Abstract
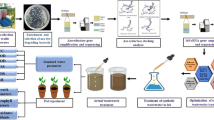
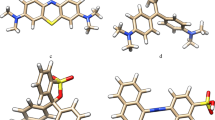
The rise of pollution due to the dye industries and textile wastes are evolving rapidly every day. The dyes are used in different trade names by the textile industries. The actual chemistry of dye is vague and difficult to understand even today though we are equipped technically. The toxic effects of the dyes and the reasons behind the acute toxicity are also an undiscovered mystery; therefore, no effective measures can be employed to degrade dyes. Deploying physical or chemical methods to pre-treat the azo dyes are expensive, extremely energy-consuming, and are not environment friendly. Hence, the use of microbes for textile dye degradation will be eco-friendly and is probably a cost-effective alternative to physicochemical methods. The present study was conducted to investigate the degradation of azo dyes isolated from textile effluent contaminated soil by employing the bacterial strains for degradation. The bacterial strains could degrade the optimum concentration of mixed azo dyes (200 mg/L) with an incubation up to 5 days. The decolourization of the dyes was expressed in terms of percentage of decolourization, and was found that about 87.35% of degradation by Bacillus subtilis strain. The enzyme responsible was analyzed as intracellular azoreductase involved in the degradation of mixed azo dyes. The enzymatic pathway and 1-phenyl-2–4(4-methyl phenyl)-diazene 1-oxide was observed as the major metabolite by GC–MS analysis. The in silico study determined the binding of mixed azo dye with azoreductase and hypothesized that their linking could be the main reason for the degradation of mixed azo dye.





Similar content being viewed by others
References
Ali S, Khatri Z, Khatri A, Tanwari A (2018) Integrated desizing–bleaching–reactive dyeing process for cotton towel using glucose oxidase enzyme. J Clean Prod 66:562–567
Ameer E, Asmaa MM Mawad, Naeima MM Yousef, Ahmed AM Shoreit (2017) Azoreductase kinetics and gene expression in the synthetic dyes-degrading Pseudomonas. EJBAS 315–322
Amoozegar M, Schumann P, Hajighasemi M, Ashengroph M, Razavi M (2008) Salinicoccus iranensis sp. nov., a novel moderate halophile. Int J Syst Evol 58(1):178–183
Anuradha NK, Umesh BJ, Jyoti PJ, Vishwas AB, Sanjay PG (2009) Biotechnological strategies for phytoremediation of the sulfonated azo dye Direct Red 5B using Blumea malcolmii Hook. Bioresour Technol 4104–4110
Aysha A, Shah I, AbuQamar S, Ashraf S (2017) Differential degradation and detoxification of an aromatic pollutant by two different peroxidases. Biomolecules 7(4):31
Bhagirath B, Reddy VR (2002) Degradation of azo and triphenylmethane dye by the bacteria isolated from local industrial waste, Environment and Accountability. EPW 37
Bürger S, Stolz A (2010) Characterization of the flavin-free oxygen-tolerant azoreductase from Xenophilus azovorans KF46F in comparison to flavin-containing azoreductases. Appl Microbiol Biotechnol 87:2067–2076
Chen BY, Chen WM, Wu FL, Chen PK, Yen CY (2008) Revealing azo-dye decolorization of indigenous Aeromonas hydrophila from fountain spring in northeast Taiwan. J Chin Inst Chem Eng 39:495–501
Claus SP, Guillou H, Ellero-Simatos S (2016) The gut microbiota: a major player in the toxicity of environmental pollutants. Npj Biofilms Microbiomes 2:16003
Dutta A, Banerjee P, Sarkar D, Bhattacharjee S, Chakrabarti S (2014) Degradation of Trypan Bluein wastewater by sunlight-assisted modified photo-Fenton reaction. Desalin Water Treat 56(6):1498–1506
Elfarash A, Mawad A, Yousef N, Shoreit A (2017) Azoreductase kinetics and gene expression in the synthetic dyes-degrading Pseudomonas. EJBAS 4(4):315–322
Grosdidier A, Zoete V, Michielin O (2011) Swiss Dock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res 39(suppl):W270–W277
Guo J, Zhou J, Wang D, Tian C, Wang P, Uddin M (2007) A novel moderately halophilic bacterium for decolorizing azo dye under high salt condition. Biodegradation 19(1):15–19
Gupta VK, Khamparia S, Tyagi I, Jaspal D, Malviya A (2015) Decolorization of mixture of dyes: a critical review. Global J Environ Sci and Manag 1:71–94
Gurulakshmi M, Sudarmani DNP, Venba R (2008) Biodegradation of leather acid dye by Bacillus subtilis. Adv Biotech 12–17
Jessica M, Wright C, John G (2012) Identification, Isolation and characterization of a novel azoreductase from Clostridium perfringens. Anaerobe 18(2):229–234
Kadam AA, Telke AA, Jagtap SS, Govindwar SP (2013) Decolorization of adsorbed textile dyes by developed consortium of Pseudomonas sp. SUK1 and Aspergillus ochraceus NCIM-1146 under solid state fermentation. J Hazard Mater 189:486–494
Kalyani DC, Telke AA, Dhanve RS, Jadhav JP (2009) Ecofriendly biodegradation and detoxification of Reactive Red 2 textile dye by newly isolated Pseudomonas sp. J Hazard Mater 163:735–742
Kumar S, Tamura K, Nei M (2004) MEGA 3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bio-inform 5:150–163
Lalnunhlimi S, Krishnaswamy V (2016) Decolorization of azo dyes (Direct Blue 151 and Direct Red 31) by moderately alkaliphilic bacterial consortium. Braz J Microbiol Publ Braz Soc Microbiol 47:39–46
Leelakriangsak M, Borisut S (2012) Characterization of the decolorizing activity of azo dyes by Bacillus subtilis azoreductase AzoR1. Songklanakarin J. Sci. Technol 34(5)
Lowry (1951) Estimation of protein
Lucas M, Peres J (2006) Decolorization of the azo dye Reactive Black 5 by Fenton and photo-Fenton oxidation. Dyes Pigm 71(3):236–244
Macwana SR, Punj S, Cooper J, Schwenk E, John GH (2010) Identification and isolation of an azoreductase from Enterococcus faecium. Curr Issues Mol Biol 12:43–48
Maier J, Kandelbauer A, Erlacher A, Cavaco Paulo A, Gubits GM (2004) A new alkali—thermostable azoreductase from Bacillus sp. Strain SF Appl Environ Microbiol 70:837–844
Misal SA, Humne VT, Lokhande PD, Gawai KR (2015) Biotransformation of nitro aromatic compounds by flavin-free NADH-azoreductase. J Bioremed Biodeg 6:2–6
Oturkar C, Nemade H, Mulik P, Patole M, Hawaldar R, Gawai K (2011) Mechanistic investigation of decolorization and degradation of Reactive Red 120 by Bacillus lentus BI377. Bioresour Technol 102(2):758–764
Phugare S, Kalyani D, Patil A, Jadhav J (2011) Textile dye degradation by bacterial consortium and subsequent toxicological analysis of dye and dye metabolites using cytotoxicity, genotoxicity and oxidative stress studies. J Hazard Mater 186(10):713–723
Pradhan P, Gireesh Babu K (2012) Isolation and identification of methanogenic bacteria from cow dung. Int J Curr Res 4(7):28–31
Purwanto A, Chen A, Shien K, Huebschmann H-J (2012) Detection, identification, and quantitation of Azo dyes in leather and textiles by GC/MS. Thermo Fisher Scientific, Singapore
Qi J, Anke MK, Szymańska K, Tischler D (2017) Immobilization of Rhodococcus opacus 1CP azoreductase to obtain azo dye degrading biocatalysts operative at acidic pH. Int Biodeterior Biodegrad 118:89–94
Ramalho PA, Cardoso MH, Cavaco-Paulo A, Ramalho MT (2004) Characterization of azo reduction activity in a novel Ascomycete yeast strain. Appl Environ Microbiol 70:2279–2288
Robinson M, Marchant R, Nigam P (2001) Remediation of dyes in textile effluent: a critical review on current treatment technologies with a proposed alternative. Bioresour Technol 77:247
Sahasrabudhe MM, Saratale RG, Saratale GD, Pathade GR (2014) Decolorization and detoxification of sulfonated toxic diazo dye C.I. Direct Red 81 by Enterococcus faecalis YZ 66. J Environ Health Sci Eng 12
Saranraj P, Stella D, Sivasakthivelan P (2014) Separation, purification and characterization of dye degrading enzyme azoreductase from bacterial isolates. Cent Eur J Exp Biol 3(2):19–25
Saratale R, Saratale G, Chang J, Govindwar S (2011) Bacterial decolorization and degradation of azo dyes: a review. J Taiwan Inst Chem Eng 42(1):138–157
Selva RD, Jaisy PR, Leena R (2011) Analysis of bacterial degradation of azo dye congo red using HPLC). J. Ind. Pollut. Control 28(1):57–68
Shah MP, Ka P, Ss N, Am D (2013) Potential effect of two Bacillus spp on decolorization of azo dye. J Bioremediation Biodegrad 4:1–4
Singh SN (2015) Microbial degradation of synthetic dyes in wastewaters. Springer, Switzerland Strain of Serratia maerascens. World J Microbiol Biotechnol 19:615
Sudha M, Bakiyaraj G, Saranya A, Sivakumar N, Selvakumar G (2018) Prospective assessment of the Enterobacter aerogenes PP002 in decolorization and degradation of azo dyes DB 71 and DG 28. J Environ Chem Eng 6(1):95–109
Telke AD, Kalyani J, Jadhav GS (2008) Kinetics and Mechanism of Reactive Red 141 Degradation by a Bacterial Isolate Rhizobium radiobacter MTCC 8161. Acta Chim Slov 55:320
Wang B, Xu M, Sun G (2010) Extracellular respiration of different amounts azo dye by Shewanella decolorationis S12 and comparative analysis of the membrane proteome. Int Biodeterior 64(4):274–280
Yu J, Ogata D, Gai Z, Taguchi S, Tanaka I, Ooi T, Yao M (2014) Structures of AzrA and of AzrC complexed with substrate or inhibitor: insight into substrate specificity and catalytic mechanism. Acta Crystallogr D Biol Crystallogr 70(2):553–564
Zahran SA, Ali-Tammam M, Hashem AM et al (2019) Azoreductase activity of dye-decolorizing bacteria isolated from the human gut microbiota. Sci Rep 9:5508
Zimmerman T, Kulla HG, Leisinger T (1982) Properties of Purified Orange II Azoreductase, the Enzyme Initiating Azo Dye Degradation by Pseudomonas KF 46. Eur J Biochem 129:197–203
Acknowledgements
We would like to thank UGC-MRP-SERO MRP-2017 (6970/16) for funding this project. We would like to thank DST-FIST CRIST LAB, Stella Maris College (Autonomous). We would like to extend our gratitude to SAIF IITM, Department of Chemistry IITM for SEM analysis and VIT, Vellore.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest for the manuscript.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Krithika, A., Gayathri, K.V., Kumar, D.T. et al. Mixed azo dyes degradation by an intracellular azoreductase enzyme from alkaliphilic Bacillus subtilis: a molecular docking study. Arch Microbiol 203, 3033–3044 (2021). https://doi.org/10.1007/s00203-021-02299-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02299-2