno sequence, and the AUG start codon, which are part of the ribosomal binding and translation initiation site. A certain amount of thermal energy, depending on the composition of base pairs, is required to melt the H-bridges in the hairpin structure. By raising the temperature, the hairpin structure gradually opens and allows ribosomes to access the translation initiation site16. In 1989, the first cis encoded regulatory RNA element determining temperature-dependent translation initiation was discovered in the cIII gene of bacteriophage λ18. Up until today several temperature-sensitive RNA structures, governing mainly the expression of heat shock and virulence genes have been reported19,20,21,22,23 but to our knowledge not leveraged for industrial applications. This study aims to provide a proof-of-concept process exploiting a temperature responsive RNA thermometer (RNAT) for bioprocess control.
ROSE-like RNA thermometer
In P. aeruginosa RL production is governed by an RNA-based temperature-sensing element an RNAT. RNATs are instrumental in an adequate response to the decisive signal of a temperature-shift from low environmental to elevated temperatures e.g. of 37 °C in a host system. The consequence after successful invasion of a host by a pathogen is the upregulation of its virulence factor production. To respond to elevated host temperatures, the pathogen P. aeruginosa, has a so termed ROSE (Repression Of heat-Shock gene Expression)-like RNAT. The RNAT is located in the 5’ UTR (untranslated region) of the rhlA mRNA, thermo-regulates the translation of the mRNA and therefore indirectly the production of the virulence factor RL. Almost all known ROSE-like RNATs govern heat shock gene expression with the exception of the ROSE-like element found to control the formation of the virulence factor RL in P. aeruginosa and in the heterologous system P. putida KT2440 pSynPro8oT_rhlAB24, 25. It was argued that at non-permissive environmental temperatures (≤ 30 °C) translation of rhlA is inhibited by the RNAT, which in turn interferes with the transcription of the rhlB gene. This phenomenon is known as polar effect24. In 2019, a ROSE-like RNAT structure was evaluated for temperature-responsivity and its ability to govern a temperature dependent expression of the rhlAB genes25. The latter study showed the potential to use RNATs as regulatory elements; however, the evaluated ROSE-like RNAT showed a limited regulatory effect on RL production. Consequently, a different temperature responsive element with a more pronounced regulatory effect on translation was investigated in this study.
FourU RNA thermometer
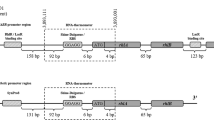
Another family of RNATs are the fourU-type RNATs. FourU RNATs located mostly in the 5’UTR, are between 40 and > 270 nucleotides in length (htrA; E. coli and shuA; Shigella dysenteriae) and consist of 1–5 hairpin structures26,27,28. The temperature responsive “hairpin 2” of the 57 nucleotides Salmonella fourU-type RNAT was used in this study. In Salmonella enterica the fourU RNAT governs the expression of the heat shock gene agsA. In vivo experiments showed that the temperature responsive wild type fourU RNAT mostly prevents translation of a subsequent bgaB gene at 30 °C but is upregulated at common mammalian host temperatures of 37 °C and above. Previous studies reported that a destabilized variant of the wild type fourU RNAT can be obtained by introducing a single point mutation into the hairpin structure. This structure is destabilized at the temperature range investigated here allowing for an earlier onset of translation initiation (at lower temperatures) than the wild type RNAT (Fig. 1)29.
Working principal, hairpin structure and RNA sequence of Salmonella wild type fourU RNAT and destabilized (G14A-C25U) mutant used in this study. Base pairs framed in red are less stable than base pairs framed in blue, grey frames indicate the fourU motive and black frames the Shine-Dalgarno sequence. (Adapted from29).
Aim of this study
The presented study was meant to be a case study on the applicability of RNAT in combination with temperature changes to govern an exemplary bulk chemical production process within the framework of an industrial biotechnology, in this case microbial RLs. A temperature-sensitive heterologous RL production system under translation control of a fourU RNAT from Salmonella29 (Fig. 1) was established. The characteristics of this system are assessed in comparison to a destabilized structure used as a control that allows translation already at lower temperatures. The use of RNATs for molecular control through partial decoupling of biomass and product formation in bioprocesses is illustrated using RL biosurfactants as an example and presented by growth respectively product formation rates and yields. Furthermore, a proof-of-concept of a bioreactor cultivation with a temperature switch was carried out to elucidate the production capacity and potential of the molecular temperature switch for bioprocess control. Hence, this work marks a fundamental step for temperature-based (molecular) process design and control by exploiting temperature responsive elements.
Results
To demonstrate the effect of temperature on RL production and cell growth, a set of experiments, comparing shake flask cultivations at different temperatures (20—42.5 °C), was conducted.
Heterologous fourU RNA thermometer thermo-regulates RL production
When cultivating P. putida KT2440 pSynPro8oT_4U_rhlAB with the functional wild type fourU RNAT a lower biomass concentration was obtained at 38 °C (1.43 g/L), compared to 20 °C (5.71 g/L). However, the lower amount of biomass at 38 °C yielded a maximum concentration of RLs (1.84 g/L) approximately 10 times higher than the maximum RL concentration (0.19 g/L) measured at 20 °C. Compared to that the cultivation of the control strain with the destabilized fourU RNAT yielded 4.55 g/L biomass with 1.64 g/L RL at 20 °C and 0.49 g/L biomass with 0.48 g/L RL at 38 °C. At 42.5 °C almost no cell growth could be observed (0.10 g/L (inoculation)—maximum of 0.15 g/L (after 6 h)). In Fig. 2 selected time courses of shake flask cultivations of P. putida KT2440 pSynPro8oT_4U_rhlAB and P. putida KT2440 pSynPro8oT_4U(G14A-C25U)_rhlAB using ModR medium with 10 g/L glucose are shown. To visualize the effect of the different temperatures tested on the strains under investigation, maximum (qmax) and mean (qavg) specific RL production rates were plotted against temperature (Fig. 3). Whereas qmax was chosen to illustrate and compare the highest rate recorded of each cultivation temperature tested and qavg to display an average rate over the cultivation period for each cultivation temperature tested. A 15-fold increase of qmax from 9 mg/(g⋅h) at 20 °C to 133 mg/(g⋅h) at 38 °C was calculated. A model describing qmax as a function of temperature (Eq. (1)) according to the group of Schoolfield30 was fitted to the experimentally determined values of qmax for the P. putida strain harboring the functional fourU RNAT (Fig. 3). The experimentally determined values for qavg respectively qmax increased from roughly 0 almost exponentially to a maximum of 67 mg/(g⋅h) (qavg) respectively 133 mg/(g⋅h) (qmax) at 38 °C. The temperature at which the highest specific RL productivity could be observed (highest qmax from model) was calculated to be 38.4 °C when using the model of Schoolfield et al. At temperatures higher than 38 °C, experimentally determined RL production rates decreased rapidly. A mathematical artifact arising from a low biomass concentration at 42.5 °C, distorts the calculated value for qmax causing a high specific production rate at that temperature. Summarizing, the temperature responsive fourU element shows a pronounced thermo regulatory effect on RL production.
Experimentally determined maximum (qmax; black circles) and mean (qavg; black triangles) specific rhamnolipid production rates plotted against temperatures tested for Pseudomonas putida KT2440 pSynPro8oT_4U_rhlAB. Maximum specific rhamnolipid production rates were described as a function of temperature by fitting a temperature model (Schoolfield et al.30; Eq. (1); black line) to the obtained data of qmax.
Increased RL-per-glucose and RL-per-biomass yields at elevated temperatures
RL-per-glucose yields (YP|S) and total RL-per-biomass yields (YP|X) are shown for different temperatures for the functional fourU RNAT in Fig. 4a. An apparent correlation of YP|S respectively YP|X, and temperature can be derived (Fig. 4a). The YP|S increased with the temperature by around 10-fold from 0.02 g/g (20 °C) to 0.20 g/g (38 °C). The YP|X value showed a similar trend with an even stronger increase when raising temperature. An almost 50-fold increase of the YP|X value between 20 °C and 38 °C from 0.03 g/g to 1.43 g/g was calculated. When temperature was raised higher than 38 °C both yield-values decreased approaching 0 at 42.5 °C. In addition to the performance parameters shown above, growth rates (µ) and biomass-to-glucose yields (YX|S) were furthermore calculated (Fig. 4b). The highest maximum specific growth rates (µmax) were reached at 32.5 °C for the strain carrying the functional fourU RNAT with a µmax of 0.41 h-1 respectively 0.33 h-1 at 35 °C for the control. YX|S remained relatively constant below and around the growth optimum (20–35 °C) with values around 0.50 g/g. Yields decreased at temperatures above 35 °C to 0.16 g/g at 38 °C before approaching almost 0 at 42.5 °C (Fig. 4b). As stated in the introduction, heterologous RL production suffers from considerable biomass production which represents a major carbon sink (roughly 50% more biomass than product)14. It is shown here that elevated temperatures, shift the RL-per-biomass yield in favor of RLs and simultaneously improve RL-per-glucose yields.
Proof-of-concept temperature switch process
To benchmark the regulatory capabilities of the investigated fourU RNAT and to assess the production capacities of the applied biological system on a technical scale, a temperature switch process was carried out in a bioreactor. To achieve a separation of growth and product formation, cells were initially grown at 25 °C for biomass generation. At this temperature, RL formation rates were low, while biomass growth was high, as indicated in Figs. 3 + 4b. Subsequently, temperature was switched to 38 °C, which represents the experimental optimum of specific RL production rates. Switching the temperature from 25 to 38 °C was performed at t = 9 h, and a stable temperature could be reached within 15 min, with an error of less than 0.05 °C. The results are shown in the figure below. As expected, RL titers were lower than 0.10 g/L at 25 °C before induction and reached around 2.28 g/L after switching the temperature to 38 °C and 48 h of total cultivation time.
For the process shown above, maximum specific RL production rates of up to 145 mg/(g h) and average specific production rates of up to 32 mg/(g h) were reached. To put this simple batch process in context, a comparison was drawn with a literature high titer heterologous RL production process. The authors used the same strain, P. putida KT2440 pSynPro8oT_rhlAB without the fourU RNAT. Their cultivation was carried out in ModR medium with glucose as carbon source as well and at constant 30 °C (table below). The following table shows further reports on (heterologous) RL production processes using Pseudomonas.
The highest titer reported to date for mono-RL production, using a heterologous host, was reached in 2016 with a two phase feeding strategy at constant 30 °C14. For the switch process shown in this study average specific RL production rates (qavg) of up to 32 mg/(g h)) were reached. This result was around 25% higher compared to the highest average specific RL production rate (qavg: 24 mg/(g h)) reported in the literature reference process for heterologous RL production using P. putida KT2440 pSynpro8oT_rhlAB14 (Table 1; Fig. 5). This finding supports the potential of temperature to be exploited as control variable in combination with a functional RNAT. However, a suitable feeding strategy would still be required in conjunction with the RNAT-driven molecular control system to allow for higher titers and achieve reduced side product formation. The presented switch process in this study is a proof-of-concept for exploiting a temperature responsive RNAT for bioprocess control on a pre-industrial scale. To our knowledge the process is the first of its kind to leverage a fourU RNAT in combination with a temperature shift to control RL production.
Bioreactor batch cultivation of Pseudomonas putida KT2440 pSynPro8oT_4U_rhlAB in ModR medium with 30 g/L glucose. Time courses of measured biomass- (circles), and glucose concentration (squares) are shown in the upper plot as well as rhamnolipid concentration (triangles) and measured temperature (red line) in the bottom plot. Biomass was grown at 25 °C until reaching an OD600 of 4 (approx. 1.4 g/L, after ~ 9 h). Then temperature was switched to 38 °C to induce rhamnolipid production as indicated by an arrow at the bottom plot.
Discussion
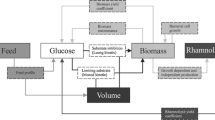
When comparing time courses of biomass and RL concentration during shake flask cultivations of P. putida KT2440 pSynPro8oT_4U_rhlAB at 20 °C and 38 °C, the final biomass concentration at 38 °C was lower while the maximum RL concentration was almost 10-times higher than what was obtained at 20 °C (Fig. 2). Consequentially a higher biomass specific productivity was reached when cultivations were performed at elevated temperatures (Fig. 3). The temperature-dependency of RL production rates using fourU RNAT was investigated in a range between 20 and 42.5 °C (Fig. 3). An almost exponential increase of RL productivity between 20 °C to an optimum at 38 °C could be observed. In the absence of limitations and by assuming that RL production rate is proportional to rhamnosyl transferase concentration, this correlation mirrors what is known for RNAT regulation behavior. It has been reported before that the opening of the RNA hairpin structure exhibits a gradual temperature-response profile and typically never entirely prevents translation34,35. The rapid decrease in productivity when the optimal temperature for RL production is exceeded could be attributed to various effects of elevated temperatures on the bacterial cell. High temperatures are for example connected to aggregation and denaturation of proteins. This may lead to deactivation of enzymes needed for RL biosynthesis. Indirect effects arising from an altered bacterial metabolism may also be considered, such as the availability of precursors necessary for RL production. A lower availability of the RL precursors HAA and dTDP-rhamnose may then present a bottleneck for RL synthesis. Low temperature results in low growth which then leads to a reduced demand for dTDP because less DNA needs to be synthesized. This would mean that more dTDP nucleotide sugars (activated sugars) for RL biosynthesis may be available, thus resulting in higher RL production rates at the respective temperature. Furthermore, the substrate binding capacity of RhlA respectively RhlB towards the previously mentioned precursors may also be negatively affected by temperatures beyond the production optimum. The total productivity of the process may be evaluated according to the average specific RL production rate which can then ultimately be used as target for process optimization as has been suggested previously25. The observed increase of average specific RL production rate over temperature may differ in prolonged processes, for example in a bioreactor cultivation with a higher initial glucose concentration. For bulk chemical production, total YP|X respectively YP|S mark important parameters for process optimization and are therefore suitable targets to assess the production capacity of the applied biological system under control of the fourU RNAT. In this study, these yields were observed to be increasing alongside with increasing maximum respectively average RL production rates when raising temperature (Figs. 3 + 4). Translating this to the design of a more efficient production process at heightened temperatures would mean both a shortening of process time (increased RL production rates q) and higher efficiency (increased product per biomass respectively per substrate yields over temperature) of RL production. The higher efficiency of RL production at elevated temperatures may be assigned to different overlapping effects. To estimate the influence of a functional fourU RNAT on the observed increase of RL productivity over temperature, the G14A-C25U mutant was used as comparison. As has been reported previously, the RNA structure of the G14A-C25U mutant is destabilized already at a lower temperatures than the wild type RNA structure29. The increase of qmax and qavg between 25 and 38 °C for the P. putida strain harboring the functional fourU RNAT was determined to be 9.3-fold respectively 11.1-fold compared to 1.4-fold for both rate changes of the G14A-C25U mutant (Fig. 3). Assuming on the one hand that the functional fourU RNAT has no significant regulatory effect on RL production rates, the increase of the rate over temperature for the G14A-C25U mutant should have been approximately the same. On the other hand, if an absence of unspecific or metabolic effects is assumed, the increase in production rate over temperature were to be solely caused by the RNATs regulatory effect. This would mean that the increase in production rate of the G14A-C25U mutant should have been close to 0. The here presented experiments revealed that besides the discussed metabolic effect, a major part of the total increase in RL production rate is to be attributed to the regulatory effect of the RNAT. The specific production rate between 25 and 38 °C for the G14A-C25U mutant increased 1.4-fold and may be due to unspecific metabolic effects (Fig. 3). Compared to that the specific production rate of the strain with the functional fourU RNAT increased by more than 9-fold on average between 25 and 38 °C, which confirms the regulatory action of the deployed fourU RNAT element in the heterologous host P. putida KT2440 (Figs. 3 + 6). The effect could be mainly assigned to the presence and regulatory effect of a functional fourU RNAT (RE, Fig. 6). The regulatory effect is derived by comparing specific productivities and time courses of cultivations with the strain carrying the functional fourU RNAT (e.g., 9.3-fold change of qmax comparing 25 to 38 °C, Fig. 6) with the control strain. In case of the control, only a slight increase in maximum specific productivity (e.g., 1.4-fold change of qmax comparing 25 to 38 °C, Fig. 6) was obtained. The observed increase in qmax for the control may be assigned to an unspecific metabolic effect (UME, Fig. 6) caused by the temperature increase itself along with elevated biochemical reaction rates. The UME accounts for 12 – 15% of the total change in average respectively maximum specific production rate over temperature as is shown below.
Comparison of increase in maximum specific rhamnolipid production rate (qmax) of Pseudomonas putida KT2440 pSynPro8oT_4U_rhlAB and control strain when grown at 25 °C and 38 °C respectively. Average differences in production rate calculated from individual biological data are shown with derived regulatory effect (RE) and unspecific metabolic effect (UME).
Different overlapping effects reported in connection to the RNATs, and their regulatory abilities have to be mentioned and considered. RNATs are often reported to have a high degree of “leakiness” respectively inefficiency when it comes to inhibition of translation36. Moreover, the regulatory potential of RNATs can only be harnessed by changing the temperature. In addition, temperature influences also the entire metabolism and individual metabolic conversion reaction rates can change by altered enzyme activity or even result in inactive enzymes due to denaturation of proteins at elevated temperatures37. This could consequently lead to an increase of substrate usage required in metabolic maintenance. An altered temperature also causes different levels of solubilized oxygen in the growth medium, with higher temperatures generally leading to a reduction in oxygen solubility. In this study, a plasmid with the rhlAB genes governed by the fourU RNAT was designed and used. The strong increase of RL production rates (> 9-fold) and yields (up to 10 respectively 50-fold for YP|S respectively YP|X) with increasing temperature indicates a high potential for dynamic temperature adjustments to be used in process control during a RL production process. Figure 6 summarizes the effect of higher biomass specific productivity at elevated temperatures and elucidates its potential for a bioreactor switch process. In the batch reactor set-up, the applicability of the molecular RNAT combined with a switch in temperature (25 °C to 38 °C) for a RL production process (Fig. 5) was shown and compared to previous results for (high titer heterologous) RL production processes (Table 1). The initial temperature of 25 °C was chosen instead of 20 °C to ensure growth at a sufficiently high growth rate while still having low RL production rates (Figs. 3 + 4b) to separate growth and production phase. In this simple batch set-up incorporating a switch from 25 °C to 38 °C after reaching an OD600 of 4, the average specific RL production rate could be increased by up to 25% compared to the literature process14 (Table 1). As such, this batch process outlines the potential of using a RNAT based temperature switch for molecular bioprocess control. Furthermore, RNAT hold several advantages over conventional chemical induction systems such as no use of chemical inducers, (reversible) induction by temperature, possible autoinduction of the system by metabolic heat and earlier onset of the induction response (mRNA is already produced). In a large-scale industrial setup, that would potentially reduce overall costs for chemical inducers, enable tighter control of product-per-biomass ratios and allow for greater flexibility of process control. Furthermore, to track metabolic changes, future studies may also include transcriptome and metabolome analysis, which could point to bottlenecks in the bacterial metabolism and provide additional key factors for optimization approaches using metabolic engineering. Future investigations should aim for a gradual adjustment of temperature over time using tailor made temperature profiles along with an optimized feeding strategy to allow for higher product titers and reduce side product formation. Process modeling may be used to facilitate the completion of this challenging task, because temperature adjustments bring about a wide range of metabolic changes and an optimum temperature profile may therefore not be apparently conceivable. A suitable modeling tool to capture the complexity of a changing temperature during a bioprocess may be an artificial neural network. The combination of a trained artificial neural network and a genetic algorithm may be a powerful strategy to compute an optimized temperature profile to achieve high RL titers. By using temperature sensitive control elements in combination with a well-directed shift in temperature a key challenge in bioprocesses can be met. Substrate carbon can be more efficiently used by reducing by-product formation in form of biomass while substrate carbon is directed into the product. As such this step contributes to the development of sustainable and efficient bioprocesses for a future bioeconomy.
Methods
Chemicals and reference substances
The chemicals used in this study were purchased from Carl Roth GmbH (Karlsruhe, Germany) if not stated otherwise. The chemicals for analytic procedures were of analytical or mass spectrometry grade. Former Hoechst AG (Frankfurt-Hoechst, Germany) kindly provided the di-RL (Rha-Rha-C10-C10) standard used for HPTLC analysis. The mono-RL (Rha-C10-C10) standard was purchased from Sigma-Aldrich Laborchemikalien GmbH (Seelze, Germany). Derivatization of RLs was carried out as described previously38 using the derivatization agents 4-bromophenacylbromide and triethylamine which were obtained from Sigma-Aldrich Laborchemikalien GmbH (Seelze, Germany).
Microorganism and plasmids
Pseudomonas putida KT2440 carrying the pSynPro8oT_rhlAB plasmid for heterologous production of mono-RLs was used as described previously13. The ROSE-like RNAT sequence on the pSynPro8oT_rhlAB plasmid as described previously25 was exchanged with either the functional fourU RNAT native to Salmonella or the destabilized sequence as control (Fig. 1)29. The sequences for the wild type fourU RNAT and the destabilized G14A-C25U mutant shown in Table 2 were ordered via Eurofins Genomics (Ebersberg, Germany) as a standard gene synthesis in a standard pEX A128 vector with ampicillin resistance for selection. Each fourU RNAT sequence variant ordered, was flanked by Flank A (Table 2) also present in the pSynPro8oT_rhlAB template plasmid containing the pSynPro8oT synthetic promotor as well as the BglII restriction site and downstream of the fourU RNAT sequence by Flank B (Table 2) coding for a part of the rhlA gene as well as the PciI restriction site.
The pEX plasmids containing the sequences described above were transferred into chemically competent Escherichia coli DH5α (New England Biolabs, Ipswich, USA) for plasmid propagation and isolation. The isolated pEX plasmids containing the different fourU RNAT variants were cut with BglII and PciI. The pSynPro8oT_rhlAB backbone isolated from P. putida KT2440 pSynPro8oT_rhlAB was cut analogously with BglII and PciI to remove the ROSE-like RNAT and to produce matching sticky ends. After dephosphorylation of digested pSynPro8oT_rhlAB backbone and ligation with digested fourU RNAT fragment from pEX plasmid using T4 DNA ligase, the plasmids pSynPro8oT_4U_rhlAB and pSynPro8oT_4U(G14A-C25U)_rhlAB containing the different fourU RNAT variants instead of the ROSE-like RNAT were transformed into electrocompetent Pseudomonas putida KT2440 wild type cells. Electrocompetent P. putida KT2440 wild type cells were prepared according to39. Electroporation of electrocompetent cells was carried out with an electroporation device (Eporator, Eppendorf AG, Hamburg, Germany) and voltage set to 2.5 kV. Strains were selected on LB agar plates containing 20 mg/L tetracycline and confirmed with subsequent sequencing (Eurofins Genomics; Ebersberg, Germany). All strains were stored as glycerol stocks at -80 °C. The strains Pseudomonas putida KT2440 pSynPro8oT_4U_rhlAB and Pseudomonas putida KT2440 pSynPro8oT_4U(G14A-C25U)_rhlAB respectively are referred to as FourU and control in the following.
Media preparation
All growth media (LB, SupM, ModR) were prepared as described previously14.
Shake flask cultivation
Cultivations were performed in a shake incubator (New Brunswick/Innova 44, chamber, Eppendorf AG, Hamburg, Germany) using temperatures between 20 and 42.5 °C and 120 rpm. The first preculture was prepared in a 250 mL baffled shake flask, by inoculating 25 mL of LB medium containing 20 mg/L tetracycline with 50 µL from a glycerol stock solution of P. putida KT2440 pSynPro8oT_4U_rhlAB or P. putida KT2440 pSynPro8oT_4U(G14A-C25U)_rhlAB respectively. The first preculture was incubated at 120 rpm and 30 °C, grown for 24 h and subsequently 1 mL was used to inoculate 100 mL of SupM medium (seed culture) in 1 L baffled shake flasks as described by14. Each seed culture was grown for 15 h at 120 rpm and 30 °C and used to inoculate the main culture in ModR medium to an OD600 of 0.3 respectively 0.5 for bioreactor cultivations.
Bioreactor cultivation
The bioreactor cultivation using a 2 L tabletop bioreactor (Minifors 4, Infors HT, Bottmingen, Switzerland) was carried out as described previously14 with minor changes. The bioreactor cultivation was carried out in duplicates and as a batch cultivation using 30 g/L glucose. Temperature was switched from 25 °C to 38 °C when reaching an OD600 of around 4 after approximately 9 h of cultivation. The antifoam SB590 (Schill + Seilacher, Hamburg, Germany) was added if needed.
Sample processing
Sample processing was carried out as previously described14. To extract RLs, culture supernatant was first acidified 1:100 (v/v) using 85% phosphoric acid. Subsequently extracted two times with 1.25:1 (v/v) ethyl acetate, then pooled and evaporated in a vacuum centrifuge at 40 °C for 40 min and 10 mbar (RVC 2-25 Cdplus, Martin Christ Gefriertrocknungsanlagen GmbH, Osterode am Harz, Germany).
Quantification of RLs with HPTLC protocol
Detection of RLs was carried out as described previously38 with a modified protocol for HPTLC40. Briefly, RL samples were re-dissolved in acetonitrile after evaporation and derivatized using a 1:1 (v/v) mixture of 135 mM bromphenacylbromid and 67.5 mM tri-ethyl-ammonium/-amin. The derivatization step was performed for 90 min at 1400 rpm and 60 °C as previously described41. Expected RL concentrations exceeding 2 g/L were diluted prior to derivatization. In a next step, derivatized samples were measured on silica gel 60 HPTLC plates with fluorescence marker F254 (Merck, Darmstadt, Germany). Measurements were carried out on a HPTLC system for quantitative analysis (CAMAG Chemie-Erzeugnisse & Adsorptionstechnik AG, Muttenz, Switzerland). Samples application was performed with the Automatic TLC Sampler 4 (ATS 4). Plate development was carried out with the Automatic Developing Chamber 2 (ADC 2), which is equipped with a 20 cm × 10 cm twin-trough chamber. Developed plates were subsequently analyzed with the TLC Scanner 4. A HPTLC imaging and data analysis software (winCATS 1.4.7.2018 software, CAMAG, Muttenz, Switzerland) was used to control the HPTLC-system. Filling speed of the syringe for sample application was set to 15 µL/s and dosage speed to 150 nL/s. The syringe was rinsed with methanol in between sample application. Application start was at 15 mm from the left and 8 mm from the lower edge and band width set to 6 mm. For plate development a mobile phase composed of 30:5:2.5:1 isopropyl acetate:ethanol:water:acetic acid was used. Tank saturation was set to 5 min and subsequent drying was carried out for 5 min with a stream of air. For scanning a deuterium (D2) lamp with slit dimensions of 3 mm × 0.3 mm was used. Plates were scanned at 263 nm at which wavelength bromphenacyl-derivatized RL congeners absorb. For data resolution and scanning speed 1 nm/step and 100 nm/s respectively were chosen.
Enzymatic assay
For quantification of glucose content in the culture supernatant of samples, an enzymatic glucose assay kit (R-Biopharm AG, Darmstadt, Germany) was used, according to the manufacturers’ instructions and scaled down to 96-well format.
Graphical analysis, regression and replicates
The scientific graphing and data analysis software (SigmaPlot, Systat Software Inc., San Jose, CA) was used to carry out regression analysis of measured data. A four-parameter logistic fit for biomass, glucose and RL concentration, was used to illustrate the time course of these variables42. Measurement results and calculated performance parameters (rates and yields) are presented as mean ± standard deviation obtained from data of duplicates from at least two independent biological experiments. To avoid mathematical artifacts caused by low biomass concentrations, specific RL production rates (q) were calculated starting at hour 3.
Parameter optimization, modeling and simulation
MATLAB 2020b (The MathWorks, Natick, MA, USA) was used to perform parameter fitting and simulation for the temperature-dependency model, shown in the results section. The Nelder-Mead numerical algorithm implemented in the embedded function “fminsearch” was used for parameter optimization. Optimization using the functions described above was performed by minimizing the error of simulation data and measured data according to a least-square error function. To demonstrate and describe temperature dependency of specific RL production rates, a mechanistic temperature model (Eq. (1)) was used for a fitting curve30.
Data availability
All obtained data has been included into the manuscript. Please turn to the corresponding author for all other requests.
References
Van Bogaert, I. N. A. et al. Microbial production and application of sophorolipids. Appl. Microbiol. Biotechnol. 76, 23–34 (2007).
Desai, J. D. & Banat, I. M. Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev. 61, 47–64 (1997).
Schramm, L. L., Stasiuk, E. N. & Marangoni, D. G. 2 Surfactants and their applications. Annu. Rep. Prog. Chem. Sect. C Phys. Chem. 99, 3–48 (2003).
Li, Y. et al. Surfactin and fengycin contribute to the protection of a Bacillus subtilis strain against grape downy mildew by both direct effect and defence stimulation. Mol. Plant Pathol. 20, 1037–1050 (2019).
Lourith, N. & Kanlayavattanakul, M. Natural surfactants used in cosmetics: glycolipids. Int. J. Cosmet. Sci. 31, 255–261 (2009).
DeSanto, K. Rhamnolipid Mechanism. US Patent 2011 0123623A1 at https://patentimages.storage.googleapis.com/b3/5c/53/42b14278e189b2/US20110123623A1.pdf (2011).
Henkel, M., Geissler, M., Weggenmann, F. & Hausmann, R. Production of microbial biosurfactants: Status quo of rhamnolipid and surfactin towards large-scale production. Biotechnol. J. 12, 1600561. https://doi.org/10.1002/biot.201600561 (2017).
Najmi, Z., Ebrahimipour, G., Franzetti, A. & Banat, I. M. In situ downstream strategies for cost-effective bio/surfactant recovery. Biotechnol. Appl. Biochem. 65, 523–532 (2018).
Müller, M. M., Hörmann, B., Syldatk, C. & Hausmann, R. Pseudomonas aeruginosa PAO1 as a model for rhamnolipid production in bioreactor systems. Appl. Microbiol. Biotechnol. 87, 167–174 (2010).
Nicas, T. I. & Iglewski, B. H. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can. J. Microbiol. 31, 387–392 (1985).
Cha, M., Lee, N., Kim, M., Kim, M. & Lee, S. Heterologous production of Pseudomonas aeruginosa EMS1 biosurfactant in Pseudomonas putida. Bioresour. Technol. 99, 2192–2199 (2008).
Ochsner, U. A., Reiser, J., Fiechter, A. & Witholt, B. Production of Pseudomonas aeruginosa rhamnolipid biosurfactants in heterologous hosts. Appl. Environ. Microbiol. 61, 3503–3506 (1995).
Beuker, J. et al. Integrated foam fractionation for heterologous rhamnolipid production with recombinant Pseudomonas putida in a bioreactor. AMB Express 6, 11. https://doi.org/10.1186/s13568-016-0183-2 (2016).
Beuker, J. et al. High titer heterologous rhamnolipid production. AMB Express 6, 124. https://doi.org/10.1186/s13568-016-0298-5 (2016).
Shapiro, R. S. & Cowen, L. E. Thermal control of microbial development and virulence: molecular mechanisms of microbial temperature sensing. MBio 3, e002381-2. https://doi.org/10.1128/mBio.00238-12 (2012).
Klinkert, B. & Narberhaus, F. Microbial thermosensors. Cell. Mol. Life Sci. 66, 2661–2676 (2009).
Sengupta, P. & Garrity, P. Sensing temperature. Curr. Biol. 23, R304–R307 (2013).
Altuvia, S., Kornitzer, D., Teff, D. & Oppenheim, A. B. Alternative mRNA structures of the cIII gene of bacteriophage λ determine the rate of its translation initiation. J. Mol. Biol. 210, 265–280 (1989).
Nocker, A. A mRNA-based thermosensor controls expression of rhizobial heat shock genes. Nucleic Acids Res. 29, 4800–4807 (2001).
Johansson, J. et al. An RNA Thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110, 551–561 (2002).
Waldminghaus, T., Gaubig, L. C. & Narberhaus, F. Genome-wide bioinformatic prediction and experimental evaluation of potential RNA thermometers. Mol. Genet. Genomics 278, 555–564 (2007).
Kortmann, J. & Narberhaus, F. Bacterial RNA thermometers: Molecular zippers and switches. Nat. Rev. Microbiol. 10, 255–265 (2012).
Wei, Y. & Murphy, E. R. Temperature-dependent regulation of bacterial gene expression by RNA thermometers. In Nucleic Acids—From Basic Aspects to Laboratory Tools (eds Larramendy, M. L. & Soloneski, S.) 157–181 (IntechOpen, 2016).
Grosso-Becerra, M. V. et al. Regulation of Pseudomonas aeruginosa virulence factors by two novel RNA thermometers. Proc. Natl. Acad. Sci. 111, 15562–15567 (2014).
Noll, P. et al. Evaluating temperature-induced regulation of a ROSE-like RNA-thermometer for heterologous rhamnolipid production in Pseudomonas putida KT2440. AMB Express 9, 154. https://doi.org/10.1186/s13568-019-0883-5 (2019).
Kouse, A. B., Righetti, F., Kortmann, J., Narberhaus, F. & Murphy, E. R. RNA-mediated thermoregulation of iron-acquisition genes in Shigella dysenteriae and pathogenic Escherichia coli. PLoS ONE 8, e63781. https://doi.org/10.1371/journal.pone.0063781 (2013).
Klinkert, B. et al. Thermogenetic tools to monitor temperature-dependent gene expression in bacteria. J. Biotechnol. 160, 55–63 (2012).
Weber, G. G., Kortmann, J., Narberhaus, F. & Klose, K. E. RNA thermometer controls temperature-dependent virulence factor expression in Vibrio cholerae. Proc. Natl. Acad. Sci. 111, 14241–14246 (2014).
Rinnenthal, J., Klinkert, B., Narberhaus, F. & Schwalbe, H. Modulation of the stability of the Salmonella fourU-type RNA thermometer. Nucleic Acids Res. 39, 8258–8270 (2011).
Schoolfield, R. M., Sharpe, P. J. H. & Magnuson, C. E. Non-linear regression of biological temperature-dependent rate models based on absolute reaction-rate theory. J. Theor. Biol. 88, 719–731 (1981).
Arnold, S. et al. Heterologous rhamnolipid biosynthesis by P. putida KT2440 on bio-oil derived small organic acids and fractions. AMB Express 9, 80. https://doi.org/10.1186/s13568-019-0804-7 (2019).
Bator, I., Karmainski, T., Tiso, T. & Blank, L. M. Killing two birds with one stone—Strain engineering facilitates the development of a unique rhamnolipid production process. Front. Bioeng. Biotechnol. 8, 899. https://doi.org/10.3389/fbioe.2020.00899 (2020).
Henkel, M., Müller, M., Hörmann, B., Syldatk, C. & Hausmann, R. Microbial rhamnolipids. In Handbook of Carbohydrate-Modifying Biocatalysts (ed. Grunwald, P.) 697–738 (Pan Stanford Series on Biocatalysis, 2016).
Rinnenthal, J., Klinkert, B., Narberhaus, F. & Schwalbe, H. Direct observation of the temperature-induced melting process of the Salmonella fourU RNA thermometer at base-pair resolution. Nucleic Acids Res. 38, 3834–3847 (2010).
Chowdhury, S., Maris, C., Allain, F. H. T. & Narberhaus, F. Molecular basis for temperature sensing by an RNA thermometer. EMBO J. 25, 2487–2497 (2006).
Chowdhury, S., Ragaz, C., Kreuger, E. & Narberhaus, F. Temperature-controlled structural alterations of an RNA thermometer. J. Biol. Chem. 278, 47915–47921 (2003).
Ross, T. & Nichols, D. S. Ecology of bacteria and fungi in foods | Influence of Temperature. In Encyclopedia of Food Microbiology (eds Batt, C. A. & Tortorello, M. L.) 602–609 (Academic Press, 2014).
Schenk, T., Schuphan, I. & Schmidt, B. High-performance liquid chromatographic determination of the rhamnolipids produced by Pseudomonas aeruginosa. J. Chromatogr. A 693, 7–13 (1995).
Choi, K. H., Kumar, A. & Schweizer, H. P. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: Application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64, 391–397 (2006).
Horlamus, F. et al. One-step bioconversion of hemicellulose polymers to rhamnolipids with Cellvibrio japonicus : A proof-of-concept for a potential host strain in future bioeconomy. GCB Bioenergy 11, 260–268 (2019).
Cooper, M. J. & Anders, M. W. Determination of long chain fatty acids as 2-naphthacyl esters by high pressure liquid chromatography and mass spectrometry. Anal. Chem. 46, 1849–1852 (1974).
Henkel, M. et al. Kinetic modeling of rhamnolipid production by Pseudomonas aeruginosa PAO1 including cell density-dependent regulation. Appl. Microbiol. Biotechnol. 98, 7013–7025 (2014).
Acknowledgements
The authors would like thank Dr. Frank Rosenau and Dr. Andreas Wittgens (Institute for Pharmaceutical Biotechnology, Ulm University, Germany) for constructive scientific discussion and for providing the initial plasmid pSynPro8.
Funding
Open Access funding enabled and organized by Projekt DEAL. This study was partially funded by the Ministry of Science, Research and the Arts of Baden-Wuerttemberg (MWK, funding code: 7533-10-5-186 B). PN and CT are members of the “BBW ForWerts” graduate program and PN received a scholarship within the frame of the Baden-Wuerttemberg Landesgraduiertenfoerderung (LGF) awarded by the Ministry of Science, Research and the Arts (MWK) of Baden-Württemberg, Germany.
Author information
Authors and Affiliations
Contributions
P.N. planned and executed the experiments, collected data, created the graphs and drafted the manuscript. C.T. and S.M. performed part of the experiments and cloning, collected and evaluated corresponding data. L.L. and R.H. contributed to evaluation of the data, design of the experiments and scientific discussion. M.H. substantially contributed to conception, and design of the conducted experiments, interpretation and evaluation of the results and drafting of the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Noll, P., Treinen, C., Müller, S. et al. Exploiting RNA thermometer-driven molecular bioprocess control as a concept for heterologous rhamnolipid production. Sci Rep 11, 14802 (2021). https://doi.org/10.1038/s41598-021-94400-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-94400-4
- Springer Nature Limited
This article is cited by
-
Sugar beet molasses bioconversion into biosurfactant: optimization and comparison with other carbon sources
Biomass Conversion and Biorefinery (2022)