Abstract
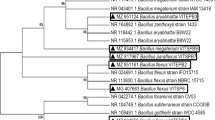
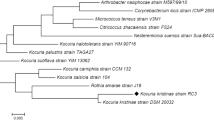
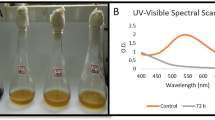
Textile industry effluent creates a negative impact to plants, animals and environment. The utilization of reactive azo dyes plays a colossal role in creating hazard to environment and water due to its toxic, carcinogenic and promote mutagenicity. The effective treatment of the effluent is mandatory to conserve nature and humans from critical risk. Microbial remediation is an eco-friendly method to degrade the dyes; specifically, bacterial consortium treatment is unique with reference to their mutual interaction. In this study, we have successfully developed an effective dye-decolourizing bacterial consortium and experimented to achieve the enhanced reactive dye decolourization in pilot scale. The culture characteristic factors influencing in bacterial consortium dye decolourization were further investigated and statistically optimised in response surface methodology (RSM) technique using central composite design (CCD) tool to maximize the decolourization process. Based on the antagonistic assay, the bacterial consortium design CD-3, consisting of Exiguobacterium aurantiacum TSL7, Bacillus firmus TSL9, and Brevibacillus laterosporus TS5 strains, demonstrated the highest RGY dye decolourization (83.78%) within 24 h. Bacterial consortium CD-3 productively decolourizes diverse dyes like magenta, yellow FG, and red (95%) in 24 h, followed by orange 3R (94%). The optimal concentration of selected factors for CD-3 dye decolourization is sodium chloride, yeast extract, initial dye concentration, inoculum size, and pH, which are 1.25%, 1.52%, 219 mg/l, 2.5%, and 6.5, respectively. The statistical analysis of tested factors, specifically dye concentration (carbon source), yeast extract (nitrogen source) and pH, has significant effects in process, and its mid-values have great influence in decolourizing results. Furthermore, the clear picture of the degraded non-toxic dye components was depicted by GC–MS result. Thus, the proposed biological treatment using bacterial consortium is effective one and can be used in the real world to address the pollution crisis.



Similar content being viewed by others
Data availability
All data are available on request.
References
Mishra BK, Kumar P, Saraswat C, Chakraborty S, Gautam A (2021) Water security in a changing environment: concept, challenges and solutions. Water 13:490
Du Plessis A, du Plessis A (2019) Schmuhl. Springer, Water as an inescapable risk
Kishor R, Purchase D, Saratale GD, Saratale RG, Ferreira LFR, Bilal M, Chandra R, Bharagava RN (2021) Ecotoxicological and health concerns of persistent coloring pollutants of textile industry wastewater and treatment approaches for environmental safety. J Environ Chem Eng 9:105012
Khan WU, Ahmed S, Dhoble Y, Madhav S (2023) A critical review of hazardous waste generation from textile industries and associated ecological impacts. J Indian Chem Soc 100:100829
RP Singh, PK Singh, R Gupta, RL Singh (2019) Treatment and recycling of wastewater from textile industry. In: Advances in biological treatment of industrial waste water and their recycling for a sustainable future, pp 225–266. https://doi.org/10.1007/978-981-13-1468-1_8
A Roy Choudhury (2014) Environmental impacts of the textile industry and its assessment through life cycle assessment. In: Roadmap to sustainable textiles and clothing: environmental and social aspects of textiles and clothing supply chain, pp 1–39. https://doi.org/10.1007/978-981-287-110-7_1
Goud BS, Cha HL, Koyyada G, Kim JH (2020) Augmented biodegradation of textile azo dye effluents by plant endophytes: a sustainable, eco-friendly alternative. Curr Microbiol 77:3240–3255
Sayyed RZ, Bhamare HM, Sapna, Marraiki N, Elgorba AM, Syed A, El Enshasy HA, Dailin JD (2020) Tree bark scrap fungus: a potential source of laccase for application in bioremediation of non textile dyes. PLOS One 15:e0229968. https://doi.org/10.1371/journal.pone.0229968
Mutiara F, Hrnawati D, Meylani V, Perveen K, Sayyed RZ (2023) Indigenous consortiumof microbial biomass effectively degrades synthetic dyes. Biomass Convers Biorefinery 9:1–15
Ngo ACR, Tischler D (2022) Microbial degradation of azo dyes: approaches and prospects for a hazard free conversion by microorganisms. Int J Environ Res Publ Health 19(4740):1–24
Kumar A (2020) Inorganic soil contaminants and their biological remediation. Plant Responses Soil Pollut 133–153. https://doi.org/10.1007/978-981-15-4964-9_8
Koehle AP, Brumwell SL, Seto EP, Lynch AM, Urbaniak C (2023) Microbial applications for sustainable space exploration beyond low Earth orbit. npj Microgravity 9:47
Ghanbari M, Dargahi A, Ahmadi M, Ghafari S, Jaafarzadeh N, Jorfi S (2023) Biodegradation of acid orange 7 dye using consortium of novel bacterial strains isolated from Persian Gulf water and soil contaminated with petroleum compounds. Biomass Conversion Biorefinery 13:13695–13706
Roy U, Manna S, Sengupta S, Das P, Datta S, Mukhopadhyay A, Bhowal A (2018) Dye removal using microbial biosorbents. Green Adsorb Pollut Removal Innov Mater 253–280. https://doi.org/10.1007/978-3-319-92162-4_8
J Singh, P Gupta, A Das (2021) Dyes from textile industry wastewater as emerging contaminants in agricultural fields. Sustain Agric Rev Emerging Contam Agric 50:109–129. https://doi.org/10.1007/978-3-030-63249-6_5
Islam T, Repon MR, Islam T, Sarwar Z, Rahman MM (2023) Impact of textile dyes on health and ecosystem: a review of structure, causes, and potential solutions. Environ Sci Pollut Res 30:9207–9242
Senthilkumar S, Ahmed Basha C, Perumalsamy M, Prabhu HJ (2012) Electrochemical oxidation and aerobic biodegradation with isolated bacterial strains for dyr wastewater combined and integrated approach. Electrochim Acta 77:171–178
Korbahti BK, Ruaf MA (2008) Applicationof response surface analysis to the photolytc degradation of basic red 2 dye. J Chem Eng 138:166–171
Palumbo JD, Baker JL, Mahoney NE (2006) Isolation of bacterial antagonists of Aspergillus flavus from almonds. Microb Ecol 52:45–52
Thiruppathi K, Rangasamy K, Ramasamy M, Muthu D (2021) Evaluation of textile dye degrading potential of ligninolytic bacterial consortia. Environ Challenges 4:100078
Krishnan J, Kishore AA, Suresh A, Madhumeetha B, Prakash DG (2017) Effect of pH, inoculum dose and initial dye concentration on the removal of azo dye mixture under aerobic conditions. Int Biodeterioration Biodegradation 11:16–27
Saha P, Sivaramakrishna A, Rao KVB (2023) Bioremediation of reactive orange 16 by industrial effluent-adapted bacterial consortium VITPBC6: process optimization using response surface methodology (RSM), enzyme kinetics, pathway elucidation, and detoxification. Environ Sci Pollut Res 30:35450–35477
Joshi S, Bajpai S, Jana S (2020) Application of ANN and RSM on fluoride removal using chemically activated D. sissoo sawdust. Environ Sci Pollut Res 27:17717–17729
Domínguez-Castillo C, Jiménez-Hidalgo M, López-Gámez J, Rodríguez-Hortal A, Alzaga-García M, Gallardo-Abárzuza M, Higueras-Milena JM, Gómez-Morón A, García-Viñas E, Bernáldez-Sánchez E (2023) High-resolution mass spectrometry identification of dye compounds and their degradation products in American cochineal from a historic shipping cargo. Dyes Pigm 216:111313
Tang D-S, Zhang L, Chen H-L, Liang Y-R, Lu J-L, Liang H-L, Zheng X-Q (2007) Extraction and purification of solanesol from tobacco:(I) Extraction and silica gel column chromatography separation of solanesol. Sep Purif Technol 56:291–295
Carolin CF, Kumar PS, Joshiba GJ (2021) Sustainable approach to decolourize methyl orange dye from aqueous solution using novel bacterial strain and its metabolites characterization. Clean Technol Environ Policy 23:173–181
Altindas C, Sher F, Cain N, Lima EC, Rashid T, IUl Hai, A Karaduman, (2023) Synergistic interaction ofmetal loaded mutifactorial nanocatalysts overbifunctional transalkylation for environmenl application. Environ Res 216:1144791–14
Canina NS, Nuhanovi M, Sulejmanovi J, Ma E, Sher F (2023) Highly effective sustainable membrne based cyanobacteria for uranium uptake from aqueous environment. Chemosphere 313(137488):1–11
Agrawal S, Bhatt A (2023) Microbial endophytes: emerging trends and biotechnological applications. Curr Microbiol 80:249
Mikesková H, Novotný Č (2012) Svobodová, Interspecific interactions in mixed microbial cultures in a biodegradation perspective. Appl Microbiol Biotechnol 95:861–870
Bhatt P, Gangola S, Bhandari G, Zhang W, Maithani D, Mishra S, Chen S (2021) New insights into the degradation of synthetic pollutants in contaminated environments. Chemosphere 268:128827
Nanjani S, Paul D, Keharia H (2021) Genome analysis to decipher syntrophy in the bacterial consortium ‘SCP’for azo dye degradation. BMC Microbiol 21:1–19
Dave SR, Patel TL, Tipre DR (2014) Bacterial degradation of azo dye containing wastes. In: Microbial degradation of synthetic dyes in wastewaters. Springer, pp 57–83. https://doi.org/10.1007/978-3-319-10942-8_3
Saratale R, Saratale G, Chang J-S, Govindwar S (2010) Decolorization and biodegradation of reactive dyes and dye wastewater by a developed bacterial consortium. Biodegradation 21:999–1015
Woutersen M, Belkin S, Brouwer B, Van Wezel AP, Heringa MB (2011) Are luminescent bacteria suitable for online detection and monitoring of toxic compounds in drinking water and its sources? Anal Bioanal Chem 400:915–929
Chakraborty J, Das S (2016) Molecular perspectives and recent advances in microbial remediation of persistent organic pollutants. Environ Sci Pollut Res 23:16883–16903
Khan R, Bhawana P, Fulekar M (2013) Microbial decolorization and degradation of synthetic dyes: a review. Rev Environ Sci Bio/Technol 12:75–97
Maniyam MN, Hari M, Yaacob NS (2020) Enhanced methylene blue decolourization by Rhodococcus strain UCC 0003 grown in banana peel agricultural waste through response surface methodology. Biocatal Agric Biotechnol 23:101486
Jasińska A, Soboń A, Góralczyk-Bińkowska A, Długoński J (2019) Analysis of decolorization potential of Myrothecium roridum in the light of its secretome and toxicological studies. Environ Sci Pollut Res 26:26313–26323
Popli S, Patel UD (2015) Destruction of azo dyes by anaerobic–aerobic sequential biological treatment: a review. Int J Environ Sci Technol 12:405–420
Krithika A, Gayathri KV, Kumar DT, Doss CGP (2021) Mixed azo dyes degradation by an intracellular azoreductase enzyme from alkaliphilic Bacillus subtilis: a molecular docking study. Arch Microbiol 203:3033–3044
Zhang C, Chen H, Xue G, Liu Y, Chen S, Jia C (2021) A critical review of the aniline transformation fate in azo dye wastewater treatment. J Clean Prod 321:128971
Bera SP, Tank S (2021) Bioremedial approach of Pseudomonas stutzeri SPM-1 for textile azo dye degradation. Arch Microbiol 203:2669–2680
Deller S, Macheroux P, Sollner S (2008) Flavin-dependent quinone reductases. Cell Mol Life Sci 65:141–160
Ajaz M, Shakeel S, Rehman A (2020) Microbial use for azo dye degradation—a strategy for dye bioremediation. Int Microbiol 23:149–159
Daims H, Taylor MW, Wagner M (2006) Wastewater treatment: a model system for microbial ecology. Trends Biotechnol 24:483–489
Mishra S, Maiti A (2018) The efficacy of bacterial species to decolourise reactive azo, anthroquinone and triphenylmethane dyes from wastewater: a review. Environ Sci Pollut Res 25:8286–8314
D.B. Vishani, A. Shrivastav (2022) Enzymatic decolorization and degradation of azo dyes. In: Development in Wastewater Treatment Research and Processes, pp 419–432. https://doi.org/10.1016/B978-0-323-85657-7.00020-1
Yagub MT, Sen TK, Afroze S, Ang HM (2014) Dye and its removal from aqueous solution by adsorption: a review. Adv Coll Interface Sci 209:172–184
Shanmugam BK, Easwaran SN, Mohanakrishnan AS, Kalyanaraman C, Mahadevan S (2019) Biodegradation of tannery dye effluent using Fenton’s reagent and bacterial consortium: a biocalorimetric investigation. J Environ Manage 242:106–113
Khalid A, Kausar F, Arshad M, Mahmood T, Ahmed I (2012) Accelerated decolorization of reactive azo dyes under saline conditions by bacteria isolated from Arabian seawater sediment. Appl Microbiol Biotechnol 96:1599–1606
Srivastava A, Parida VK, Majumder A, Gupta B, Gupta AK (2021) Treatment of saline wastewater using physicochemical, biological, and hybrid processes: insights into inhibition mechanisms, treatment efficiencies and performance enhancement. J Environ Chem Eng 9:105775
Pereira L, Alves M (2012) Dyes—environmental impact and remediation. In: Environmental protection strategies for sustainable development, pp 111–162. https://doi.org/10.1007/978-94-007-1591-2_4
Fanin N, Mooshammer M, Sauvadet M, Meng C, Alvarez G, Bernard L, Bertrand I, Blagodatskaya E, Bon L, Fontaine S (2022) Soil enzymes in response to climate warming: Mechanisms and feedbacks. Funct Ecol 36:1378–1395
Hatfield JL, Prueger JH (2015) Temperature extremes: Effect on plant growth and development. Weather Clim Extremes 10:4–10
Barua S, Sohag Miah M, Nuruddin Mahmud MM, Rahman, (2023) Isolation and identification of naturally occurring textile effluent degrading bacteria and evaluation of their ability to inhibit potentially toxic elements. Results in Engineering 17:100967
Patel H, Yadav VK, Yadav KK, Choudhary N, Kalasariya H, Alam MM, Gacem A, Amanullah M, Ibrahium HA (2022) A recent and systemic approach towards microbial degradation of dyes from textile industries. Water 14:1–24. https://doi.org/10.3390/w14193163
Bera SP, Shah MP, Godhaniya M (2022) Microbial remediation of textile dye acid orange by a novel bacterial consortium SPB92. Frontier in Environmental science 10:1–8
Mnif I, Bouassida M, Ayed L (2023) DhouhaGhirbi, Optimization of textile effluent bacterial treatment and improvement of the process efficiency through SPB1 biosurfactant addition. Water Sci Technol 87:1765–1778
Haque M, Hossen N, Rahman A, Roy J, Talukder R, Ahmed M (2024) Ahiduzzaman, AmdadulHaque, Decolorization, degradation and detoxification, of mutagenic dye methyl orange by novel biofilm producing plant growth promoting rhizobacteria. Chemosphere 346:1–13
Desai C, Jain KR, Boopathy R, Van Hullebusch ED, Madamwar D (2021) Eco-sustainable bioremediation of textile dye waters: Innovative microbial treatment technologies and mechanistic insight of textile dye biodegradation. Front Microbiol 12:707083
Haque M, Haque A, Mosharaf K (2021) Polash Kisku Marcus, Decolorization, degradation and detoxification of carcinogenic sulfonated azo dye methyl orange by newly developed biofilm consortia. Saudi J Biol Sci 28:793–804
Mohanty SS, Kumar A (2021) Enhanced degradation of anthraquinone dyes by microbial monoculture and developed consortium through the production of specific enzymes. Sci Rep 7678:1–15
Sun Su, Liu P, Ullah M (2023) Efficient azo dye biodecolorization system using lignin co cultured white rot fungus. J Fungi 9:1–13
Aryanti PTP, Nugroho FA, Prasetyo BH, Rahayu FS, Kadier A, Sher F (2022) Integrated electrocogulation tight ultrafiltration of river waterdecontamination: the influence of electrode configuration and operating pressure cleaner. Eng Technol 9:1–10
Prajapati D, Bhatt A, Gupte A, Gupte S (2022) Fungi: a sustainable and versatile tool for transformation, detoxification, and degradation of environmental pollutants. In: Progress in Mycology: Biology and Biotechnological Applications. Springer, pp 593–619. https://doi.org/10.1007/978-981-16-3307-2_20
Zafar S, Bukhari AD, Rehman A (2022) Azo dyes degradation by microorganisms- an efficient sustainable approach. Saudi J Biol Sci 29:1–9
Tian Y, Kangil Wu, Lin S, Shi M, Yang Liu XuSu, Islam R (2024) Biodegradation and decolorization of crystal violet dye by cocultivation with fungi and bacteria. ACS Omega 9:7668–7678
Acknowledgements
The authors are sincerely thankful to Vice Chancellor and Registrar of Periyar University, Salem, for the financial support through University Research Fellowship (URF).
Funding
This project was supported by Researchers Supporting Project number (RSP2024R216) King Saud University, Riyadh, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
R. Palanivelan and S. Ramya: conceptualization, investigation, and writing—original draft. S. Aradhana, Amal Abdullah A. Sabour, and Ranganathan Muthusamy: writing and reviewing and editing, P.M. Ayyasamy and Mathiyazhagan Narayanan: supervision, writing, and reviewing and editing.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Palanivelan, R., Ramya, S., Aradhana, S. et al. Bioprospective decolourization of reactive azo dyes at pilot scale by a developed bacterial consortium using the RSM and CCD model. Biomass Conv. Bioref. (2024). https://doi.org/10.1007/s13399-024-05701-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13399-024-05701-3