Abstract
Background
Tsetse flies are the biological vectors of African trypanosomes, the causative agents of sleeping sickness in humans and nagana in animals. The tsetse endosymbiont Sodalis glossinidius has been suggested to play a role in tsetse susceptibility to infection. Here we investigate the prevalence of African trypanosomes within tsetse from the Luambe National Park, Zambia and if there is an association between S. glossinidius and presence of trypanosomes within the tsetse examined.
Methods
Tsetse representing three species (Glossina brevipalpis, Glossina morsitans morsitans and Glossina pallidipes), were sampled from Luambe National Park, Zambia. Following DNA extraction, PCR was used to examine the tsetse for presence of trypanosomes and the secondary endosymbiont S. glossinidius.
Results
S. glossinidius infection rates varied significantly between tsetse species, with G. brevipalpis (93.7%) showing the highest levels of infection followed by G. m. morsitans (17.5%) and G. pallidipes (1.4%). ITS-PCR detected a wide variety of trypanosomes within the tsetse that were analysed. Significant differences were found in terms of trypanosome presence between the three tsetse species. A high proportion of G. m. morsitans were shown to carry T. brucei s.l. DNA (73.7%) and of these around 50% were positive for Trypanosoma brucei rhodesiense. T. vivax, T. godfreyi, T. simiae, T. simiae Tsavo and T. congolense were also detected. No association was found between the occurrence of S. glossinidius and the presence of trypanosome DNA in any of the three tsetse species tested.
Conclusion
The current work shows that T. b. rhodesiense was circulating in Luambe National Park, representing a risk for people living in the park or surrounding area and for tourists visiting the park. The differences in trypanosome DNA presence observed between the different tsetse species tested may indicate host feeding preferences, as the PCR will not discriminate between a fly with an active/resident infection compared to a refractory fly that has fed on an infected animal. This makes it difficult to establish if S. glossinidius may play a role in the susceptibility of tsetse flies to trypanosome infection.
Similar content being viewed by others
Background
Tsetse flies are the biological vector of African trypanosomes, the causative agents of sleeping sickness in humans and nagana in animals. They host three symbiotic bacteria, Wigglesworthia glossinidia, Sodalis glossinidius and Wolbachia [1]. W. glossinidia is the primary symbiont and is found in all tsetse flies, its main role is in the provision of B vitamins that are absent in the tsetse diet of vertebrate blood [2]. The other two bacteria are generally found in laboratory tsetse, however, their prevalence within wild tsetse populations is variable. S. glossinidius, originally classified as a rickettsia-like organism [3], has no clear role within the tsetse but is thought to play a part in susceptibility to trypanosome infection [4–6]. Wolbachia has recently been shown to be involved in cytoplasmic incompatibility in tsetse [7].
There have been many studies over the years investigating natural rates of trypanosome infection in tsetse flies. In the past, microscopy was used to assess infection. Flies were commonly dissected using the method described by Lloyd and Johnson [8], where infection was ascertained by position of trypanosomes in the tsetse. However, infection rates in wild flies were often low, for instance Okoth and Kapaata [9] found less than 1% of flies from Uganda with a salivary gland infection. More recently PCR has been used to examine tsetse flies for trypanosome infections. This has the advantage of being able to identify trypanosomes to species/sub species level and also identify mixed infections [10]. However, PCR may over estimate tsetse infection rates, as it will not discriminate between an active infection of the fly or simply trypanosomes that have been imbibed with a bloodmeal [11].
Just over thirty years ago, Maudlin [12] established that tsetse susceptibility to midgut infection by trypanosomes was a maternally inherited characteristic. Later work associated susceptibility with the presence of S. glossinidius and it was suggested that the action of a chitinase, produced by S. glossinidius, broke down chitin during the pupation period, causing the build up of N-acetyl-D-glucosamine [4]. This compound was thought to inhibit tsetse lectins that killed incoming trypanosomes when tsetse fed on an infected host. More recently alternative trypanocidal mechanisms have been put forward including antimicrobial peptides [13] or oxidative stress [14]. A role for W. glossinidia in tsetse immunity has also been suggested, where older flies devoid of the primary symbiont show greater susceptibility to trypanosome infection than flies with the symbiont [15].
Although there have been several field studies of tsetse flies over the past thirty years looking at trypanosome infections, very little is known about the prevalence of secondary symbionts within tsetse, with very few species being investigated. Several techniques have been used to screen tsetse flies for S. glossinidius infection, these include electron microscopy [16, 17], dot blots with radioactive probes [18, 19] and more recently PCR [20–23]. In studies using electron microscopy [16] and PCR [21, 24], all laboratory tsetse tested positive for S. glossinidius. A selective advantage has been suggested for S. glossinidius presence based on increased puparial survival in laboratory tsetse [25]. G. m. morsitans from the University of Bristol colony showed a prevalence of 80% for S. glossinidius while samples taken from Zimbabwe, where these flies originated showed a prevalence of 20% [25].
One of the first studies to investigate the prevalence of S. glossinidius in wild tsetse flies was undertaken in Liberia [19]. Using radioactive dot blots to diagnose infection, S. glossinidius prevalence was found to be 85% (95% CI = 66.3-95.8%), 31% (95% CI = 11.0%-58.7%) and 9.3% (95% CI = 6.9%-12.2%) in Glossina nigrofusca, Glossina pallicera and Glossina palpalis palpalis respectively. An association was shown between S. glossinidius presence and trypanosome infection (diagnosed by microscopy) in G. p. palpalis with symbiont positive flies six times more likely to be trypanosome positive. In more recent years several authors have tried to show an association between S. glossinidius presence in wild tsetse flies and trypanosome presence using molecular techniques. Farikou et al. [22] showed a link between the presence of S. glossinidius and trypanosome infection in wild flies in Cameroon. PCR analysis showed that 54.9% (95% CI = 50.2%-59.6%) of flies tested were positive for S. glossinidius infection, of these 75% were also positive for trypanosome infection. This association between trypanosome susceptibility and S. glossinidus was also shown in Glossina pallidipes in Kenya, however, there were very few flies which were both S. glossinidius and trypanosome infected so no definitive conclusion could be made [23].
In the current work we have used PCR to detect S. glossinidius and an ITS-PCR [26] to detect trypanosomes in tsetse from Zambia. The latter primer set detects the following trypanosomes, Trypanozoon (T. brucei brucei. T. brucei gambiense, T. brucei rhodesiense T. evansi and T. equiperdum), T. congolense savannah, T. congolense forest, T. congolense kilifi, T. simiae, T. simiae Tsavo, T. godfreyi (all Nannomonas) and T. vivax (Duttonella). The majority of these trypanosomes establish within the midgut of the tsetse before moving to either the salivary glands or proboscis where they mature into the mammalian infective stage. The exceptions are T. vivax where the lifecycle is restricted to the mouthparts of the fly, T. evansi, which is mechanically transmitted and T. equiperdum, which is sexually transmitted.
Methods
Tsetse fly collection
Tsetse flies were surveyed using a stratified random sampling design. Using a land cover classification of Luambe National Park, Zambia and the surrounding area [27], five vegetation types considered the most suitable tsetse habitat were identified (riverine woodland, thicket, mopane woodland, mopane scrub woodland and Combretum-Terminalia woodland). A 500 m grid was placed over the study area and squares selected with a percentage composition of 55% or above for the selected vegetation classes. For mopane scrub woodland and riverine woodland lower thresholds of 50% and 10% respectively were used due to lack of larger areas of homogenous woodland for these classes. Ten trap sites, two for each vegetation type, were then randomly selected from the candidate list using a random number generator. The central point for the selected 500 m square was used to locate the trap site on the ground and assess its suitability. One trap was placed in the most suitable location as close as possible to the central point for each grid square. A second trap was then placed 200 m from the first in a direction that was perpendicular to the prevailing wind direction so that both traps were within the same grid square.
Standard Epsilon traps (Bonar Industries (Pvt) Ltd, Zimbabwe) were used to sample the tsetse flies. Traps were set once daily at 0600 and flies collected the following day at the same time. Traps were generally set without odour baits, but baits were used on each trap for a period during the study (two sachets (5 cm × 5 cm, 150 μm thick) containing 3-n-propylphenol, octenol and 4 methylphenol in the ratio 1:6:12 and an open 500 ml bottle containing methylethylketone (MEK) at the entrance to the traps). The study ran from June to October, 2006 with sampling occurring during one week of each month during that period.
During October, when ambient temperatures were at their greatest, three artificial refuges were created to sample G. brevipalpis. They were sited in well-shaded areas of riverine woodland within 50 m of the Luangwa River. A 200 litre metal drum with one open end was spilt in half longitudinally and buried in soil and leaf litter as described by Vale [28]. Flies were sampled over several days towards the end of the study period by placing a net over the open end of the refuge during the hottest period of the day. The tsetse sampled consisted of 419 G. pallidipes (Combretum-Terminalia woodland: 67; mopane scrub: 72; mopane woodland: 73; artificial refuge: 44; riverine woodland: 74), 137 G. m. morsitans (Combretum-Terminalia woodland: 38; mopane scrub: 28; mopane woodland: 29; riverine woodland: 21; thicket: 21) and 55 G. brevipalpis (artificial refuge: 54; riverine woodland: 1). Flies were stored individually in acetone and transported to Edinburgh University where they were stored at -20°C.
DNA extraction and PCR
Frozen tsetse flies were thawed at room temperature. Flies were washed using 5% sodium hypochlorite and phosphate buffered saline (g/l NaCl 8.0, KCl 0.2, Na2HPO4 1.15, KH2PO4 0.2, pH 7.3) (OXOID) to remove any external sources of DNA. Flies were then crushed with pestles in individual 1.5 ml microcentrifuge tubes; pestles were thoroughly cleaned with bleach and then autoclaved between uses to prevent contamination. DNA was extracted from the tissue using the QIAGEN® DNeasy® blood and tissue kit following the manufacture’s instructions.
To test the integrity of the samples, all DNA extracts were tested for amplification in 25 μl reactions using 1 μl of template DNA for W. glossinidia DNA using primers targeting the 16S [29] or fliC gene [30]. Three G. pallidipes failed to show amplification for W. glossinidia DNA and were removed from the study set. All other tsetse extracts showed positive amplification for W. glossinidia and were subjected to PCR for S. glossinidius, targeting the GroEL gene [31] and African trypanosomes targeting the ITS region, which is present at around 200 copies per cell [26]. For the latter, the species of trypanosome was determined by the size of the band produced, as described by Njiru et al. [26]. Flies testing positive for T. brucei s.l. DNA using the ITS-PCR were further evaluated to examine if they were T. b. rhodesiense by using the PLC-SRA-PCR described by Picozzi et al. [32]. This multiplex PCR tests for both the single copy SRA gene (found in T. b. rhodesiense) and the single copy PLC gene (common to all Trypanozoon). If a sample is SRA positive then it is diagnosed as T. b. rhodesiense, a fly positive for PLC but not SRA suggests that the sample is not T. b. rhodesiense, however, there is enough DNA to amplify a single copy trypanosome gene if it were present. Where there is no amplification (either PLC or SRA) then there is not enough DNA to amplify a single copy gene, as such it is not possible to diagnose the infection, in this case these flies were discounted when determining the prevalence of T. b. rhodesiense. On occasion an artefact will occur that is indicative of a VSG gene, this occurrence will not speciate the trypanosome infection, but is indicative of the presence of parasitic material. Therefore, flies that returned a VSG gene amplification in isolation were discounted from further calculations regarding the prevalence of T. b. rhodesiense.
All PCRs were repeated at least twice, and in all PCRs a positive control (DNA known to test positive for that specific PCR) and a negative control (distilled water) were run. A DNA engine DYAD™ Peltier thermal cycler was used to run the PCRs, and PCR products were separated by electrophoresis on 1.5% molecular grade agarose (BIOLINE) gel stained with 0.04_ μl/ml of GelRed nucleic acid stain (BIOTIUM). Separated products were then viewed under ultraviolet light in a transilluminator (BIO-RAD), and band sizes analysed compared to exACTGene low range plus DNA ladder (Fisher Scientific International Inc.). No T. congolense kilifi was detected, and as it was not possible to differentiate between T. congolense savannah and T. congolense forest, any band of 700 bp was classified as T. congolense.
Statistical analysis
Fisher’s exact test was used for all analyses in the study. Firstly, the prevalence of S. glossinidius was examined for differences between the three species of tsetse studied in the current work. Secondly, the data was examined to assess if detection of S. glossinidius was associated with trypanosome presence. Finally, the prevalence of trypanosomes was examined to look for statistical differences between and within tsetse species. As T. vivax completes its lifecycle exclusively within the proboscis of the fly it was excluded from the analysis. All analysis was carried out in WinPepi (version 11.32) and statistical significance was accepted at the 95% confidence level throughout.
Ethical statement
All activities in protected areas were fully approved by the Zambian Wildlife Authority (permit numbers 316295 and 323947) as part of a wider study investigating trypanosomes in the Luangwa Valley [33].
Results
Prevalence of S. glossinidius in wild fly populations from Zambia
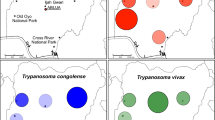
The prevalence of S. glossinidius in the various species of tsetse is shown in Figure 1. The prevalence of S. glossinidius in G. brevipalpis was significantly higher than in G. m. morsitans (p < 0.001) and G. pallidipes (p < 0.001). The prevalence of S. glossinidius in G. m. morsitans was significantly higher than in G. pallidipes (p < 0.001). A significant difference (p = 0.032) was found between male and female G. pallidipes in relation to S. glossinidius infection, with males (11.1% 95% CI 1.4-34.7%) having a higher prevalence than females (1.2% 95% CI 0.4-.2.9%). No difference (p = 0.566) was found between male (12.0% 95% CI 2.5-31.2%) and female G. m. morsitans (18.8% 95% CI 12.0-.27.2%). Only female G. brevipalpis were caught so it was not possible to compare them to male flies.
Prevalence of S. glossinidius in three species of tsetse from Luambe National Park in Zambia using PCR. The prevalence was significantly lower in G. pallidipes (1.4% 95% CI 0.8-1.5, n = 419) when compared to G. m. morsitans (17.5% 95% CI 12.1-24.7, n = 137) and G. brevipalpis (93.7% 95% CI 82.7-97.1, n = 55). The prevalence in G. m. morsitans was significantly lower than in G. brevipalpis. Error bars show 95% confidence intervals.
Prevalence of trypanosomes in individual species of tsetse
The prevalence of trypanosomes detected by PCR is summarised in Table 1 and illustrated in Figure 2.
G. brevipalpis
Of the 55 flies examined, 56.4% (95% CI 42.3%-69.8%) were positive for trypanosome DNA. Flies that were T. brucei s.l. positive were further evaluated to examine if they contained the SRA gene. Of the 21 G. brevipalpis (38.2%) that were positive, two were shown to be T. b. rhodesiense, six flies were PLC positive, the remaining 13 flies showed no amplification, suggesting that there was not enough DNA present to make a definitive diagnosis. Discounting the 13 flies that did not amplify, this gives an overall prevalence for T. b. rhodesiense of 4.8% (95% CI 0.6-16.2%).
DNA from a single species of trypanosome (51.6%) was found in the majority of positive flies. All the flies where two species of trypanosomes were detected (38.7%) involved T. brucei s.l. in partnership with T. simiae, T. simiae Tsavo, T. congolense and T. vivax. There were two flies (6.5%) where DNA from three trypanosome species was amplified, both involved T. brucei s.l. and T. vivax with one fly being positive for T. simiae and the other being positive for T. simiae Tsavo. In one fly (3.2%) four trypanosome species were detected (T. b. brucei s.l., T. congolense, T simiae Tsavo and T. vivax).
G. m. morsitans
Of the 137 flies examined, 83.9% (95% CI 76.7%-89.7%) were positive for trypanosome DNA. Flies that were T. brucei s.l. positive were further evaluated to see if they contained the SRA gene. Of the 101 G. m. morsitans (73.7%) that were T. brucei s.l. positive, 42 were shown to be T. b. rhodesiense, 41 flies were PLC positive while the remaining 18 flies showed no amplification (n = 10) or were VSG positive (n = 8). Discounting these 18 flies, this gives an overall prevalence for T. b. rhodesiense of 35.3% (95% CI 26.0-43.7%).
DNA from a single species of trypanosome (53.9%) was found in the majority of positive flies. With the exception of two flies where T. congolense and T. vivax was detected, all flies with two species of trypanosomes detected (38.3%) involved T. brucei s.l. in partnership with T. vivax, T. congolense, T. godfreyi T. simiae Tsavo or T. simiae. There were nine flies (7.8%) where DNA from three trypanosome species was amplified, all of which were positive for T. vivax, T. brucei s.l. and T. congolense DNA.
G. pallidipes
Of the 419 flies examined, 29.1% (95% CI 24.8%-33.7%) were positive for trypanosome DNA. Of the 53 G. pallidipes that were T. brucei s.l. positive (12.6%), two were shown to be T. b. rhodesiense, 24 flies were PLC positive while the remaining 27 flies showed no amplification (n = 25) or were VSG positive (n = 2). Discounting these 27 flies, this gives an overall prevalence for T. b. rhodesiense of 0.5% (95% CI 0.06-1.8%).
DNA from a single species of trypanosome (68.9%) was found in the majority of positive flies. However, DNA was amplified from two trypanosome species in thirty samples (24.6%). In the majority of these cases this involved T. brucei s.l. in partnership with T. godfreyi, T. congolense, T. vivax and T. simiae. Other flies in which two species of trypanosome were amplified included T. godfreyi in partnership with T. congolense, T. vivax, T. simiae and T. simiae Tsavo, there were also flies positive for T. vivax and T. simiae Tsavo, T. vivax and T. congolense and T. congolense and T. simiae. There were seven flies (5.4%) where DNA from three trypanosome species was amplified, the majority of these flies amplified T. brucei s.l. and T. godfreyi in partnership with T. simiae, T. congolense, T. simiae Tsavo and T. vivax. The two other flies, where three species of trypanosomes were amplified, contained T. vivax, T. brucei s.l. and T. congolense and T. vivax, T. godfreyi and T. simiae. There was one fly where four trypanosome species were amplified. This fly contained DNA from T. vivax, T. godfreyi T. simiae and T.simiae Tsavo.
Comparison of trypanosomes found in tsetse
T. brucei s.l. DNA was identified significantly more frequently in G. m. morsitans (73.7%) when compared to G. brevipalpis (38.1%, p < 0.001) and G. pallidipes (12.6%, p < 0.001). The difference between G. brevipalpis and G. pallidipes was also significant (p < 0.001). T. b. rhodesiense DNA was identified significantly more frequently in G. m. morsitans (35.3%) when compared to G. brevipalpis (4.8%, p < 0.001) and G. pallidipes (0.5%, p < 0.001). The difference between G. brevipalpis and G. pallidipes was also significant (p = 0.026).
T. vivax DNA was amplified significantly more frequently in G. m. morsitans (32.8%) when compared to G. brevipalpis (14.5%, p = 0.012) and G. pallidipes (12.6%, p < 0.001). The difference between G. brevipalpis and G. pallidipes was not significant (p = 0.066).
T. godfreyi DNA was identified significantly more frequently in G. pallidipes (12.6%) when compared to G. brevipalpis (0%, p = 0.009) and G. m. morsitans (2.2%, p = 0.003). The difference between G. brevipalpis and G. m. morsitans was not significant (p = 0.559).
T. simiae DNA was identified in significantly more samples from G. brevipalpis (18.2%) when compared to G. m. morsitans (2.2%%, p < 0.001) and G. pallidipes (3.1%, p < 0.001). The difference between G. m. morsitans and G. pallidipes was not significant (p = 0.772).
T. simiae Tsavo DNA was identified in significantly more samples from G. brevipalpis (12.6%) when compared to G. pallidipes (1.9%, p = 0.009). There was no significant difference between G. brevipalpis and G. m. morsitans (3.6%, p = 0.153) and G. m. morsitans and G. pallidipes (p = 0.195).
T. congolense DNA was found significantly more frequently in G. m. morsitans (14.6%) when compared to G. pallidipes (6.0%, p = 0.003). There was no significant difference between G. m. morsitans and G. brevipalpis (10.9%, p = 0.642) and G. brevipalpis and G. pallidipes (p = 0.155).
With the exception of T. simiae Tsavo in G. m. morsitans where males were more likely to be positive than females there was no difference between trypanosome infection and gender (data not shown).
Association between S. glossinidius and trypanosome infection
No association was found between the presence of S. glossinidius and trypanosomes that undergo development in the midgut in G. brevipalpis (p = 1), G. m. morsitans (p = 0.103) or G. pallidipes (p = 0.686). There was also no association detected at the individual trypanosome species level for any of the fly species examined (data not shown).
Discussion
S. glossinidius prevalence
Significant differences were detected between the three species of tsetse examined in the current work. Almost all the G. brevipalpis flies were infected with S. glossinidius while very few G. pallidipes were infected. There are very few publications on S. glossinidius prevalence in wild tsetse, however, the results obtained for G. brevipalpis are similar to those from Tanzania where 100% (95% CI 80.5-100%) of flies were found to be infected [31]. For G. m. morsitans the prevalence is similar to that obtained for flies in Zimbabwe where 28.5% (95% CI 19.6-39%) of flies were positive [31]. For G. pallidipes the prevalence of S. glossinidius is much lower than that obtained in Kenya [23], Tanzania and Zimbabwe [31] where rates of infection were 15.9% (95% CI: 12–20.5%), 83.3% (95% CI 74–90.4%) and 17.6% (95% CI 11.3-25.7%) respectively. Studies on other species of tsetse have shown varying rates of infection, for example Farikou et al. [22] reported a prevalence of 55% in G. p. palpalis in Cameroon, while no S. glossinidius were found in Glossina fuscipes fuscipes from Kenya [34] or Uganda [35].
Although in the current work we found a significant difference between genders in terms of S. glossinidius infection in G. pallidipes, this was most likely due to the low numbers of male flies caught. Higher levels of S. glossinidius were reported in female G. austeni by Wamwiri et al. [23], however, there was no difference between genders for G. pallidipes. Matthew [31] found no significant difference between genders for S. glossinidius infection in G. m. morsitans and G. pallidipes from Zimbabwe and G. pallidipes from Tanzania.
Trypanosome detection in tsetse flies
A wide variety of trypanosomes were detected in the three species of tsetse sampled in the current work. We were surprised to find such a high T. brucei s.l. prevalence within G. m. morsitans, this could suggest that they are quite susceptible to infection by these trypanosomes or could be a reflection of tsetse feeding habits. As DNA was extracted from the whole fly it is impossible to tell if the infections had matured, if they represented immature infections only or were dying trypanosomes from a residual bloodmeal. During collection of the tsetse used in this study, Anderson [27] undertook dissection studies to estimate infection rates in G. pallidipes and G. m. morsitans. Results showed that very few flies had mature infections and no T. brucei s.l. species were detected during the study. Of the 1293 G. pallidipes dissected, the prevalence of Duttonella species was 2.86% (95% CI 2.02-3.92) and Nannomonas species was 1.31% (95% CI 0.77-2.10). A further 266 G. m. morsitans flies were dissected with a Duttonella prevalence of 4.89% (95% CI 2.63-8.21) and Nannomonas prevalence of 3.38% (95% CI 1.56-6.33). A total of eight immature infections were detected in G. pallidipes and four in G. m. morsitans and these could have been either Nannonomas or T. brucei s.l. Even if all these infections had been T. brucei s.l. infections, the infection rates would only be 0.62% (95% CI 0.27-1.22) and 1.50% (95% CI 0.41-3.81), which are 20 and 49 fold lower than that diagnosed by PCR. Although PCR is more sensitive than microscopical techniques, it is unclear in this case if the results presented here are actual infections or just the PCR amplifying trypanosome DNA from a recent bloodmeal that will ultimately not lead to an active midgut infection. Given that Anderson [27] found very few active infections in tsetse during dissection studies in the study area this might suggest that the high prevalence is a result of high prevalence in the local wildlife. Similar results were obtained by Malele et al. [36], with dissection studies showing low levels of infection (T. brucei s.l. = 2.5%) and PCR showing much higher levels of infection (T. brucei s.l. = 55%), a 22 fold difference. Therefore, further work might be required in order to verify the use of PCR to diagnose trypanosome infections in wild tsetse.
The variation between species might be due to tsetse host preference. Although no bloodmeal analysis was carried out on the current samples, previous work had shown that G. brevipalpis normally feed on hippopotamus [37]. G. m. morsitans normally feed on Suidae (mostly warthog and bushpig), hippopotamus and ruminants (cattle, bushbuck, buffalo and other wild ruminants). Samples from the Luangwa Valley, Zambia, showed these tsetse primarily fed on vervet monkey, baboon, man, side-striped jackal, domestic dog, lion, elephant, black rhinoceros, Burchell’s zebra, bushpig, warthog, hippopotamus, giraffe, buffalo, bushbuck, kudu, duiker, waterbuck, roan antelope, hartebeest, impala and domestic ox [37]. However, there are no domesticated animals found in this part of the Luangwa Valley, due largely to the high trypanosomiasis challenge. G. pallidipes normally feed on ruminants and Suidae, however, although samples from Zambia were analysed no specific information on them is provided [37]. The high prevalence of T. b. rhodesiense in G. m. morsitans was surprising and further supports different feeding habits by the three tsetse species sampled in the current work. Work by Anderson et al. [33] found that the majority of T. brucei s.l. infections in wildlife were concentrated in four species (bushbuck, waterbuck, lion and leopard). T. b. rhodesiense was identified in wildlife from the nearby game management areas of Musalangu and Lower Lupande. The overall prevalence for T. b. rhodesiense was 0.5% (95% CI 0.06–1.72%) in the Luangwa Valley with the animals positive for T. b. rhodesiense being a bushbuck and a buffalo. Also of interest in this study is the low detection rate of T. godfreyi in G. m. morsitans in comparison with G. pallidipes. As the warthog is considered the definitive host for this parasite [38] and G. m. morsitans are believed to feed predominantly on suidae it may suggest a wider host range for this parasite. Bloodmeal analysis on the tsetse flies analysed in the current work may shed some light on what the three species of tsetse feed on and why the rates vary widely between tsetse species.
There have been very few papers where tsetse flies have been investigated for T. b. rhodesiense. Auty et al. [11] analysed 133 samples by PCR that were positive for trypanosomes by microscopy, the overall prevalence of T. brucei s.l. was 0.8% (95% CI: 0.05-1.2%) and 0.7% (95% CI: 0.4-1.2%) for Glossina swynnertoni and G. pallidipes respectively. Of the T. brucei s.l. positive flies, around 10% of flies were positive for SRA, giving an overall prevalence of 0.01% (95% CI: 0–0.54) for G. swynnertoni and 0.0085% (95% CI 0–0.059) for G. pallidipes. Further work in Tanzania found similar levels of T. brucei s.l. positive G. swynnertoni (0.7%, 95% CI 0.2-1.7%), however, in this case all were also positive for SRA [39]. These results are much lower than what we found in the current work in Zambia and might be related to trypanosomes found in mammals in the area where the flies were captured. In the current work the proportion of PLC positive to T. b. rhodesiense in terms of G. brevipalpis was 0.25 (2 from 8), for G. m. morsitans it was 0.51 (42 from 83) and 0.08 (2 from 24) for G. pallidipes. Although it is not possible to tell if flies that were positive for T. b. rhodesiense were also positive for T. b. brucei the ratio of infection for the 3 fly species was 0.3, 1.02 and 0.09 for G. brevipalpis, G. m. morsitans and G. pallidipes respectively. There is little published work looking at T. b. rhodesiense in Zambia, however, recently Lisulo et al. [40] and Namangala et al. [41] have found human infective parasites in dogs in the Luangwa valley and there have been reports of both tourists [42, 43] and locals being infected with T. b. rhodesiense[44], suggesting there is active transmission in the area.
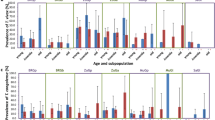
When the distribution of trypanosome infections is considered by vegetation type, no clear pattern emerges. Although discreet vegetation units occur in Luambe National Park, the mobility of both hosts and vectors means that variation in infection rates due to habitat influences may not be evident at this scale. However, of interest in this study is the high infection rate with T. b. rhodesiense in G. m. morsitans in both Combretum-Terminalia woodland and thicket (Figure 3).
Bar chart showing the distribution of trypanosomes in G. m. morsitans and G. pallidipes by vegetation type. Key: CW, Combretum-Terminalia woodland; MS, mopane scrub woodland; MW, mopane woodland; RW, riverine woodland; T, thicket; Gmm, G. m. morsitans; Gp, G. pallidipes; T, T. brucei s.l.; Tbr, T. b. rhodesiense; Tv, T. vivax; Tg, T. godfreyi; Ts, T. simiae; TsT, T. simiae; Tc, T. congolense. Error bars show 95% confidence intervals.
Association between S. glossinidius and trypanosome infection
In the current work no association was detected between the presence of S. glossinidius and presence of trypanosomes in any of the three tsetse species examined. This is in disagreement with recent work published by Farikou et al. [22] and Wamwiri et al. [23] for G. p. palpalis and G. pallidipes respectively, but does agree with work undertaken on G. austeni[23]. In terms of G. pallidipes this could be due to the low prevalence of S. glossinidius detected in the current work, with the exception of T. godfreyi, G. pallidipes in many cases had significantly fewer trypanosomes present than G. m morsitans and G. brevipalpis. However, almost all G. brevipalpis flies (92.7%) were infected with S. glossinidius, despite this, this species of tsetse did not contain the most trypanosome positive flies (50.9% of flies with a trypanosome infection that establishes in the midgut). Instead it was G. m. morsitans where the prevalence of S. glossinidius was 17.5%, where the most trypanosome infected flies was observed (78.1% with midgut trypanosomes). As described above, a number of reasons could explain the detection of trypanosomes in these samples. For example host preference and detection of DNA from residual bloodmeals. Tsetse susceptibility has also been linked to the teneral phenomenon, where tsetse are most susceptible to infection on their first feed [45], as such if a susceptible fly does feed on an uninfected host, it will therefore become refractory to infection at future bloodmeals.
Molecular methods of detection are more sensitive than traditional dissection techniques and enable the assessment of mixed infections, but for various reasons it may be difficult to assess an association between S. glossinidius and trypanosome infection on field collected tsetse, particularly when flies are analysed only by molecular methods. Although the study by Farikou et al. [22] combined both techniques, this approach also has limitations. For example, trypanosome-susceptible flies that fed on an animal not infected with trypanosomes would become refractory to later infection (as susceptibility to infection is largely a teneral phenomenon [45]). In contrast, if the same fly had fed on a trypanosome positive animal it would have become infected, so despite being a susceptible fly, using their approach it would be classified as a refractory fly. Although in the current work we could not discriminate between established infections in the tsetse and residual bloodmeal contamination, one way to investigate this further would be to trap live flies and then feed them on a clean bloodmeal source for period of at least a week. This would allow any residual DNA to be cleared from the fly meaning that only established infections would be present. However, as mentioned above, this would still allow susceptible flies that have become refractory to infection by feeding on an uninfected host to be included. When investigating establishment of trypanosome infections in tsetse, Harley [46, 47] showed that the majority of the tsetse emerging from pupae collected from the wild, which were then fed on infected rats were refractory to infection. Therefore, the best method to investigate if S. glossinidius does have an impact on tsetse susceptibility would be to collect tsetse pupae from the wild and examine flies for trypanosomes and S. glossinidius after an infective bloodmeal was allowed to establish in the tsetse. This would be the only way to control for both susceptible flies becoming refractory following a feed on a non-infected animal and residual blood contamination through feeding on infected hosts.
Conclusion
The current work shows that prevalence of S. glossinidius varies within species of tsetse caught within the same area, and that these tsetse were positive for a variety of trypanosomes. Whether or not this is linked to the susceptibility of these tsetse flies to certain trypanosome species, or is a function of host preference is unclear. The high trypanosome prevalence (particularly T. brucei s.l. in G. m. morsitans) detected by PCR in the current work but low prevalence detected by microscopy [27] might suggest a high level of infection in the local wildlife, however, the fly population might be highly refractory to infection.
Finally, no link was found between presence of S. glossinidius and presence of trypanosome DNA; flies were diagnosed trypanosome positive by PCR rather than by microscopy and therefore this might wrongly diagnose a fly as trypanosome positive due to it being recently fed. The discrepancy between dissection results and PCR analysis requires further analysis and therefore trying to associate S. glossinidius presence and trypanosome susceptibility should be interpreted with caution.
References
Aksoy S: Tsetse–A haven for microorganisms. Parasitol Today. 2000, 16: 114-118. 10.1016/S0169-4758(99)01606-3.
Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, Aksoy S: Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet. 2002, 32: 402-407. 10.1038/ng986.
Dale C, Maudlin I: Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int J Syst Bacteriol. 1999, 49: 267-275. 10.1099/00207713-49-1-267.
Welburn SC, Arnold K, Maudlin I, Gooday GW: Rickettsia-like organisms and chitinase production in relation to transmission of trypanosomes by tsetse flies. Parasitology. 1993, 107: 141-145. 10.1017/S003118200006724X.
Welburn SC, Maudlin I: Tsetse-trypanosome interactions: rites of passage. Parasitol Today. 1999, 15: 399-403. 10.1016/S0169-4758(99)01512-4.
Hamidou Soumana I, Loriod B, Ravel S, Tchicaya B, Simo G, Rihet P, Geiger A: The transcriptional signatures of Sodalis glossinidius in the Glossina palpalis gambiensis flies negative for Trypanosoma brucei gambiense contrast with those of this symbiont in tsetse flies positive for the parasite: possible involvement of a Sodalis-hosted prophage in fly Trypanosoma refractoriness?. Infect Genet Evol. 2014, 24: 41-56.
Alam U, Medlock J, Brelsfoard C, Pais R, Lohs C, Balmand S, Carnogursky J, Heddi A, Takac P, Galvani A, Aksoy S: Wolbachia symbiont infections induce strong cytoplasmic incompatibility in the tsetse fly Glossina morsitans. PLoS Pathog. 2011, 7: e1002415-10.1371/journal.ppat.1002415.
Lloyd L, Johnson WB: The trypanosome infections of tsetse flies in northern Nigeria and a new method of estimation. Bull Entomol Res. 1924, 14: 265-288.
Okoth JO, Kapaata R: Trypanosome infection rates in Glossina fuscipes fuscipes Newst. in the Busoga sleeping sickness focus, Uganda. Ann Trop Med Parasitol. 1986, 80: 459-461.
Masiga DK, McNamara JJ, Laveissière C, Truc P, Gibson WC: A high prevalence of mixed trypanosome infections in tsetse flies in Sinfra, Côte d’Ivoire, detected by DNA amplification. Parasitology. 1996, 112: 75-80. 10.1017/S0031182000065094.
Auty HK, Picozzi K, Malele I, Torr SJ, Cleaveland S, Welburn S: Using molecular data for epidemiological inference: assessing the prevalence of Trypanosoma brucei rhodesiense in tsetse in Serengeti, Tanzania. PLoS Negl Trop Dis. 2012, 6: e1501-10.1371/journal.pntd.0001501.
Maudlin I: Inheritance of susceptibility to Trypanosoma congolense infection in Glossina morsitans. Ann Trop Med Parasitol. 1982, 76: 225-227.
Hao Z, Kasumba I, Lehane MJ, Gibson WC, Kwon J, Aksoy S: Tsetse immune responses and trypanosome transmission: implications for the development of tsetse-based strategies to reduce trypanosomiasis. Proc Natl Acad Sci U S A. 2001, 98: 12648-12653. 10.1073/pnas.221363798.
MacLeod ET, Maudlin I, Darby AC, Welburn SC: Antioxidants promote establishment of trypanosome infections in tsetse. Parasitology. 2007, 134: 827-831. 10.1017/S0031182007002247.
Pais R, Lohs C, Wu Y, Wang J, Aksoy S: The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl Environ Microbiol. 2008, 74: 5965-5974. 10.1128/AEM.00741-08.
Maudlin I, Dukes P: Extrachromosomal inheritance of susceptibility to trypanosome infection in tsetse flies: I. selection of susceptible and refractory lines of Glossina morsitans morsitans. Ann Trop Med Parasitol. 1985, 79: 317-324.
Moloo SK, Shaw MK: Rickettsial infections of midgut cells are not associated with susceptibility of Glossina morsitans centralis to Trypanosoma congolense infection. Acta Trop. 1989, 46: 223-227. 10.1016/0001-706X(89)90022-3.
Welburn SC, Gibson WC: Cloning of a repetitive DNA from the rickettsia-like organisms of tsetse flies (Glossina spp.). Parasitology. 1989, 98: 81-84. 10.1017/S0031182000059710.
Maudlin I, Welburn SC, Mehlitz D: The relationship between rickettsia-like-organisms and trypanosome infections in natural populations of tsetse in Liberia. Trop Med Parasitol. 1990, 41: 265-267.
O’Neill SL, Gooding RH, Aksoy S: Phylogenetically distant symbiotic microorganisms reside in Glossina midgut and ovary tissues. Med Vet Entomol. 1990, 7: 377-383.
Geiger A, Ravel S, Frutos R, Cuny G: Sodalis glossinidius (Enterobacteriaceae) and vectorial competence of Glossina palpalis gambiensis and Glossina morsitans morsitans for Trypanosoma congolense savannah type. Curr Microbiol. 2005, 51: 35-40. 10.1007/s00284-005-4525-6.
Farikou O, Njiokou F, Mbida Mbida JA, Njitchouang GR, Djeunga HN, Asonganyi T, Simarro PP, Cuny G, Geiger A: Tripartite interactions between tsetse flies, Sodalis glossinidius and trypanosomes--an epidemiological approach in two historical human African trypanosomiasis foci in Cameroon. Infect Genet Evol. 2010, 10: 115-121. 10.1016/j.meegid.2009.10.008.
Wamwiri FN, Alam U, Thande PC, Aksoy E, Ngure RM, Aksoy S, Ouma JO, Murilla GA: Wolbachia, Sodalis and trypanosome co-infections in natural populations of Glossina austeni and Glossina pallidipes. Parasit Vectors. 2013, 6: 232-10.1186/1756-3305-6-232.
MacLeod ET: PhD Thesis. Factors Affecting Transmission of Trypanosomes Through Tsetse Flies. 2005, The University of Edinburgh, College of Medicine and Veterinary Medicine
Baker RD, Maudlin I, Milligan PJ, Molyneux DH, Welburn SC: The possible role of Rickettsia-like organisms in trypanosomiasis epidemiology. Parasitology. 1990, 100: 209-217. 10.1017/S0031182000061217.
Njiru ZK, Constantine CC, Guya S, Crowther J, Kiragu JM, Thompson RC, Dávila AM: The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res. 2005, 95: 186-192. 10.1007/s00436-004-1267-5.
Anderson NE: PhD thesis. An Investigation into the Ecology of Trypanosomiasis in Wildlife of the Luangwa Valley, Zambia. 2009, University of Edinburgh, College of Medicine and Veterinary Medicine
Vale GA: Artificial refuges for tsetse flies (Glossina spp.). Bull Entomol Res. 1971, 61: 331-350. 10.1017/S0007485300057874.
Chen X, Li S, Aksoy S: Concordant evolution of a symbiont with its host insect species: molecular phylogeny of genus Glossina and its bacteriome-associated endosymbiont, Wigglesworthia glossinidia. J Mol Evol. 1999, 48: 49-58. 10.1007/PL00006444.
Hamidou Soumana I, Berthier D, Tchicaya B, Thevenon S, Njiokou F, Cuny G, Geiger A: Population dynamics of Glossina palpalis gambiensis symbionts, Sodalis glossinidius, and Wigglesworthia glossinidia, throughout host-fly development. Infect Genet Evol. 2013, 13: 41-48.
Matthew CZ: PhD Thesis. Biological and Molecular Aspects of Sodalis glossinidius. 2007, The University of Edinburgh, College of Medicine and Veterinary Medicine
Picozzi K, Carrington M, Welburn SC: A multiplex PCR that discriminates between Trypanosoma brucei brucei and zoonotic T. b. rhodesiense. Exp Parasitol. 2008, 118: 41-46. 10.1016/j.exppara.2007.05.014.
Anderson NE, Mubanga J, Fevre EM, Picozzi K, Eisler MC, Thomas R, Welburn SC: Characterisation of the wildlife reservoir community for human and animal trypanosomiasis in the Luangwa Valley, Zambia. PLoS Negl Trop Dis. 2011, 5: e1211-10.1371/journal.pntd.0001211.
Lindh JM, Lehane MJ: The tsetse fly Glossina fuscipes fuscipes (Diptera: Glossina) harbours a surprising diversity of bacteria other than symbionts. Antonie Van Leeuwenhoek. 2011, 99: 711-720. 10.1007/s10482-010-9546-x.
Alam U, Hyseni C, Symula RE, Brelsfoard C, Wu Y, Kruglov O, Wang J, Echodu R, Alioni V, Okedi LM, Caccone A, Aksoy S: Implications of microfauna-host interactions for trypanosome transmission dynamics in Glossina fuscipes fuscipes in Uganda. Appl Environ Microbiol. 2012, 78: 4627-4637. 10.1128/AEM.00806-12.
Malele II, Magwisha HB, Nyingilili HS, Mamiro KA, Rukambile EJ, Daffa JW, Lyaruu EA, Kapange LA, Kasilagila GK, Lwitiko NK, Msami HM, Kimbita EN: Multiple trypanosoma infections are common amongst Glossina species in the new farming areas of Rufiji district, Tanzania. Parasit Vectors. 2011, 4: 217-10.1186/1756-3305-4-217.
Clausen PH, Adeyemi I, Bauer B, Breloeer M, Salchow F, Staak C: Host preferences of tsetse (Diptera: Glossinidae) based on bloodmeal identifications. Med Vet Entomol. 1998, 12: 169-180. 10.1046/j.1365-2915.1998.00097.x.
Auty H, Anderson NE, Picozzi K, Lembo T, Mubanga J, Hoare R, Fyumagwa RD, Mable B, Hamill L, Cleaveland S, Welburn SC: Trypanosome diversity in wildlife species from the Serengeti and Luangwa Valley ecosystems. PLoS Negl Trop Dis. 2012, 6: e1828-10.1371/journal.pntd.0001828.
Malele II, Kinung’hi SM, Nyingilili HS, Matemba LE, Sahani JK, Mlengeya TD, Wambura M, Kibona SN: Glossina dynamics in and around the sleeping sickness endemic Serengeti ecosystem of northwestern Tanzania. J Vector Ecol. 2007, 32: 263-268. 10.3376/1081-1710(2007)32[263:GDIAAT]2.0.CO;2.
Lisulo M, Sugimoto C, Kajino K, Hayashida K, Mudenda M, Moonga L, Ndebe J, Nzala S, Namangala B: Determination of the prevalence of African trypanosome species in indigenous dogs of Mambwe district, eastern Zambia, by loop-mediated isothermal amplification. Parasit Vectors. 2014, 7: 19-10.1186/1756-3305-7-19.
Namangala B, Oparaocha E, Kajino K, Hayashida K, Moonga L, Inoue N, Suzuki Y, Sugimoto C: Preliminary investigation of trypanosomosis in exotic dog breeds from Zambia’s Luangwa and Zambezi valleys using LAMP. Am J Trop Med Hyg. 2013, 89: 116-118. 10.4269/ajtmh.13-0078.
Cottle LE, Peters JR, Hall A, Bailey JW, Noyes HA, Rimington JE, Beeching NJ, Squire SB, Beadsworth MB: Multiorgan dysfunction caused by travel-associated African trypanosomiasis. Emerg Infect Dis. 2012, 18: 287-289. 10.3201/eid1802.111479.
Richter J, Göbels S, Göbel T, Westenfeld R, Müller-Stöver I, Häussinger D: A returning traveller with fever, facial swelling, and skin lesions. BMJ. 2012, 344: e2092-10.1136/bmj.e2092.
Namangala B, Hachaambwa L, Kajino K, Mweene AS, Hayashida K, Simuunza M, Simukoko H, Choongo K, Chansa P, Lakhi S, Moonga L, Chota A, Ndebe J, Nsakashalo-Senkwe M, Chizema E, Kasonka L, Sugimoto C: The use of loop-mediated isothermal amplification (LAMP) to detect the re-emerging Human African trypanosomiasis (HAT) in the Luangwa and Zambezi valleys. Parasit Vectors. 2012, 5: 282-10.1186/1756-3305-5-282.
Welburn SC, Maudlin I: The nature of the teneral state in Glossina and its role in the acquisition of trypanosome infection in tsetse. Ann Trop Med Parasitol. 1992, 86: 529-536.
Harley JM: Comparison of the susceptibility of infection with trypanosoma rhodesiense of Glossina pallidipes, G. morsitans, G. fuscipes and G. brevipalpis. Ann Trop Med Parasitol. 1971, 55: 185-189.
Harley JM: The influence of the age of the fly at the time of the infecting feed on infection of Glossina fuscipes with Trypanosoma rhodesiense. Ann Trop Med Parasitol. 1971, 65: 191-196.
Acknowledgments
We would like to thank John Silutongwe, John Mashili and Lyson Phiri (Kakumbi Field Research Station, Department of Veterinary and Tsetse Control Services, Zambia) for technical support and Kepson Chansa (Zambian Wildlife Authority) for logistical support. We would also like to thank Professor Michael Thrusfield (University of Edinburgh) for his statistical advice and Professor Ian Maudlin (University of Edinburgh) for discussions on the manuscript. Simon Durkin acknowledges the Wolfson Foundation Intercalated Award for support during is studies on the Infectious Diseases Honours programme at the University of Edinburgh. Neil Anderson would like to thank the Royal Zoological Society of Scotland for financial and logistical support and the United Kingdom Department for International Development Animal Health Programme (DFID-AHP) for financial support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JWD and SMD analysed G. pallidipes and JEHD analysed G. m. morsitans and G. brevipalpis for symbiont and trypanosome presence using ITS-PCR. SD and LCH carried out SRA-PCR. NEA collected the flies from Luambe National Park. ETM carried out the statistical analysis. NEA and ETM designed the study and wrote the manuscript. All the authors have read and approved the final manuscript.
Jonny W Dennis, Simon M Durkin, Jemima E Horsley Downie contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Dennis, J.W., Durkin, S.M., Horsley Downie, J.E. et al. Sodalis glossinidius prevalence and trypanosome presence in tsetse from Luambe National Park, Zambia. Parasites Vectors 7, 378 (2014). https://doi.org/10.1186/1756-3305-7-378
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-3305-7-378