Abstract
Profiling of wild and laboratory tsetse populations using 16S rRNA gene amplicon sequencing allowed us to examine whether the “Wigglesworthia-Sodalis-Wolbachia dogma” operates across species and populations. The most abundant taxa, in wild and laboratory populations, were Wigglesworthia (the primary endosymbiont), Sodalis and Wolbachia as previously characterized. The species richness of the microbiota was greater in wild than laboratory populations. Spiroplasma was identified as a new symbiont exclusively in Glossina fuscipes fuscipes and G. tachinoides, members of the palpalis sub-group, and the infection prevalence in several laboratory and natural populations was surveyed. Multi locus sequencing typing (MLST) analysis identified two strains of tsetse-associated Spiroplasma, present in G. f. fuscipes and G. tachinoides. Spiroplasma density in G. f. fuscipes larva guts was significantly higher than in guts from teneral and 15-day old male and female adults. In gonads of teneral and 15-day old insects, Spiroplasma density was higher in testes than ovaries, and was significantly higher density in live versus prematurely deceased females indicating a potentially mutualistic association. Higher Spiroplasma density in testes than in ovaries was also detected by fluorescent in situ hybridization in G. f. fuscipes.
Similar content being viewed by others
Introduction
Tsetse (Glossina spp.; Diptera: Glossinidae) are viviparous, obligate blood feeding flies found in sub-Saharan Africa. They are the only cyclical vectors of African trypanosomes, responsible for human African trypanosomosis (HAT) and animal African trypanosomosis (AAT)1, 2. Tsetse larvae feed on milk produced in the milk glands of their mothers, pupariating less than an hour after birth. Adult flies of both sexes feed exclusively on largely sterile blood meals.
The microbiota of tsetse flies is of interest because of their unique lifestyle, highlighted by their bilateral transmission, and reproductive strategy, including the elicitation of phenotypes like cytoplasmic incompatibility, as well as its potential for vector and disease control3,4,5. So far, it is known that tsetse flies harbour three main symbiotic microbes: Wigglesworthia, Sodalis and Wolbachia. These three symbionts form the tsetse symbiosis dogma. The primary mutualist symbiont Wigglesworthia provides dietary supplements that are necessary for host fecundity as well as supporting larval development and the maturation process of the adult immune system6,7,8,9. The facultative symbiont Sodalis is present in tsetse populations with a putative role in the ability to transmit trypanosomes10. Finally, Wolbachia has been found in natural populations of tsetse flies with some species exhibiting up to 100% infection rate11, 12, while others have been found to be free of Wolbachia, like G. p. palpalis (Gpp)12. In addition, the Wolbachia strain present in Glossina morsitans morsitans (Gmm) can induce cytoplasmic incompatibility under laboratory conditions11.
There have been a limited number of culture-dependent and culture-independent studies aiming to characterize the microbiota associated with tsetse flies. Using classical microbiological approaches, Geiger and colleagues isolated Acinetobacter, Enterobacter, Enterococcus, Providencia, Sphingobacterium, Chryseobacterium, Lactococcus, Staphylococcus and Pseudomonas species from the guts of field collected Gpp in Cameroon13,14,15,16. They also isolated a new bacterial species, Serratia glossinae, from the midgut of G. palpalis gambiensis (Gpg) collected in Burkina Faso14. A screen for both cultivable and non-cultivable bacteria in whole G. fuscipes fuscipes (Gff) was performed with flies collected in Kenya17. Firmicutes, and particularly members of the Bacillus genus, were identified as the most dominant group while Paenibacillus, Staphylococcus and Exiguobacterium spp. were also isolated at lower density. Gammaproteobacteria were also present, mainly members of the Enterobacteriaceae family like Morganella and Providencia and to a lesser degree Pseudomonas spp., while Burkholderia was the only member of Betaproteobacteria detected in this study17. Using a culture independent approach, beyond the mutualist symbiont Wigglesworthia, only Bacillus and Serratia spp. were additionally detected17. Aksoy and colleagues sampled guts of Ugandan Gff, Gmm, and G. pallidipes (Gpal) tsetse flies, and profiled the microbiota using Illumina amplicon sequencing18. Wigglesworthia was the dominant taxon, while Sodalis was generally detected at low density (<0.05%). However, a small number of flies harboured high levels of Sodalis and Serratia spp. Νon-Wigglesworthia Enterobacteriaceae together with Halomonas spp. were also found at lower abundance at all field sites studied, with some bacterial taxa being unique to a sample site.
Spiroplasma is a genus of wall-less bacteria belonging to the class Mollicutes and it has been associated with diverse plants and arthropods19,20,21,22. Spiroplasma is grouped into three major clades as has been shown by 16S rRNA gene-based as well as multi locus sequence typing (MLST) studies23,24,25,26,27,28,29,30. Spiroplasma exhibits a dual life, with capacity to live intracellularly in a variety of tissues and systemically in the haemolymph31. Spiroplasma has developed a wide range of symbiotic associations, producing diverse effects on insect evolution, ecology, reproduction and sex determination. Spiroplasma has been found to confer protection against a nematode in Drosophila neotestacea 32, against fungi in the pea aphid (Acyrthosiphon pisum)33, and against a parasitoid wasp in Drosophila hydei 34. Spiroplasma can also be pathogenic in plants35, insects36,37,38 and crustaceans39,40,41,42,43,44. Moreover, several species of Spiroplasma have been associated with reproductive alterations such as male killing29, 45,46,47,48. Except Spiroplasma, other reproductive parasites that have been associated with insects are Arsenophonus, Cardinium, and Rickettsia. Arsenophonus is known to establish diverse symbiotic interactions with around 5% of insect species, with the most profound phenotype induced being the son-killer trait49, 50. Cardinium has been found exclusively to Hymenoptera, Hemiptera, Diptera, and Acari and it is known to induce cytoplasmic incompatibility and feminization51, 52. Finally, Rickettsia has been associated with regulating insect growth and immunity to pathogenic fungi53,54,55.
In this study we employed high throughput sequencing of the 16S rRNA gene to unravel the diversity of tsetse associated bacteria in a wider variety of species, field and laboratory populations than any previous tsetse microbiota study. We asked whether the “Wigglesworthia-Sodalis-Wolbachia dogma” applies across species and populations, and whether the microbiota varies between laboratory and field individuals of the same tsetse species. Spiroplasma was identified as a novel symbiont of Gff and G. tachinoides (Gt), and infection prevalence was surveyed in laboratory and natural populations. Quantitative PCR was used to characterize its density in different developmental stages and tissues, and to quantify infection levels in collapsing mass-rearing tsetse fly colonies. Fluorescent in situ hybridization (FISH) was used to localize the newly identified symbiont in tissues including the gonads.
Results
16S rRNA gene amplicon sequencing reveals novel interspecific diversity in natural populations of tsetse flies
Microbial community composition and diversity of thirty-two whole insects from G. medicorum (Gmed), G. morsitans submorsitans (Gms), G. p. gambiensis (Gpg), and G. tachinoides (Gt) collected in Folonzo, Burkina Faso were investigated by 16S rRNA gene amplicon sequencing, producing 5,761,899 reads after quality filtering. These reads were combined with a total of 8,300,515 quality-filtered reads generated from 124 whole guts of Gff, Gmm, Gpal from a previous study18, which used an identical technical approach for amplicon generation and sequencing. Including the data from the above mentioned study18 provided additional Wigglesworthia/co-divergence context to our dataset due to the increased host diversity. This approach enabled us to characterize low-frequency, high-abundance taxa. Whole insect samples from Gpg, and Gt were the most bacterial species-rich samples containing higher numbers of unique OTUs (Supplementary Table 1).
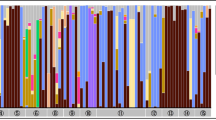
The primary nutritional endosymbiont of tsetse flies Wigglesworthia glossinidia was the most abundant taxon in all samples, and constituted between 71 and 99% of the total community in each individual. Variation in the relative abundance of W. glossinidia was due to the heterogeneous distribution of secondary taxa, which varied in infection frequency and abundance between individuals in both an intra- and inter-specific fashion (Fig. 1a). Secondary taxa included the facultative symbionts S. glossinidius and Wolbachia, alongside Spiroplasma, which have not previously been reported in tsetse flies. The relative abundance of secondary taxa was highly variable (from <0.01% to 28%) depending upon the genus of the bacterium and the species of Glossina (Fig. 1a). This contributed to the variation in bacterial community composition between Glossina species. Clustering by species is illustrated in Fig. 1b, where Principal Component 1 and Principal Component 2 describe 58.53% and 10.84% of the variance respectively. Clustering can be partly attributed to the co-diversification of Wigglesworthia, which is the main component of the community, with its tsetse host56. For this reason, outliers are conspicuous, as is observed with the two individuals infected with Spiroplasma and Rickettsia at 13.15% and 23.72% relative abundance respectively (Fig. 1b).
(a) Relative abundance of Klebsiella, Rickettsia, Sodalis, and Spiroplasma in whole wild tsetse flies. (Gmed: G. medicorum; Gms: G. morsitans submorsitans; Gpg: G. p. gambiensis; Gt: G. tachinoides). (b) Weighted Unifrac Principal Component Analysis of 16 S rRNA gene MiSeq data. Each data point represents an individual tsetse fly and is coloured according to Glossina species. Convergent evolution between the primary endosymbiont Wigglesworthia and its host due to direct vertical transmission generates a tsetse species-clustering pattern that simplifies the detection of emergent taxa such as Spiroplasma and Rickettsia. All gut samples originated from the study by Aksoy et al. in 2014, and whole samples were collected in Burkina Faso. (Gmm: Glossina morsitans morsitans; Gff: G. fuscipes fuscipes; Gmed: G. medicorum; Gms: G. morsitans submorsitans; Gpal: G. pallidipes; Gpg: G. palpalis gambiensis; Gt: G. tachinoides).
Sodalis was found at higher frequency and relative abundance in whole Gmed and Gpal guts (Figs 2 and Supplementary Figure 1). In the other Glossina species it has been detected but to a much lower abundance with a relative abundance of 0.5% or less, with Gms exhibiting the lowest abundance. Wolbachia infections were found infrequently and with low relative abundance of up to 0.04% in any wild sample, with Gmed exhibiting the highest infection prevalence (Supplementary Figure 1).
Sodalis relative abundance in each tsetse species. Boxes denote the interquartile range, the line within the box is the median, and whiskers extend to the most extreme value within 1.5 *interquartile range. Outliers are indicated as circles. Gff: G. fuscipes fuscipes (n = 76); Gmed: G. medicorum (n = 8); Gmm: G. morsitans morsitans (n = 6); Gms: G. morsitans submorsitans (n = 8); Gpal: G. pallidipes (n = 42); Gpg: G. p. gambiensis (n = 8); Gt: G. tachinoides (n = 8).
In addition, several other taxa previously associated with tsetse flies were detected including multiple members of the Enterobacteriaceae, such as Klebsiella, Erwinia, Trabulsiella, Pantoea, and Serratia. These infections occurred at low relative abundance, excluding those with Klebsiella, which was found to be dominant in one Gpg and one Gms whole fly at a relative abundance of 24.3% and 15.3% respectively (Fig. 1a). Amplicon profiling was also able to detect taxa that had not previously been associated with the tsetse fly. Several wild individuals of Gff and Gt, which belong to the palpalis subgroup of the Glossina genus, were infected with Spiroplasma. Relative abundances were generally low (<1%) (Supplementary Table 2), but were found to be as high as 13.2% in one Gt whole fly from Burkina Faso (Fig. 1a).
16S rRNA gene amplicon sequencing of laboratory reared tsetse flies
Gff, Gmm, and Gpal tissue samples from three developmental stages were sequenced, producing 2,445,369 reads after quality filtering. Similarly to wild populations, the three known taxa (Wigglesworthia, Sodalis and Wolbachia) were found in the laboratory flies. However, additional bacterial species were also detected, with members of Flavobacterium, Propiniobacterium, Brevundimonas, Aeromonas, and Rhodospirillales identified in Gmm, Gff, and Gpal. Sequences related to Acinetobacter and Pantoea were identified in Gmm and Gpal. Additionally, sequences related to Streptococcus were found in Gmm, and Gff, while sequences related to Shewanella, and Pedobacter were discovered only in Gmm. Relative abundance was influenced by tissue sample type with gut tissues being enriched for Wigglesworthia while reproductive tissues were characterized by the presence of Wolbachia and Sodalis.
For Gpal, the most bacterial species-rich samples were those associated with gonads of teneral flies while gut samples were less species-rich based on both Chao1 and ACE indices (Supplementary Table 3). Gut samples of teneral males and females displayed lower species richness (Supplementary Table 3). The same trend was observed for Gff. Gut samples of teneral flies exhibited the lowest species diversity and richness indices, which increased over time (Supplementary Table 3). Conversely, gonads presented a higher diversity and richness index in teneral flies and decreased in aged flies. This pattern was not observed in Gmm. Finally, the natural populations exhibited a statistically significant higher species-rich index (Chao1) when compared with the laboratory populations (p < 0.016).
We observed variation in the frequency and relative abundance of Wolbachia in lab populations. The mean relative abundance of Wolbachia was significantly higher in Gmm flies compared with those from the Gff or Gpal populations (ANOVA, p ≤ 0.01) (Supplementary Table 2). This was due to increased relative abundance of Wolbachia in reproductive tissues compared to larval or gut tissues within the Gmm population (ANOVA, p ≤ 0.01).
Bacterial communities were strongly clustered according to the tissue of origin separating the bacterial communities from guts from those from reproductive tissues (Fig. 3a). This factor explained 81.3% of the total variance. Canonical analysis of principal coordinates (CAP), revealed distinct clustering within the gonadal tissue (Fig. 3b). The bacterial communities associated with the gonadal tissue also seem to be statistically affected by the host; Gmm, Gff, and Gpal bacterial communities associated with the reproductive organs clustered separately (Fig. 3b), with Spiroplasma driving the Gff cluster and Wolbachia the Gmm. CAP ordinations were supported by significant traceQ_m’HQ_m statistics (0.9598; p < 0.05).
(a) Principal coordinate analysis (PCοΑ) of bacterial communities based on relative abundances of OTUs with ordinations from laboratory populations of gut, ovaries, testes and larvae. Variance explained by each PCοΑ axis is given in parentheses. (b) Canonical analysis of principal coordinates (CAP) ordinations of gonadal bacterial communities based on relative abundances of OTUs from the laboratory populations. The constrained ordinations show maximized differences among the different Glossina species, (Gmm: Glossina morsitans morsitans, Gff: Glossina fuscipes fuscipes, Gpal: Glossina pallidipes). (traceQ_m’HQ_m (0.9598; p < 0.05)).
Spiroplasma infection status assessed by PCR screening of natural and laboratory tsetse populations
We used PCR-based screening methods to assay for the presence of four insect reproductive parasites: Spiroplasma, Arsenophonous, Rickettsia, and Cardinium, in four Glossina species from the laboratory, Gmm (n = 19), Gff (n = 76), Gpal (n = 20), and Gpg (n = 19) and wild Gff (n = 98). Of the four examined Glossina species, Spiroplasma infections were found only in Gff with an infection ranging from 6.7 to 80% (Table 1), while none of the four tsetse species examined were infected with Arsenophonus, Rickettsia or Cardinium.
To examine the distribution of Spiroplasma, six additional Glossina species were PCR-screened for Spiroplasma infection. Only Gt and Gpp were positive for Spiroplasma, and showed an infection rate of 26.7% and 12.5% respectively (Table 1). The PCR screening for Spiroplasma infection was further extended to 327 historical and contemporary samples from wild and laboratory colonies representing 10 species of tsetse fly (Table 1). Only members of the palpalis subgroup were found infected with Spiroplasma, including Gff, Gpp and Gt, with a prevalence ranging from 6% to 80%. Notably, the prevalence was higher in laboratory colonies than natural populations, and some populations demonstrated a disparity in infection between sexes (Table 1).
Genotyping of Spiroplasma strains
Spiroplasma strains from Gff flies of both sexes from laboratory colonies, a natural population from Uganda and from one natural population of Gt flies from Burkina Faso were genotyped by MLST analysis. Four laboratory and one field sample of Gff harbour Spiroplasma strains with identical sequences for all loci studied (Supplementary Table 4). Interestingly, the Spiroplasma strain present in Gt is distinct from the Gff Spiroplasma strain with sequence polymorphisms detected in all loci examined. Eight polymorphisms were observed in fruR, seven in the region 16S rRNA-23S rRNA-5S rRNA, four in 16S rRNA, three in dnaA, two in ftsZ, and one in rpoB and parE. Both strains belong to the citri clade, which is mostly composed of plant pathogens (Fig. 4 and Supplementary Figures 2–7). Most of the pathogenic Spiroplasma species belong to the Citri clade57 with prominent examples including S. kunkelii that causes the corn stunt disease21, S. phoeniceum that infects periwinkle58, and S. penaei that infects Pacific white shrimp42. The closest relatives of the tsetse Spiroplasma strains are S. insolitum and S. atrichopogonis, which were isolated from a fall flower and a biting midge (Diptera: Ceratopogonidae) respectively59, 60. Neither S. insolitum or S. atrichopogonis have been reported to be pathogenic to plants or midges.
Bayesian inference phylogeny based on the 16S rRNA gene sequence: The topology resulting from the Maximum Likelihood (ML) method was similar. Bayesian posterior probabilities and ML bootstrap values based on 1000 replicates are given at each node, with the posterior probabilities given first followed by the ML bootstrap values (only values >50% are indicated), respectively. Asterisks indicate support values lower than 50%. The Spiroplasma strains present in Gff and Gt are indicated in bold letters. For each Spiroplasma species the GenBank accession number is given to the left of the name.
Spiroplasma density across developmental stages
qPCR was used to assess the density of the Spiroplasma infection in larval guts, and in guts and gonads of males and females collected at two developmental stages: (a) teneral and (b) 15-day-old adults. Spiroplasma infection levels were significantly higher in larval guts compared to the guts of teneral or 15-day-old adults (Fig. 5a). There was no significant difference in the infection levels between testes of teneral and 15-day-old adults (Supplementary Figure 8). In a similar way no significant difference was observed between ovaries of teneral and 15-day-old adults (Supplementary Figure 9). However, there was a significant difference in Spiroplasma infection level between testes and ovaries from teneral flies (Fig. 5b).
Quantification of Spiroplasma titre in terms of the symbiont dnaA gene copies normalized by the tsetse β-tubulin gene. (a) Gff gut from larvae, male and female teneral and 15-day old tsetse flies (n = 3, each sample is a poοl of five) p < 0.005, (b) gonads from male and female teneral tsetse flies (n = 3, each sample is a poοl of five), p < 0.05 (Anova test was performed; statistical significant differences are indicated with an asterisk*).
Spiroplasma density was also examined in a mass-rearing colony where mortality was high and the colony was on the verge of collapse. Examination of live and dead insects indicated that in males Spiroplasma density was similar, whereas in females density was higher in live insects than in those that had recently perished (Fig. 6a and b). When we examined exclusively females carrying a larva, we found that the live females with a larva had a higher titre of Spiroplasma than gravid females that died prematurely (Fig. 6c). The prevalence of Wolbachia, Arsenophonus, Cardinium, and Rickettsia was also examined in whole tsetse flies from the collapsing colony. None of the 34 individuals tested were found to harbour any of the above mentioned symbionts.
Quantification of Spiroplasma titre as Spiroplasma dnaA gene copy number normalized to the tsetse β-tubulin gene. (a) Gff whole insects from healthy/live males and prematurely dead males from the mass-rearing facility in Ethiopia (n = 6), (b) Gff whole insects from healthy/live females and prematurely dead females from the mass-rearing facility in Ethiopia (n = 9), p < 0.05. (c) Gff whole insects from healthy/live females carrying a larvae and prematurely dead females carrying a larva from the mass-rearing facility in Ethiopia (n = 6), p < 0.05. (ANOVA test was performed; statistical significant differences are indicated with an asterisk *).
In situ hybridization of Spiroplasma
Dissected ovaries and testes of teneral adults from a Gff laboratory colony were subjected to FISH using a Spiroplasma specific probe. Spiroplasma detection was sparse and sporadic in ovaries (Fig. 7a), while in testes it was observed at high densities (Fig. 7b).
Localization of Spiroplasma in the male and female reproductive system of Gff. In fluorescent in situ hybridization (FISH) images blue and yellow indicate insect nuclear DNA and Spiroplasma respectively. (a) FISH on dissected ovaries (O), (b) FISH on dissected testes (T) with an inset showing a testis at a higher magnification.
Discussion
The present study showed that the bacterial communities associated with tsetse flies are more complex than previously reported, thus challenging the Wigglesworthia-Sodalis-Wolbachia dogma3, 61, 62. Using 16S rRNA gene-based sequencing approaches, several additional bacterial genera with broad phylogenetic origins were discovered to be associated with the tsetse fly including Klebsiella, Rickettsia and Spiroplasma. The prevalence and infection levels observed in some tsetse species, particularly those of Spiroplasma, were similar to those seen for Sodalis, suggesting that they may play an important role in the biology and ecology of tsetse flies. The question is where these symbionts come from, and what factors determine the structure of the symbiotic communities of tsetse flies.
Previous studies have shown that the microbiota of tsetse flies is characterized by the presence of Wigglesworthia, Sodalis and Wolbachia. All three symbionts are maternally transmitted, while Sodalis can also be transmitted paternally, and colonize during the early juvenile stages: Wigglesworthia and Sodalis through milk gland secretions as larvae, and Wolbachia through the germ line during embryogenesis3, 63, 64. As larvae are intrauterine, the only bacteria that they encounter prior to pupation originate from within the adult female tsetse fly. Due to the obligate requirement of Wigglesworthia, there is high fidelity in vertical transmission from mother to offspring65. This makes it difficult for other bacteria to invade, as microbes occupy many of the available niches within the host from the early stages of development. Conversely, this also means that the tsetse immune system has evolved to accommodate bacteria, which could facilitate colonization by environmental microbes able to exploit deficits in the immune system. Due to the unique biology of tsetse flies, there is only a short time window for colonization between larval deposition and pupation in the soil. In addition, the colonizers would have to survive metamorphosis in order to persist.
Until recently, there was the notion that tsetse flies feed exclusively on blood, which is mostly sterile and therefore should not serve as a source of microbes. There is now evidence that Gpg flies deprived of a blood meal can feed on water or sugar water, and that sugar residues are detectable in wild-caught flies66. Therefore, it is possible that these previously unrecognized feeding habits could be a source of environmental microbes, and could be the origin of the low-frequency high-abundance infections observed in multiple individuals in this study.
Spiroplasma was detected in members of the palpalis sub-group (Gff, Gpp and Gt), whereas Sodalis was significantly more prevalent in Gmed (fusca group). Previous studies have also shown that Sodalis infection is more prevalent in G. brevipalpis (fusca group) than in Gmm and Gpal (both morsitans group)67. However, the relationship of Spiroplasma with the palpalis subgroup seems to be more exclusive than that of Sodalis, since the latter has previously been identified in individuals belonging to all tsetse sub-groups18, 67, 68.
A key approach to detecting invasive taxa is to sample whole insects rather than individual tissues such as the gut, where Wigglesworthia is dominant and will therefore obscure the detection of lower-abundance taxa. A broad phylogenetic range of host species is important to encompass the available diversity, as there seems to be variation between sub-groups, species, and even individuals within the same species.
For example, Rickettsia was discovered at high abundance in just one individual, despite the profiling of hundreds of insects by amplicon and PCR profiling. Rickettsia has been also identified in a previous study using an amplicon sequencing approach18 but also to G. morsitans from Senegal during a PCR screen69.
Spiroplasma infection was more prevalent in laboratory colonies with both males and females harbouring Spiroplasma, whereas in natural populations prevalence was lower and only females were infected. The lack of infection in wild individuals may be due to insufficient sampling effort, or could be due to the differences in population dynamics between laboratory-reared and wild-caught flies. It has been reported, for example, that some symbionts may be present in such low abundances that they are undetectable by conventional PCR screens70. MLST indicated that the strain found in wild Gff from Uganda was identical, based on the loci examined, to that in the colonized flies (originating from the Central African Republic), suggesting the association between Spiroplasma and Gff may be ancient. Although there have been no direct studies on the relative transmission rate of tsetse symbionts in the laboratory and field, paternal transmission during mating can occur for the secondary symbiont Sodalis 64. While this study only detected Spiroplasma infection in palpalis group flies, screening more specimens from the morsitans and fusca groups should provide more detailed information on the dynamics and spread of Spiroplasma infection in natural populations.
Another potential explanation for the absence of Spiroplasma in the morsitans and fusca groups is their frequent infection with Wolbachia 12, 71. In the morsitans group the prevalence of Wolbachia can vary between 9.5 and 100%, while in the fusca group it can vary from 0 to 15.6%12, 71. An existing Wolbachia infection may have led to the development of competitive exclusion with Spiroplasma, though it is not yet clear whether they share an ecological niche within the host, and whether co-occurrence could create evolutionary pressure strong enough to drive competitive exclusion72. In D. melanogaster, coinfections between Wolbachia and Spiroplasma were asymmetrical: Spiroplasma negatively affected the titre of Wolbachia, whereas Wolbachia density did not affect Spiroplasma titre73. Similarly to Spiroplasma in Gff, tissue tropism was observed in D. melanogaster infected with Spiroplasma, with the ovaries showing the highest density73. Competitive inter- and intraspecific microbial interactions have also been observed in mosquito vector species where mutual exclusion between Asaia and Wolbachia has been observed in the reproductive organs while native gut microbiota seems to prevent the vertical transmission of Wolbachia in Anopheles mosquitoes74, 75. Gff has previously been shown to harbor Wolbachia, though prevalence in natural populations is very heterogeneous, with an average infection rate of 44.3%76. Spiroplasma, on the other hand, is found at much lower frequency in natural populations, but is found at higher density per individual when compared with Wolbachia.
MLST analysis indicated that the Spiroplasma strains detected in Gff and Gt populations, albeit different, both belong to the citri clade. Prominent examples of taxa from this clade include S. kunkelii, S. phoeniceum, and S. citri, all of which are plant pathogens21, 58, 77. S. poulsonii, which has been shown to have a protective effect against parasitic wasps in D. melanogaster, is also a member of this clade20.
When examining gut tissues, Spiroplasma titre was highest in larvae, and gradually decreased in both males and females over the course of adulthood. High larval titre indicates vertical transmission from mother to offspring, possibly via the milk gland; a mechanism already exploited by Wigglesworthia and Sodalis. High larval density is an abnormal trait in the context of other insect-associated Spiroplasma species. Multiple strains of Spiroplasma infect a number of species of Drosophila and are able to induce a variety of phenotypes in their insect host ranging from parasitic reproductive manipulators to protective symbionts20, 24, 78. In D. hydei and D. melanogaster, Spiroplasma titre steadily increases during larval and adult development with no differentiation between males and females73, 79. Interestingly, Drosophila male killing Spiroplasma strains exhibit a very high titre in the haemolymph78, a pattern not observed in the Gff Spiroplasma strain (data not shown). In addition, Spiroplasma titre in Gff is much lower than that described for Drosophila male killing strains29, 78. Wolbachia is the only other maternally inherited endosymbiont found in Drosophila, and is also found in tsetse flies. Wolbachia confers density-dependent protection against insect viruses at different developmental stages in several Drosophila species80,81,82,83. Based on the above, it is possible that high Spiroplasma density may also play a role in larval fitness. This warrants further study, as protection against viral or bacterial pathogens during intrauterine larval development would constitute a rare phenotype for a bacterial endosymbiont. Recent studies in D. melanogaster showed that Wolbachia and Spiroplasma can affect immune signalling pathways in the presence of both insect pathogenic and non-pathogenic bacteria84.
Gut infection was maintained into adulthood, particularly in males. This suggests that Spiroplasma is either able to maintain infection during metamorphosis, possibly due to extracellular proliferation73, or that it can rapidly re-colonize upon reformation of the gut. Spiroplasma density was also significantly higher in the testes of teneral males than in the ovaries of teneral females. Localization to the testes suggests that Spiroplasma may be sexually transmitted from males to females, as has already been observed with Sodalis in tsetse flies, and Asaia in Anopheles stephensi 64, 85. The above properties can be exploited in paratransgenic approaches in a similar way to those currently being explored for Sodalis 64, 86 and Asaia 87.
In a collapsing colony of Gff flies, live females had a higher Spiroplasma density than prematurely dead females. This was true of both gravid and non-gravid females, and indicates that Spiroplasma may contribute to adult female fitness. It is therefore possible that Spiroplasma could play a protective role, as has been observed in other facultative strains of Spiroplasma 20, 34, 88 and/or a nutritional role.
Materials and Methods
Insect specimen collection and DNA isolation
All natural populations of Glossina specimens were collected in four countries, Burkina Faso, Uganda, United Republic of Tanzania, and South Africa (Table 1 and Supplementary Table 5). All wild flies were collected using biconical traps and collection intervals were four hours. Upon collection, flies were transferred to the main collection point and were placed in 100% acetone and stored at room temperature. Upon arrival in the lab, DNA was extracted immediately using the CTAB method (Cetyl trimethylammonium bromide)89. Laboratory populations were also analysed in a similar way. Samples of Gff suffering high mortality were collected from the mass rearing facility in Kality, Ethiopia. For a detailed description of the analysis performed see Supplementary Information.
Multiplex Illumina MiSeq Sequencing, data, and statistical analysis
The V4 region of the 16 S rRNA gene was amplified using fusion primers F515 (5′-GTGCCAGCMGCCGCGGTAA-3′), and 805R (5′-GACTACCAGGGTATCTAAT-3′) from individual wild flies of G. medicorum (Gmed), G. m. submorsitans (Gms), G. p. gambiensis (Gpg), and G. tachinoides (Gt) collected in Burkina Faso. Data generated from the wild flies were combined with the data generated from 124 whole guts of Gff, Gmm, Gpal from a previous study18, which used an identical technical approach for amplicon generation and sequencing.
The V3-V4 region of the 16S rRNA gene was amplified using fusion primers U341F (5′-CCTACGGGRSGCAGCAG-3′), and 805 R (5′-GACTACCAGGGTATCTAAT-3′) from pools of tissues from larvae and adults of laboratory populations of Gmm, Gff, and Gpal (Supplementary Table 5).
For a detailed description of the PCR conditions please see Supplementary Information. The gene sequences reported in this study have been deposited in NCBI under Bioproject numbers PRJNA345319, and PRJNA345350-52. Statistical analyses was performed using Unifrac distances, PCoA analyses, CAP, ANOVA and Tukey-Kramer post-hoc tests as described in the Supplementary Information.
PCR screening and Spiroplasma multi locus genotyping
Gmm, Gff, Gpg, and Gpal were assayed for the presence of Spiroplasma, Arsenophonus, Cardinium, and Rickettsia symbionts by PCR. An additional six species of Glossina (G. austeni (Ga), G. brevipalpis (Gb), G. m. centralis (Gmc), Gms, G. p. palpalis (Gpp) and Gt were screened for Spiroplasma only. The primer sequences used to detect each symbiont along with their target genes, product sizes, conditions, and annealing temperatures are listed in the Supplementary Information.
The Spiroplasma strains present in Glossina species were genotyped with a multi-locus sequence typing (MLST) approach using five marker genes (rpoB, parE, dnaA, ftsZ and fruR) and a 4,702 bp region spanning the 16S rRNA-23S rRNA-5S rRNA region. Details of the conditions used are presented in the Supplementary Information. Sequencing was performed as described previously90. All gene sequences generated in this study have been deposited into at GenBank under accession numbers KX159363-KX159393.
Phylogenetic analysis
All nucleotide sequences were manually edited with Geneious 7.1.2. Multiple alignments were generated by MUSCLE91 and ClustalW92 by Geneious 7.1.2, and adjusted by eye. Phylogenetic analyses were conducted for all analysed Spiroplasma sequences (16 S rRNA, rpoB, dnaA, parE, ftsZ and fruR genes, and the region 16 S rRNA-23S rRNA-5S rRNA region) separately by two methods: Bayesian Inference (BI) and Maximum Likelihood (for a detailed description see Supplementary Information).
Quantitative Real Time-PCR and Fluorescent in situ Hybridization (FISH)
Spiroplasma density was quantified by qPCR using the dnaA Spiroplasma specific primers FqdnaA/RqdnaADoud for 35 cycles at 56 °C and normalized to the host β-tubulin gene. Primers and a detailed description used for the qPCR experiments are presented in Supplementary Table 6. qPCR data were analysed using a one-way ANOVA method, as described previously93 using the XLSTAT program.
Gff specimens from the Seibersdorf laboratory colony were used for FISH. Teneral male and female flies were dissected in PBS 2–3 days after eclosion. Dissected tissues were dried on poly-L-lysine-coated glass slides (Sigma, UK) for 20 min at 65 °C and kept at 4 °C until further use. Tissue samples were fixed in freshly prepared 4% paraformaldehyde solution for 30 min at 4 °C. A detailed description of tissue processing and image capture is included in the Supplementary Information.
References
Aksoy, S. Sleeping sickness elimination in sight: Time to celebrate and reflect, but not relax. PLoS Negl. Trop. Dis. 5, e1008 (2011).
Welburn, S. C., Fèvre, E. M., Coleman, P. G., Odiit, M. & Maudlin, I. Sleeping sickness: a tale of two diseases. Trends Parasitol. 17, 19–24 (2001).
Wang, J., Weiss, B. L. & Aksoy, S. Tsetse fly microbiota: form and function. Front. Cell. Infect. Microbiol. 3, 69 (2013).
Abd-Alla, A. M. M. et al. Improving Sterile Insect Technique (SIT) for tsetse flies through research on their symbionts and pathogens. J. Invertebr. Pathol. 112, S2–10 (2013).
Bourtzis, K., Lees, R. S., Hendrichs, J. & Vreysen, M. J. B. More than one rabbit out of the hat: Radiation, transgenic and symbiont-based approaches for sustainable management of mosquito and tsetse fly populations. Acta Trop. 157, 115–130 (2016).
Aksoy, S. Wigglesworthia gen. nov. and Wigglesworthia glossinidia sp. nov., taxa consisting of the mycetocyte-associated, primary endosymbionts of tsetse flies. Int. J. Syst. Bacteriol. 45, 848–851 (1995).
Weiss, B. & Aksoy, S. Microbiome influences on insect host vector competence. Trends Parasitol. 27, 514–522 (2011).
Weiss, B. L., Wang, J. & Aksoy, S. Tsetse immune system maturation requires the presence of obligate symbionts in larvae. PLoS Biol. 9, e1000619 (2011).
Weiss, B. L., Maltz, M. & Aksoy, S. Obligate symbionts activate immune system development in the tsetse fly. J. Immunol. 188, 3395–3403 (2012).
Farikou, O. et al. Tripartite interactions between tsetse flies, Sodalis glossinidius and trypanosomes - An epidemiological approach in two historical human African trypanosomiasis foci in Cameroon. Infect. Genet. Evol. 10, 115–121 (2010).
Alam, U. et al. Wolbachia symbiont infections induce strong cytoplasmic incompatibility in the tsetse fly Glossina morsitans. PLoS Pathog. 7, e1002415 (2011).
Doudoumis, V. et al. Detection and characterization of Wolbachia infections in laboratory and natural populations of different species of tsetse flies (genus Glossina). BMC Microbiol. 12, 1–13 (2012).
Geiger, A. et al. First isolation of Enterobacter, Enterococcus, and Acinetobacter spp. as inhabitants of the tsetse fly (Glossina palpalis palpalis) midgut. Inf. Genet. Evol 9, 1364–1370 (2009).
Geiger, A., Fardeau, M.-L., Falsen, E., Ollivier, B. & Cuny, G. Serratia glossinae sp. nov., isolated from the midgut of the tsetse fly Glossina palpalis gambiensis. Int. J. Syst. Evol. Microbiol. 60, 1261–1265 (2010).
Geiger, A. et al. Bacterial diversity associated with populations of Glossina spp. from Cameroon and distribution within the Campo sleeping sickness focus. Microb. Ecol. 62, 632–643 (2011).
Geiger, A., Fardeau, M.-L., Njiokou, F. & Ollivier, B. Glossina spp. gut bacterial flora and their putative role in fly-hosted trypanosome development. Front. Cell. Infect. Microbiol. 3, 34 (2013).
Lindh, J. M. & Lehane, M. J. The tsetse fly Glossina fuscipes fuscipes (Diptera: Glossina) harbours a surprising diversity of bacteria other than symbionts. Antonie van Leeuwenhoek 99, 711–720 (2011).
Aksoy, E. et al. Analysis of multiple tsetse fly populations in Uganda reveals limited diversity and species-specific gut microbiota. Appl. Environ. Microbiol. 80, 4301–4312 (2014).
Bastian, F. O., Elzer, P. H. & Wu, X. Spiroplasma spp. biofilm formation is instrumental for their role in the pathogenesis of plant, insect and animal diseases. Exp. Mol. Pathol. 93, 116–128 (2012).
Paredes, J. C. et al. Genome sequence of the Drosophila melanogaster male-killing Spiroplasma strain MSRO endosymbiont. mBio 6, e02437–14 (2015).
Whitcomb, R. F. et al. Spiroplasma kunkelii sp. nov.: characterization of the etiological agent of corn stunt disease. Int. J. Syst. Evol. Microbiol. 36, 170–178 (1986).
Gasparich, G. E. The genus Spiroplasma and its non-helical descendants: phylogenetic classification, correlation with phenotype and roots of the Mycoplasma mycoides clade. Int. J. Syst. Evol. Microbiol. 54, 893–918 (2004).
Nai, Y.-S. et al. A new spiroplasma isolate from the field cricket (Gryllus bimaculatus) in Taiwan. J. Invertebr. Pathol. 120, 4–8 (2014).
Haselkorn, T. S., Cockburn, S. N., Hamilton, P. T., Perlman, S. J. & Jaenike, J. Infectious adaptation: potential host range of a defensive endosymbiont in Drosophila. Evolution 67, 934–945 (2013).
Heres, A. & Lightner, D. V. Phylogenetic analysis of the pathogenic bacteria Spiroplasma penaei based on multilocus sequence analysis. J. Invertebr. Pathol. 103, 30–35 (2010).
Haselkorn, T. S., Markow, T. A. & Moran, N. A. Multiple introductions of the Spiroplasma bacterial endosymbiont into Drosophila. Mol. Ecol 18, 1294–1305 (2009).
Bi, K., Huang, H., Gu, W., Wang, J. & Wang, W. Phylogenetic analysis of Spiroplasmas from three freshwater crustaceans (Eriocheir sinensis, Procambarus clarkia and Penaeus vannamei) in China. J. Invertebr. Pathol. 99, 57–65 (2008).
Montenegro, H., Solferini, V. N., Klaczko, L. B. & Hurst, G. D. D. Male-killing Spiroplasma naturally infecting Drosophila melanogaster. Insect. Mol. Biol 14, 281–287 (2005).
Anbutsu, H. & Fukatsu, T. Population dynamics of male-killing and non-male-killing Spiroplasmas in Drosophila melanogaster. Appl. Environ. Microbiol. 69, 1428–1434 (2003).
Fukatsu, T., Tsuchida, T., Nikoh, N. & Koga, R. Spiroplasma Symbiont of the Pea Aphid, Acyrthosiphon pisum (Insecta: Homoptera). Appl. Environ. Microbiol. 67, 1284–1291 (2001).
Schwarz, R. S. et al. Honey bee colonies act as reservoirs for two Spiroplasma facultative symbionts and incur complex, multiyear infection dynamics. Microbiol. Open 3, 341–355 (2014).
Jaenike, J., Unckless, R., Cockburn, S. N., Boelio, L. M. & Perlman, S. J. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329, 212–215 (2010).
Łukasik, P., Guo, H., van Asch, M., Ferrari, J. & Godfray, H. C. J. Protection against a fungal pathogen conferred by the aphid facultative endosymbionts Rickettsia and Spiroplasma is expressed in multiple host genotypes and species and is not influenced by co-infection with another symbiont. J. Evol. Biol. 26, 2654–2661 (2013).
Xie, J., Vilchez, I. & Mateos, M. Spiroplasma bacteria enhance survival of Drosophila hydei attacked by the parasitic wasp Leptopilina heterotoma. PLoS ONE 5, e12149 (2010).
Gasparich, G. E. Spiroplasmas and phytoplasmas: Microbes associated with plant hosts. Biologicals 38, 193–203 (2010).
Mouches, C., Bové, J. M. & Albisetti, J. Pathogenicity of Spiroplasma apis and other spiroplasmas for honey-bees in southwestern France. Ann. Microbiol. (Paris) 135A, 151–155 (1984).
Clark, T. B. et al. Spiroplasma melliferum, a new species from the honeybee (Apis mellifera). Int. J. Syst. Evol. Microbiol. 35, 296–308 (1985).
Humphery-Smith, I., Grulet, O., Le, Goff, F. & Chastel, C. Spiroplasma (Mollicutes: Spiroplasmataceae) pathogenic for Aedes aegypti and Anopheles stephensi (Diptera: Culicidae). J. Med. Entomol. 28, 219–222 (1991).
Wang, W. et al. A spiroplasma associated with tremor disease in the Chinese mitten crab (Eriocheir sinensis). Microbiology 150, 3035–3040 (2004).
Wang, W. et al. A novel Spiroplasma pathogen causing systemic infection in the crayfish Procambarus clarkii (Crustacea: Decapod), in China. FEMS Microbiol. Lett. 249, 131–137 (2005).
Wang, W. et al. Spiroplasma eriocheiris sp. nov., associated with mortality in the Chinese mitten crab, Eriocheir sinensis. Int. J. Syst. Evol. Microbiol. 61, 703–708 (2011).
Nunan, L. M., Lightner, D. V., Oduori, M. A. & Gasparich, G. E. Spiroplasma penaei sp. nov., associated with mortalities in Penaeus vannamei, Pacific white shrimp. Int. J. Syst. Evol. Microbiol. 55, 2317–2322 (2005).
Liang, T. et al. Identification and isolation of a spiroplasma pathogen from diseased freshwater prawns, Macrobrachium rosenbergii, in China: A new freshwater crustacean host. Aquaculture 318, 1–6 (2011).
Ding, Z. et al. Classification of circulating hemocytes from the red swamp crayfish Procambarus clarkii and their susceptibility to the novel pathogen Spiroplasma eriocheiris in vitro. Aquaculture 356–357, 371–380 (2012).
Williamson, D. L. et al. Spiroplasma poulsonii sp. nov., a new species associated with male-lethality in Drosophila willistoni, a neotropical species of fruit fly. Int. J. Syst. Bacteriol. 49(Pt 2), 611–618 (1999).
Tabata, J. et al. Male Killing and incomplete inheritance of a novel Spiroplasma in the moth Ostrinia zaguliaevi. Microb. Ecol. 61, 254–263 (2011).
Hurst, G. D. & Jiggins, F. M. Male-killing bacteria in insects: mechanisms, incidence, and implications. Emerg. Infect. Dis. 6, 329–336 (2000).
Hayashi, M., Watanabe, M., Yukuhiro, F., Nomura, M. & Kageyama, D. A nightmare for males? A maternally transmitted male-killing bacterium and strong female bias in a green lacewing population. PLoS ONE 11, e0155794 (2016).
Duron, O. et al. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 6, 27 (2008).
Ferree, P. M., Avery, A., Azpurua, J., Wilkes, T. & Werren, J. H. A bacterium targets maternally inherited centrosomes to kill males in. Nasonia. Curr. Biol. 18, 1409–1414 (2008).
Zhang, Y. K., Chen, Y. T., Yang, K. & Hong, X.-Y. A review of prevalence and phylogeny of the bacterial symbiont Cardinium in mites (subclass: Acari). Systematic and Applied Acarology 21, 978–990 (2016).
Giorgini, M., Monti, M. M., Caprio, E., Stouthamer, R. & Hunter, M. S. Feminization and the collapse of haplodiploidy in an asexual parasitoid wasp harboring the bacterial symbiont Cardinium. Heredity (Edinb) 102, 365–371 (2009).
Hendry, T. A., Hunter, M. S. & Baltrus, D. A. The facultative symbiont Rickettsia protects an invasive whitefly against entomopathogenic Pseudomonas syringae strains. Appl. Environ. Microbiol. 80, 7161–7168 (2014).
Liu, Y., Shen, D., Zhou, F., Wang, G. & An, C. Identification of immunity-related genes in Ostrinia furnacalis against entomopathogenic fungi by RNA-seq analysis. PLoS ONE 9, e86436 (2014).
Himler, A. G. et al. Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science 332, 254–256 (2011).
Chen, X., Li, S. & Aksoy, S. Concordant evolution of a symbiont with its host insect species: molecular phylogeny of genus Glossina and its bacteriome-associated endosymbiont, Wigglesworthia glossinidia. J. Mol. Evol. 48, 49–58 (1999).
Lo, W.-S., Chen, L.-L., Chung, W.-C., Gasparich, G. E. & Kuo, C.-H. Comparative genome analysis of Spiroplasma melliferum IPMB4A, a honeybee-associated bacterium. BMC Genomics 14, 22 (2013).
Saillard, C. et al. Spiroplasma phoeniceum sp. nov., a new plant-pathogenic species from Syria. Int. J. Syst. Evol. Microbiol. 37, 106–115 (1987).
Hackett, K. J. et al. Spiroplasma insolitum sp. nov., a new species of group I Spiroplasma with an unusual DNA base composition. Int. J. Syst. Evol. Microbiol. 43, 272–277 (1993).
Koerber, R. T., Gasparich, G. E., Frana, M. F. & Grogan, W. L. Spiroplasma atrichopogonis sp. nov., from a ceratopogonid biting midge. Int. J. Syst. Evol. Microbiol. 55, 289–292 (2005).
Geiger, A., Ponton, F. & Simo, G. Adult blood-feeding tsetse flies, trypanosomes, microbiota and the fluctuating environment in sub-Saharan Africa. ISME J. 9, 1496–1507 (2015).
Wamwiri, F. N. et al. Wolbachia, Sodalis and trypanosome co-infections in natural populations of Glossina austeni and Glossina pallidipes. Parasit. Vectors 6, 232 (2013).
Balmand, S., Lohs, C., Aksoy, S. & Heddi, A. Tissue distribution and transmission routes for the tsetse fly endosymbionts. J. Invertebr. Pathol. 112, S116–S122 (2013).
Vooght, L. D., Caljon, G., Hees, J. V. & Abbeele, J. V. D. Paternal transmission of a secondary symbiont during mating in the viviparous tsetse fly. Mol. Biol. Evol. 32, 1977–1980 (2015).
Attardo, G. M. et al. Analysis of milk gland structure and function in Glossina morsitans: Milk protein production, symbiont populations and fecundity. J. Insect Physiol. 54, 1236–1242 (2008).
Solano, P. et al. Do tsetse flies only feed on blood? Infect. Genet. Evol. 36, 184–189 (2015).
Dennis, J. W. Sodalis glossinidius prevalence and trypanosome presence in tsetse from Luambe National Park, Zambia. Parasit. Vectors 7, 378 (2014).
Geiger, A., Cuny, G. & Frutos, R. Two Tsetse Fly Species, Glossina palpalis gambiensis and Glossina morsitans morsitans, carry genetically distinct populations of the secondary symbiont Sodalis glossinidius. Appl. Environ. Microbiol. 71, 8941–8943 (2005).
Mediannikov, O., Audoly, G., Diatta, G., Trape, J.-F. & Raoult, D. New Rickettsia sp. in tsetse flies from Senegal. Comp. Immunol. Microbiol. Infect. Dis. 35, 145–150 (2012).
Schneider, D. I., Klasson, L., Lind, A. E. & Miller, W. J. More than fishing in the dark: PCR of a dispersed sequence produces simple but ultrasensitive Wolbachia detection. BMC Microbiol. 14, 121 (2014).
Doudoumis, V. et al. Tsetse-Wolbachia symbiosis: Comes of age and has great potential for pest and disease control. J. Invertebr. Pathol. 112, S94–103 (2013).
Charlat, S. et al. Competing selfish genetic elements in the butterfly Hypolimnas bolina. Curr. Biol. 16, 2453–2458 (2006).
Goto, S., Anbutsu, H. & Fukatsu, T. Asymmetrical interactions between Wolbachia and Spiroplasma endosymbionts coexisting in the same insect host. Appl. Environ. Microbiol. 72, 4805–4810 (2006).
Rossi, P. et al. Mutual exclusion of Asaia and Wolbachia in the reproductive organs of mosquito vectors. Parasit. Vectors 8, 278 (2015).
Hughes, G. L. et al. Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc. Natl. Acad. Sci. USA 111, 12498–12503 (2014).
Alam, U. et al. Implications of microfauna-host interactions for trypanosome transmission dynamics in Glossina fuscipes fuscipes in Uganda. Appl. Environ. Microbiol. 78, 4627–4637 (2012).
Saglio, P. et al. Spiroplasma citri gen. and sp. n.: A Mycoplasma-Like organism associated with ‘Stubborn’ disease of citrus. Int. J. Syst. Evol. Microbiol. 23, 191–204 (1973).
Anbutsu, H. & Fukatsu, T. Spiroplasma as a model insect endosymbiont. Environ. Microbiol. Rep. 3, 144–153 (2011).
Kageyama, D. et al. Prevalence of a non-male-killing spiroplasma in natural populations of Drosophila hydei. Appl. Environ. Microbiol. 72, 6667–6673 (2006).
Teixeira, L., Ferreira, Á. & Ashburner, M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 6, e-1000002 (2008).
Hedges, L. M., Brownlie, J. C., O’Neill, S. L. & Johnson, K. N. Wolbachia and virus protection in insects. Science 322, 702–702 (2008).
Martinez, J. et al. Symbionts commonly provide broad spectrum resistance to viruses in insects: A comparative analysis of Wolbachia strains. PLoS Pathog. 10, e-1004369 (2014).
Stevanovic, A. L., Arnold, P. A. & Johnson, K. N. Wolbachia-mediated antiviral protection in Drosophila larvae and adults following oral infection. Appl. Environ. Microbiol. 81, 8215–8223 (2015).
Shokal, U. et al. Effects of co-occurring Wolbachia and Spiroplasma endosymbionts on the Drosophila immune response against insect pathogenic and non-pathogenic bacteria. BMC Microbiol. 16, 16 (2016).
Damiani, C. et al. Paternal transmission of symbiotic bacteria in malaria vectors. Curr. Biol. 18, R1087–R1088 (2008).
Van den Abbeele, J. et al. Enhancing tsetse fly refractoriness to trypanosome infection – A new IAEA coordinated research project. J. Invertebr. Pathol. 112(Supplement 1), S142–S147 (2013).
Mancini, M. V. et al. Paratransgenesis to control malaria vectors: a semi-field pilot study. Parasit. Vectors 9, 140 (2016).
Xie, J., Butler, S., Sanchez, G. & Mateos, M. Male killing Spiroplasma protects Drosophila melanogaster against two parasitoid wasps. Heredity 112, 399–408 (2014).
Doyle, J. J. & Doyle, J. L. Isolation of plant DNA from fresh tissue. Focus 12, 13–15 (1990).
Tsiamis, G. et al. Olive-mill wastewater bacterial communities display a cultivar specific profile. Curr. Microbiol. 64, 197–203 (2012).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Wang, J., Brelsfoard, C., Wu, Y. & Aksoy, S. Intercommunity effects on microbiome and GpSGHV density regulation in tsetse flies. Journal of Invertebrate Pathology 112, S32–S39 (2013).
Acknowledgements
This research was partially supported by: (a) FAO/IAEA Research Contract No 17752, (b) GRST grant MIS423722 “Detection and characterization of the role of symbiotic bacteria in tsetse flies under mass-rearing conditions”, (c) by the Slovak Research and Development Agency under the contract No. APVV-15-0604, and (d) ACD IP BBSRC projects BB/J017698/1 and BB/K501773/1.
Author information
Authors and Affiliations
Contributions
Conceived and design the study: A.D., K.B., G.T. Conducted the experiments and analysed the results: D.V., B.F., A.S., A.A., D.A., G.I., S.P., R.B., T.P., M.S., P.A., A.b.A., G.T. Drafted the manuscript: D.V., B.F., D.A., K.B., G.T. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Doudoumis, V., Blow, F., Saridaki, A. et al. Challenging the Wigglesworthia, Sodalis, Wolbachia symbiosis dogma in tsetse flies: Spiroplasma is present in both laboratory and natural populations. Sci Rep 7, 4699 (2017). https://doi.org/10.1038/s41598-017-04740-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-04740-3
- Springer Nature Limited