Abstract
Fungal diversity notes is one of the important journal series of fungal taxonomy that provide detailed descriptions and illustrations of new fungal taxa, as well as providing new information of fungal taxa worldwide. This article is the 11th contribution to the fungal diversity notes series, in which 126 taxa distributed in two phyla, six classes, 24 orders and 55 families are described and illustrated. Taxa in this study were mainly collected from Italy by Erio Camporesi and also collected from China, India and Thailand, as well as in some other European, North American and South American countries. Taxa described in the present study include two new families, 12 new genera, 82 new species, five new combinations and 25 new records on new hosts and new geographical distributions as well as sexual-asexual reports. The two new families are Eriomycetaceae (Dothideomycetes, family incertae sedis) and Fasciatisporaceae (Xylariales, Sordariomycetes). The twelve new genera comprise Bhagirathimyces (Phaeosphaeriaceae), Camporesiomyces (Tubeufiaceae), Eriocamporesia (Cryphonectriaceae), Eriomyces (Eriomycetaceae), Neomonodictys (Pleurotheciaceae), Paraloratospora (Phaeosphaeriaceae), Paramonodictys (Parabambusicolaceae), Pseudoconlarium (Diaporthomycetidae, genus incertae sedis), Pseudomurilentithecium (Lentitheciaceae), Setoapiospora (Muyocopronaceae), Srinivasanomyces (Vibrisseaceae) and Xenoanthostomella (Xylariales, genera incertae sedis). The 82 new species comprise Acremonium chiangraiense, Adustochaete nivea, Angustimassarina camporesii, Bhagirathimyces himalayensis, Brunneoclavispora camporesii, Camarosporidiella camporesii, Camporesiomyces mali, Camposporium appendiculatum, Camposporium multiseptatum, Camposporium septatum, Canalisporium aquaticium, Clonostachys eriocamporesiana, Clonostachys eriocamporesii, Colletotrichum hederiicola, Coniochaeta vineae, Conioscypha verrucosa, Cortinarius ainsworthii, Cortinarius aurae, Cortinarius britannicus, Cortinarius heatherae, Cortinarius scoticus, Cortinarius subsaniosus, Cytospora fusispora, Cytospora rosigena, Diaporthe camporesii, Diaporthe nigra, Diatrypella yunnanensis, Dictyosporium muriformis, Didymella camporesii, Diutina bernali, Diutina sipiczkii, Eriocamporesia aurantia, Eriomyces heveae, Ernakulamia tanakae, Falciformispora uttaraditensis, Fasciatispora cocoes, Foliophoma camporesii, Fuscostagonospora camporesii, Helvella subtinta, Kalmusia erioi, Keissleriella camporesiana, Keissleriella camporesii, Lanspora cylindrospora, Loratospora arezzoensis, Mariannaea atlantica, Melanographium phoenicis, Montagnula camporesii, Neodidymelliopsis camporesii, Neokalmusia kunmingensis, Neoleptosporella camporesiana, Neomonodictys muriformis, Neomyrmecridium guizhouense, Neosetophoma camporesii, Paraloratospora camporesii, Paramonodictys solitarius, Periconia palmicola, Plenodomus triseptatus, Pseudocamarosporium camporesii, Pseudocercospora maetaengensis, Pseudochaetosphaeronema kunmingense, Pseudoconlarium punctiforme, Pseudodactylaria camporesiana, Pseudomurilentithecium camporesii, Pseudotetraploa rajmachiensis, Pseudotruncatella camporesii, Rhexocercosporidium senecionis, Rhytidhysteron camporesii, Rhytidhysteron erioi, Septoriella camporesii, Setoapiospora thailandica, Srinivasanomyces kangrensis, Tetraploa dwibahubeeja, Tetraploa pseudoaristata, Tetraploa thrayabahubeeja, Torula camporesii, Tremateia camporesii, Tremateia lamiacearum, Uzbekistanica pruni, Verruconis mangrovei, Wilcoxina verruculosa, Xenoanthostomella chromolaenae and Xenodidymella camporesii. The five new combinations are Camporesiomyces patagoniensis, Camporesiomyces vaccinia, Camposporium lycopodiellae, Paraloratospora gahniae and Rhexocercosporidium microsporum. The 22 new records on host and geographical distribution comprise Arthrinium marii, Ascochyta medicaginicola, Ascochyta pisi, Astrocystis bambusicola, Camposporium pellucidum, Dendryphiella phitsanulokensis, Diaporthe foeniculina, Didymella macrostoma, Diplodia mutila, Diplodia seriata, Heterosphaeria patella, Hysterobrevium constrictum, Neodidymelliopsis ranunculi, Neovaginatispora fuckelii, Nothophoma quercina, Occultibambusa bambusae, Phaeosphaeria chinensis, Pseudopestalotiopsis theae, Pyxine berteriana, Tetraploa sasicola, Torula gaodangensis and Wojnowiciella dactylidis. In addition, the sexual morphs of Dissoconium eucalypti and Phaeosphaeriopsis pseudoagavacearum are reported from Laurus nobilis and Yucca gloriosa in Italy, respectively. The holomorph of Diaporthe cynaroidis is also reported for the first time.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Table of contents
Phylum Ascomycota R.H. Whittaker
Subphylum Pezizomycotina O.E. Erikss. & Winka
Class Dothideomycetes O.E. Erikss. & Winka
Subclass Dothideomycetidae P.M. Kirk et al.
Capnodiales Woron.
Dissoconiaceae Crous & de Hoog
1151.Dissoconium eucalypti Crous & Carnegie, in Crous et al., Fungal Divers. 26(1): 157 (2007), new record of the sexual morph (contributed by Rekhani H. Perera)
Mycosphaerellaceae Lindau
1152.Pseudocercospora maetaengensis J.F. Li & Phookamsak, sp. nov. (contributed by Junfu Li and Rungtiwa Phookamsak)
Subclass Pleosporomycetidae C.L. Schoch et al.
Hysteriales Lindau
Hysteriaceae Chevall.
1153.Hysterobrevium constrictum (N. Amano) E. Boehm & C.L. Schoch, in Boehm et al., Stud. Mycol. 64: 64 (2009), new record for Yunnan, China (contributed by Anusha H. Ekanayaka)
1154.Rhytidhysteron camporesii Ekanayaka & K.D. Hyde, sp. nov. (contributed by Anusha H. Ekanayaka)
1155.Rhytidhysteron erioi Ekanayaka & K.D. Hyde, sp. nov. (contributed by Anusha H. Ekanayaka)
Pleosporales Luttrell ex M.E. Barr
Amorosiaceae Thambug. & K.D. Hyde
1156.Angustimassarina camporesii D. Pem, Doilom & K.D. Hyde, sp. nov. (contributed by Dhandevi Pem and Mingkwan Doilom)
Camarosporidiellaceae Wanas. et al.
1157.Camarosporidiella camporesii Tibpromma & K.D. Hyde, sp. nov. (contributed by Saowaluck Tibpromma)
Coniothyriaceae W.B. Cooke
1158.Foliophoma camporesii D. Pem, Doilom & K.D. Hyde, sp. nov. (contributed by Dhandevi Pem and Mingkwan Doilom)
Dictyosporiaceae Boonmee & K.D. Hyde
1159.Dendryphiella phitsanulokensis N.G. Liu & K.D. Hyde, in Hyde et al., Mycosphere 9(2): 287 (2018), new host record (contributed by Kasun M. Thambugala and Naruemon Huanraluek)
1160.Dictyosporium muriformis N.G. Liu, K.D. Hyde & J.K. Liu, sp. nov. (contributed by Ning-Guo Liu and Jian-Kui (Jack) Liu)
Didymellaceae Gruyter et al.
1161.Ascochyta medicaginicola Qian Chen & L. Cai, Stud. Mycol. 82: 187 (2015), new record for Italy (contributed by Pranami D. Abeywickrama)
1162.Ascochyta pisi Lib., Pl. crypt. Arduenna, fasc. (Liège) 1(nos 1-100): no. 59 (1830), new host record (contributed by Subodini N. Wijesinghe)
1163.Didymella camporesii Manawasinghe & K.D. Hyde, sp. nov. (contributed by Ishara S. Manawasinghe)
1164.Didymella macrostoma (Mont.) Qian Chen & L. Cai, in Chen et al., Stud. Mycol. 82: 177 (2015), new host record (contributed by Pranami D. Abeywickrama)
1165.Neodidymelliopsis camporesii D. Pem, Doilom & K.D. Hyde, sp. nov. (contributed by Dhandevi Pem and Mingkwan Doilom)
1166.Neodidymelliopsis ranunculi W.J. Li & K.D. Hyde, in Hyde et al., Fungal Divers.: https://doi.org/10.1007/s13225-016-0373-x, [41] (2016), new host record (contributed by Pranami D. Abeywickrama)
1167.Nothophoma quercina (Syd. & P. Syd.) Qian Chen & L. Cai, Stud. Mycol. 82: 213 (2015), new host record (contributed by Napalai Chaiwan)
1168.Xenodidymella camporesii D. Pem, Doilom & K.D. Hyde, sp. nov. (contributed by Dhandevi Pem and Mingkwan Doilom)
Didymosphaeriaceae Munk
1169.Kalmusia erioi Samarak., Thambugala & K.D. Hyde, sp. nov. (contributed by Milan C. Samarakoon)
1170.Montagnula camporesii Phukhams. & K.D. Hyde, sp. nov. (contributed by Chayanard Phukhamsakda)
1171.Neokalmusia kunmingensis H.B. Jiang, Phookamsak & K.D. Hyde, sp. nov. (contributed by Hong-Bo Jiang and Rungtiwa Phookamsak)
1172.Pseudocamarosporium camporesii Q. Tian & K.D. Hyde, sp. nov. (contributed by Qing Tian)
1173.Tremateia lamiacearum Samarak. & K.D. Hyde, sp. nov. (contributed by Milan C. Samarakoon)
1174.Tremateia camporesii Samarak. & K.D. Hyde, sp. nov. (contributed by Milan C. Samarakoon)
Fuscostagonosporaceae Jayasiri, Camporesi & K.D. Hyde
1175.Fuscostagonospora camporesii Tennakoon & K.D. Hyde, sp. nov. (contributed by Danushka S. Tennakoon)
Halotthiaceae Ying Zhang et al.
1176.Brunneoclavispora camporesii Boonmee & Phookamsak, sp. nov. (contributed by Saranyaphat Boonmee and Rungtiwa Phookamsak)
Lentitheciaceae Yin. Zhang et al.
1177.Keissleriella camporesiana Phukhams. & K.D. Hyde, sp. nov. (contributed by Chayanard Phukhamsakda)
1178.Keissleriella camporesii C.G. Lin & K.D. Hyde, sp. nov. (contributed by Chuan-Gen Lin)
1179.Pseudomurilentithecium Mapook & K.D. Hyde, gen. nov. (contributed by Ausana Mapook)
1180.Pseudomurilentithecium camporesii Mapook & K.D. Hyde, sp. nov. (contributed by Ausana Mapook)
Leptosphaeriaceae M.E. Barr
1181.Plenodomus triseptatus S.N. Wijesinghe, Bulgakov & K.D. Hyde, sp. nov. (contributed by Subodini N. Wijesinghe)
Lophiostomataceae Sacc.
1182.Neovaginatispora fuckelii (Sacc.) A. Hashim., K. Hiray. & Kaz. Tanaka, in Hashimoto et al., Stud. Mycol. 90: 188 (2018), new host record (contributed by Shi-Ke Huang)
Macrodiplodiopsidaceae Voglmayr et al.
1183.Pseudochaetosphaeronema kunmingense D.P. Wei, Wanas. & K.D. Hyde, sp. nov. (contributed by Deping Wei)
Melanommataceae G. Winter
1184.Camposporium appendiculatum D.F. Bao, Z.L. Luo, K.D. Hyde & H.Y. Su, sp. nov. (contributed by Dan-Feng Bao)
1185.Camposporium lycopodiellae (Crous & R.K. Schumach.) Tibpromma & K.D. Hyde, comb. nov. (contributed by Saowaluck Tibpromma)
1186.Camposporium multiseptatum D.F. Bao, Z.L. Luo, K.D. Hyde & H.Y. Su, sp. nov. (contributed by Dan-Feng Bao)
1187.Camposporium pellucidum (Grove) S. Hughes, Mycol. Pap. 36: 9 (1951), new record for Yunnan, China (contributed by Dan-Feng Bao)
1188.Camposporium septatum N.G. Liu, J.K. Liu & K.D. Hyde, sp. nov. (contributed by Ning-Guo Liu)
1189.Uzbekistanica pruni Chaiwan, Wanas., Bulgakov & K.D. Hyde, sp. nov. (contributed by Napalai Chaiwan)
Occultibambusaceae D.Q. Dai & K.D. Hyde
1190.Occultibambusa bambusae D.Q. Dai & K.D. Hyde, in Dai et al., Fungal Divers.: https://doi.org/10.1007/s13225-016-0367-8, [26] (2016), new host record from Taiwan (contributed by Anuruddha Karunarathna and Ruvishika S. Jayawardena)
Parabambusicolaceae Kaz. Tanaka & K. Hiray.
1191.Paramonodictys N.G. Liu, K.D. Hyde & J.K. Liu, gen. nov. (contributed by Ning-Guo Liu and Jian-Kui (Jack) Liu)
1192.Paramonodictys solitarius N.G. Liu, K.D. Hyde & J.K. Liu, sp. nov. (contributed by Ning-Guo Liu and Jian-Kui (Jack) Liu)
Periconiaceae (Sacc.) Nann.
1193.Periconia palmicola J.F. Li & Phookamsak, sp. nov. (contributed by Junfu Li and Rungtiwa Phookamsak)
Phaeosphaeriaceae M.E. Barr
1194.Bhagirathimyces S.M. Singh & S.K. Singh, gen. nov. (contributed by Sanjay K. Singh and S.M. Singh)
1195.Bhagirathimyces himalayensis S.M. Singh & S.K. Singh, sp. nov. (contributed by Sanjay K. Singh and Shiv Mohan Singh)
1196.Loratospora arezzoensis Bundhun, Wanas., Jeewon & K.D. Hyde, sp. nov. (contributed by Digvijayini Bundhun)
1197.Neosetophoma camporesii Q. Tian & K.D. Hyde, sp. nov. (contributed by Qing Tian)
1198.Paraloratospora Bundhun, Tennakoon, Phookamsak & K.D. Hyde, gen. nov. (contributed by Digvijayini Bundhun and Rungtiwa Phookamsak)
1199.Paraloratospora camporesii Bundhun, Jeewon & K.D. Hyde, sp. nov. (contributed by Digvijayini Bundhun)
1200.Paraloratospora gahniae (Crous) Thiyagaraja, Bundhun & K.D. Hyde, comb. nov. (contributed by Vinodhini Thiyagaraja)
1201.Phaeosphaeria chinensis K.K. Zhang, Hongsanan, Tennakoon & N. Xie, Phytotaxa. 419(1): 32 (2019), new host record from Taiwan (contributed by Anuruddha Karunarathna and Ruvishika S. Jayawardena)
1202.Phaeosphaeriopsis pseudoagavacearum Crous & Y. Marín, in Marin-Felix et al., Stud. Mycol. 94: 63 (2019), new record of the sexual morph (contributed by Rungtiwa Phookamsak)
1203.Septoriella camporesii Goonas. & K.D. Hyde, sp. nov. (contributed by Ishani D. Goonasekara)
1204.Wojnowiciella dactylidis (Wijayaw., Camporesi & K.D. Hyde) Hern.-Restr. & Crous, Sydowia 68: 221 (2016), new host record (contributed by Danushka S. Tennakoon)
Tetraplosphaeriaceae Kaz. Tanaka & K. Hiray.
1205.Ernakulamia tanakae Rajeshkumar & K.D. Hyde, sp. nov. (contributed by Kunhiraman C. Rajeshkumar)
1206.Pseudotetraploa rajmachiensis Rajeshkumar, K.D. Hyde & S. Lad, sp. nov. (contributed by Kunhiraman C. Rajeshkumar and Sneha S. Lad)
1207.Tetraploa dwibahubeeja Rajeshkumar, K.D. Hyde & S. Lad, sp. nov. (contributed by Kunhiraman C. Rajeshkumar and Sneha S. Lad)
1208.Tetraploa pseudoaristata Rajeshkumar, K.D. Hyde & G. Anand, sp. nov. (contributed by Kunhiraman C. Rajeshkumar and Garima Anand)
1209.Tetraploa thrayabahubeeja Rajeshkumar, K.D. Hyde & G. Anand, sp. nov. (contributed by Kunhiraman C. Rajeshkumar and Garima Anand)
1210.Tetraploa sasicola (Kaz. Tanaka & K. Hiray.) Kaz. Tanaka & K. Hiray., Fungal Divers. 63: 253 (2013), new host record from Taiwan (contributed by Anuruddha Karunarathna and Ruvishika S. Jayawardena)
Torulaceae Corda
1211.Torula camporesii Phookamsak, E.F. Yang & K.D. Hyde, sp. nov. (contributed by Rungtiwa Phookamsak and Er-Fu Yang)
1212.Torula gaodangensis J. Yang & K.D. Hyde, in Hyde et al., Fungal Divers. 87: 113 (2017), new host record for Yunnan, China (contributed by Er-Fu Yang and Rungtiwa Phookamsak)
Trematosphaeriaceae K.D. Hyde et al.
1213.Falciformispora uttaraditensis Boonmee, Huanraluek & K.D. Hyde, sp. nov. (contributed by Saranyaphat Boonmee and Naruemon Huanraluek)
Dothideomycetes, order incertae sedis
Botryosphaeriales C.L. Schoch et al.
Botryosphaeriaceae Theiss. & H. Syd.
1214.Diplodia mutila (Fr.) Mont., Annls Sci. Nat., Bot., sér. 2 1: 302 (1834), new host record (contributed by Pranami D. Abeywickrama)
1215.Diplodia seriata De Not., Mém. R. Accad. Sci. Torino, Ser. 2 7: 26 (1845), new host record (contributed by Ishara S. Manawasinghe)
Muyocopronales Mapook, Boonmee & K.D. Hyde
Muyocopronaceae K.D. Hyde
1216.Setoapiospora Mapook & K.D. Hyde, gen. nov. (contributed by Ausana Mapook)
1217.Setoapiospora thailandica Mapook & K.D. Hyde, sp. nov. (contributed by Ausana Mapook)
Tubeufiales Boonmee & K.D. Hyde
Tubeufiaceae M.E. Barr
1218.Camporesiomyces D.P. Wei & K.D. Hyde, gen. nov. (contributed by De-Ping Wei and Dhanushaka N. Wanasinghe)
1219.Camporesiomyces mali D.P. Wei & K.D. Hyde, sp. nov. (contributed by De-Ping Wei and Dhanushaka N. Wanasinghe)
1220.Camporesiomyces patagoniensis (R.M. Sánchez, A.N. Mill. & Bianchin) D.P. Wei & K.D. Hyde, comb. nov. (contributed by De-Ping Wei and Dhanushaka N. Wanasinghe)
1221.Camporesiomyces vaccinii (Carris) D.P. Wei & K.D. Hyde, comb. nov. (contributed by De-Ping Wei and Dhanushaka N. Wanasinghe)
Venturiales Y. Zhang ter, C.L. Schoch & K.D. Hyde
Sympoventuriaceae Y. Zhang ter, C.L. Schoch & K.D. Hyde
1222.Verruconis mangrovei Devadatha, V.V. Sarma & E.B.G. Jones, sp. nov. (contributed by Bandarupalli Devadatha, V. Venkateswara Sarma and E.B. Gareth Jones)
Dothideomycetes, familyincertae sedis
1223.Eriomycetaceae Huanraluek & K.D. Hyde, fam. nov. (contributed by Kasun M. Thambugala and Naruemon Huanraluek)
1224.Eriomyces Huanraluek, Thambugala & K.D. Hyde, gen. nov. (contributed by Kasun M. Thambugala and Naruemon Huanraluek)
1225.Eriomyces heveae Huanraluek, Thambugala & K.D. Hyde, sp. nov. (contributed by Kasun M. Thambugala and Naruemon Huanraluek)
Class Lecanoromycetes O.E. Erikss. & Winka
Subclass Lecanoromycetidae P.M. Kirk et al. ex Miadl. et al.
Caliciales Bessey
Caliciaceae Chevall.
1226.Pyxine berteriana (Fée) Imshaug, Trans. Am. Microsc. 76: 254 (1957), new host record (contributed by Vinodhini Thiyagaraja)
Class Leotiomycetes O.E. Erikss. & Winka
Helotiales Nannf.
Heterosphaeriaceae Rehm
1227.Heterosphaeria patella (Tode) Grev., Scott. crypt. fl. (Edinburgh) 2: 103 (1823), new host reord (contributed by Kunthida Phutthacharoen)
Ploettnerulaceae Kirschst.
1228.Rhexocercosporidium microsporum (Ekanayaka & K.D. Hyde) Phutthacharoen & K.D. Hyde, comb. nov. (contributed by Kunthida Phutthacharoen)
1229.Rhexocercosporidium senecionis Phutthacharoen, Ekanayaka & K.D. Hyde, sp. nov. (contributed by Kunthida Phutthacharoen)
Vibrisseaceae Korf
1230.Srinivasanomyces S. Rana & S.K. Singh, gen. nov. (contributed by Sanjay K. Singh and Shiwali Rana)
1231.Srinivasanomyces kangrensis S. Rana & S.K. Singh, sp. nov. (contributed by Sanjay K. Singh and Shiwali Rana)
Class Pezizomycetes O.E. Erikss. & Winka
Pezizales J. Schröt.
Helvellaceae Fr.
1232.Helvella subtinta M. Zeng, Q. Zhao & K.D. Hyde, sp. nov. (contributed by Ming Zeng and Qi Zhao)
Pyronemataceae Corda
1233.Wilcoxina verruculosa M. Zeng, Q. Zhao & K.D. Hyde, sp. nov. (contributed by Ming Zeng and Qi Zhao)
Class Sordariomycetes O.E. Erikss. & Winka
Subclass Diaporthomycetidae Senan. et al.
Diaporthales Nannf.
Cryphonectriaceae Gryzenh. & M.J. Wingf.
1234.Eriocamporesia R.H. Perera, Samarak. & K.D. Hyde, gen. nov. (contributed by Rekhani H. Perera and Milan C. Samarakoon)
1235.Eriocamporesia aurantia R.H. Perera, Samarak. & K.D. Hyde, sp. nov. (contributed by Rekhani H. Perera and Milan C. Samarakoon)
Cytosporaceae
1236.Cytospora fusispora M. Niranjan & V.V. Sarma, sp. nov. (contributed by M. Niranjan and V. Venkateswara Sarma)
1237.Cytospora rosigena Chaiwan, Wanas., Bulgakov & K.D. Hyde, sp. nov. (contributed by Napalai Chaiwan)
Diaporthaceae
1238.Diaporthe camporesii Manawasinghe & K.D. Hyde, sp. nov. (contributed by Ishara S. Manawasinghe and Indunil C. Senanayake)
1239.Diaporthe cynaroidis Marinc., M.J. Wingf. & Crous, CBS Diversity Ser. (Utrecht) 7: 39 (2008), new record of the sexual-asexual connection (contributed by Indunil C. Senanayake)
1240.Diaporthe foeniculina (Sacc.) Udayanga & Castl., in Udayanga et al., Persoonia 32: 95 (2014), new host record from Italy (contributed by Pranami D. Abeywickrama and Indunil C. Senanayake)
1241.Diaporthe nigra Brahmanage & K.D. Hyde, sp. nov. (contributed by Rashika S. Brahmanage)
Myrmecridiales Crous
Myrmecridiaceae Crous
1242.Neomyrmecridium guizhouense N.G. Liu, K.D. Hyde & J.K. Liu, sp. nov. (contributed by Ning-Guo Liu)
Phomatosporales Senan. et al.
Phomatosporaceae Senan. & K.D. Hyde
1243.Lanspora cylindrospora Devadatha, V.V. Sarma & E.B.G. Jones, sp. nov. (contributed by Bandarupalli Devadatha, V. Venkateswara Sarma and E.B. Gareth Jones)
Diaporthomycetidae, genus incertae sedis
1244.Pseudoconlarium N.G. Liu, K.D. Hyde & J.K. Liu, gen. nov. (contributed by Ning-Guo Liu and Jian-Kui (Jack) Liu)
1245.Pseudoconlarium punctiforme N.G. Liu, K.D. Hyde & J.K. Liu, sp. nov. (contributed by Ning-Guo Liu and Jian-Kui (Jack) Liu)
Subclass Hypocreomycetidae O.E. Erikss. & Winka
Glomerellales Chadef. ex Réblová et al.
Glomerellaceae Locq. ex Seifert & W. Gams
1246.Colletotrichum hederiicola Jayaward. & K.D. Hyde, sp. nov. (contributed by Ruvishika S. Jayawardena)
Hypocreales Lindau
Bionectriaceae Samuels & Rossman
1247.Acremonium chiangraiense J.F. Li, R.H. Perera & Phookamsak, sp. nov. (contributed by Junfu Li, Rekhani H. Perera and Rungtiwa Phookamsak)
1248.Clonostachys eriocamporesiana R.H. Perera & K.D. Hyde, sp. nov. (contributed by Rekhani H. Perera)
1249.Clonostachys eriocamporesii R.H. Perera & K.D. Hyde, sp. nov. (contributed by Rekhani H. Perera)
Nectriaceae Tul. & C. Tul.
1250.Mariannaea atlantica A.L. Alves, A.C.S Santos & P.V. Tiago, sp. nov. (contributed by Amanda Lucia Alves, Ana Carla da Silva Santos and Patricia Vieira Tiago)
Subclass Savoryellomycetidae Hongsanan et al.
Conioscyphales Réblová & Seifert
Conioscyphaceae Réblová & Seifert
1251.Conioscypha verrucosa J. Yang & K.D. Hyde, sp. nov. (contributed by Jing Yang)
Pleurotheciales Réblová & Seifert
Pleurotheciaceae Réblová & Seifert
1252.Neomonodictys Y.Z. Lu, C.G. Lin & K.D. Hyde, gen. nov. (contributed by Yong-Zhong Lu and Chuan-Gen Lin)
1253.Neomonodictys muriformis Y.Z. Lu, C.G. Lin & K.D. Hyde, sp. nov. (contributed by Yong-Zhong Lu and Chuan-Gen Lin)
Savoryellales Boonyuen et al.
Savoryellaceae Jaklitsch & Réblová
1254.Canalisporium aquaticium J. Yang & K.D. Hyde, sp. nov. (contributed by Jing Yang)
Subclass Sordariomycetidae O.E. Erikss & Winka
Coniochaetales Huhndorf et al.
Coniochaetaceae Malloch & Cain
1255.Coniochaeta vineae S.K. Huang & K.D. Hyde, sp. nov. (contributed by Shi-Ke Huang)
Pseudodactylariales Crous
Pseudodactylariaceae Crous
1256.Pseudodactylaria camporesiana W. Dong, Doilom & K.D. Hyde, sp. nov. (contributed by Wei Dong and Mingkwan Doilom)
Chaetosphaeriales , genera incertae sedis
1257.Neoleptosporella camporesiana R.H. Perera & K.D. Hyde, sp. nov. (contributed by Rekhani H. Perera)
Subclass Xylariomycetidae O.E. Erikss & Winka
Amphisphaeriales D. Hawksw. & O.E. Erikss.
Apiosporaceae K.D. Hyde et al.
1258.Arthrinium marii Larrondo & Calvo, Mycologia 82 (3): 397 (1990), new host record from Italy (contributed by Kasun M. Thambugala)
Pseudotruncatellaceae Crous
1259.Pseudotruncatella camporesii Goonas. & K.D. Hyde, sp. nov. (contributed by Ishani D. Goonasekara)
Sporocadaceae Corda
1260.Pseudopestalotiopsis theae (Sawada) Maharachch., K.D. Hyde & Crous, in Maharachchikumbura et al., Stud. Mycol. 79: 183 (2014), new record for Guangdong, China (contributed by Indunil C. Senanayake)
Xylariales Nannf.
Diatrypaceae Nitschke
1261.Diatrypella yunnanensis Brahmanage, Thyagaraja & K.D. Hyde, sp. nov. (contributed by Rashika S. Brahmanage)
1262.Fasciatisporaceae S.N. Zhang, K.D. Hyde & J.K. Liu, fam. nov. (contributed by Sheng-Nan Zhang and Jian-Kui (Jack) Liu)
1263.Fasciatispora cocoes S.N. Zhang, K.D. Hyde & J.K. Liu, sp. nov. (contributed by Sheng-Nan Zhang and Jian-Kui (Jack) Liu)
Xylariaceae Tul. & C. Tul.
1264.Astrocystis bambusicola R.H. Perera & K.D. Hyde, in Hyde et al., Index Fungorum 347: 1 (2017), new record from Yunnan, China (contributed by Hong-Bo Jiang and Rungtiwa Phookamsak)
Xylariales , genera incertae sedis
1265.Melanographium phoenicis S.N. Zhang, K.D. Hyde & J.K. Liu, sp. nov. (contributed by Sheng-Nan Zhang and Jian-Kui (Jack) Liu)
1266.Xenoanthostomella Mapook & K.D. Hyde, gen. nov. (contributed by Ausana Mapook)
1267.Xenoanthostomella chromolaenae Mapook & K.D. Hyde, sp. nov. (contributed by Ausana Mapook)
Subphylum Saccharomycotina O.E. Erikss. & Winka
Class Saccharomycetes O.E. Erikss. & Winka
Saccharomycetales, genusincertae sedis
1268.Diutina bernali Haelew., Pfliegler, Horváth & Imre, sp. nov. (contributed by Walter P. Pfliegler and Enikő Horváth)
1269.Diutina sipiczkii Pfliegler, Haelew., Horváth & Imre, sp. nov. (contributed by Alexandra Imre and Danny Haelewaters)
Phylum Basidiomycota R.T. Moore
Subphylum Agaricomycotina Doweld
Class Agaricomycetes Doweld
Subclass Agaricomycetidae Parmasto
Agaricales Underw.
Cortinariaceae R. Heim ex Pouzar
1270.Cortinarius ainsworthii Liimat. & Niskanen, sp. nov. (contributed by Kare Liimatainen and Tuula Niskanen)
1271.Cortinarius aurae Niskanen & Liimat., sp. nov. (contributed by Kare Liimatainen and Tuula Niskanen)
1272.Cortinarius britannicus Liimat. & Niskanen, sp. nov. (contributed by Kare Liimatainen and Tuula Niskanen)
1273.Cortinarius heatherae Overall, sp. nov. (contributed by Andy Overall)
1274.Cortinarius scoticus Niskanen & Liimat., sp. nov. (contributed by Tuula Niskanen and Kare Liimatainen)
1275.Cortinarius subsaniosus Liimat. & Niskanen, sp. nov. (contributed by Kare Liimatainen and Tuula Niskanen)
Subclass Auriculariomycetidae Jülich
Auriculariales J. Schröt.
Auriculariaceae Fr.
1276.Adustochaete nivea Alvarenga, sp. nov. (contributed by Renato Lúcio Mendes Alvarenga and Tatiana Baptista Gibertoni)
Introduction
Fungi have been under studied for more than 40 years and species concepts are still confused. This was mainly because of a lack of reliable methods to resolve species taxonomy and reliance on morphological characters and therefore rather subjective classifications (Dayarathne et al. 2016). This meant that when identifying a collection of a taxon, the preferred option meant clumping a species into an existing name (see Than et al. 2008). It is only in the past 15 years that molecular data has provided a better understanding of a species, and this has resulted in a great increase in the number of new species described in recent years (Hyde et al. 2018b, c). Secondly, fungi are neither plants nor animals and thus not studied by many researchers, nor are they common organisms investigated by microbiologists (Hawksworth 1981) and therefore the study of mycology has seen a decline since the 1980s. Thirdly, most previous research was in temperate countries and was on the decline (e.g. at Exeter and Portsmouth Universities and IMI in the UK). In the tropics and many temperate countries, funding for most areas of science was minimal. However, with the rise of Asian and other world economies, more funding has been placed in science in these countries, resulting in an expansion in research, including mycology. This has resulted in many more scientific publications coming out of Asia (see Dai et al. 2015) and Brazil and relatively less from Europe and the USA.
In plant pathology, fungal species were identified to what are now known as species complexes (Jayawardena et al. 2016), however, with the use of molecular data it has become easier to define a species in pathogenic genera (Hyde et al. 2014; Nilsson et al. 2014). Hyde and Alcorn (1993) and Hyde and Philemon (1994) published checklists of the pathogens of northern Australia and the Western Province of Papua New Guinea based solely on morphology and we now suspect that many of the taxa identified, were wrongly named. For example, in the case of Colletotrichum, Hyde and Alcorn (1993) identified several Colletotrichum species, including C. gloeosporioides, which have now been shown to be species complexes. Hyde et al. (2009) and Cai et al. (2009) published papers which were the start of a complete change to the taxonomic understanding of Colletotrichum species predominantly based on sequence data. Therefore, Phoulivong et al. (2010) was able to show that C. gloeosporioides did not occur in the tropics. Subsequent publications (e.g. Cannon et al. 2012; Damm et al. 2013; Huang et al. 2014) resulted in drastic changes in the understanding of the genus. Hyde et al. (2014) provided a “One Stop Shop” for identifying species in the genus and this was updated by Jayawardena et al. (2019a, b, c). Similar advances have taken places in all other plant pathogenic genera, such as Botryosphaeriales (Dissanayake et al. 2016; Phillips et al. 2019), Diaporthe (Dissanayake et al. 2017b), Fusarium (Jayawardena et al. 2019b) and Phyllosticta (Wikee et al. 2013) and it is now essential to use molecular data to identify species in the majority of plant pathogenic genera (Jayawardena et al. 2019a, b, c).
Similarly, many asexual hyphomycetes and coelomycetes have now been linked to the sexual counterparts and renamed with the use of molecular data (Shenoy et al. 2006, 2007, 2010; Wijayawardene et al. 2016a, b; Li et al. 2020). In the past, it was only possible to link asexual morphs if one isolated an ascomycete and it formed a hyphomycete or coelomycete in culture or vice versa (Wijayawardene et al. 2017b). Some links were concluded based on the asexual and sexual morphs growing alongside each other, but these links were not often verified. It also meant that the related sexual and asexual morph had different names in the dual nomenclature system (Hawksworth 2011). Molecular data now allows us to link the asexual and sexual morphs based on comparison of gene sequences. This has led to not only a large number of sexual and asexual morphs being linked, but the Botanical Congress introducing a (at the time revolutionary and disputed) new rule that one fungus species can only have one name (Hawksworth 2011).
With such vast advances in our understanding of fungi and species, it has been possible to classify the fungi in reliable classification schemes. The first outlines for the Ascomycota were provided by Eriksson (1982), periodically updated (Eriksson and Hawksworth 1991, 1993), Myconet, Outlines by Lumbsch and Huhndorf (2007, 2010), and more recently Notes on Ascomycota (Wijayawardene et al. 2017a) and the Outline of Ascomycota (Wijayawardene et al. 2018a). Most recently, outlines of basal fungi (Wijayawardene et al. 2018b) and Basidiomycota (He et al. 2019) have been published and will finally culminate in the first outline of the Kingdom fungi (Wijayawardene et al. in press).
The next major question to be researched and resolved is what “is a species”. Previously, morphology (Hyde et al. 2011), chemotaxonomy (Kuhnert et al. 2017), analysis of ITS sequence data, analysis of multi-genes (rDNA and protein coding genes) and more recently whole genomes have advanced the understanding of species concepts. However, the final decision as to introduce a new species or not, is still mainly subjective and relies on genes available, morphology, taxon sampling in phylogenetic tree and interpretation of nucleotide differences (Jeewon and Hyde 2016). As more and more molecular data becomes available and obtaining whole genomes becomes cheaper, we will have more data than we can possibly handle. Therefore, selected studies are needed to establish what “is a species”, but the results are unlikely to establish strict rules across all groups of fungi, but only recommendations for good science.
Although fungi play vital roles in all ecosystems, as decomposers, epiphytes, endophytes, symbionts of plants, as well as animal and plant pathogens, they have been relatively understudied (Hyde et al. 2018a, c). As plant and human pathogens they seriously impact on our daily lives (Hyde et al. 2018b) and require enhanced study. Fungi also have an important place in biotechnological applications as has been shown in the recent paper of Hyde et al. (2019b), which demonstrated 50 application areas of the fungi. As mycorrhizae, biocontrol agents and food and beverage components, more research is needed on these important organisms. Similarly, Asian culture and the demand for consuming different mushrooms from those consumed in Europe and the USA, as well as medicinal mushroom products (Thongbai et al. 2015; Bandara et al. 2019; Jumbam et al. 2019), means there is a great potential for industrial utilization and profit from any mushroom research (Hyde et al. 2019b). Several recent reviews have shown that consumption of medicinal mushrooms is likely to have beneficial impacts on health and thus important as medicinal products (De Silva et al. 2012a, b; Wisitrassameewong et al. 2012).
The knowledge of species, genera and higher taxa will only increase if we collect, isolate, sequence and provide new data on the world’s fungi. By providing more data on a species it will be possible to resolve what is a species, genus or higher taxon. By providing isolates it will be possible to carry out assays to establish any potential benefit or novel compounds produced. By collecting plant pathogens it will be possible to better resolve species and make recommendations for quarantine and plant breeding. The Fungal Diversity Notes series (e.g. Tibpromma et al. 2018), Fungal Planet series (Crous et al. 2017), Mycosphere notes series (Hyde et al. 2018b; Pem et al. 2019b), Cryptogamie Mycologie series (Buyck et al. 2017), Botanica marina series (Jones et al. 2019a), Fungal Systematics and Evolution series (Song et al. 2019) and Asian Journal of Mycology notes series (Hyde et al. in press) do all of the above and will significantly help to improve our knowledge of the fungi.
The scientific community is moving towards a web-based provision of knowledge and in this regard mycologists are developing websites that are keeping abreast of developments. The early databases (Index Fungorum and MycoBank) provide a nomenclature for the fungi, while recent additions provide data on the fungal groups, genera and species (Jones et al. 2019b, Monkai et al. 2019; Pem et al. 2019a). This trend will continue and expand so that more information becomes digital.
The present paper is the 11th in the series of Fungal Diversity Notes with entries 1151–1276. The paper introduces new taxa, new data, and other taxonomic contributions on various groups of fungi. This special issue of Fungal Diversity, in its 100th volume, is a tribute to Erio Camporesi, a prolific collector of fungi in Italy, where many of the earlier studies on fungi were initiated (Saccardo 1912).
Materials and methods
Materials and methods follow the previous fungal diversity notes (Hyde et al. 2016; Tibpromma et al. 2017; Wanasinghe et al. 2018; Phookamsak et al. 2019). Taxa described in this study were mainly collected from Italy and some Asian countries viz. China, India and Thailand, as well as in some other European countries (Belgium, Denmark, Estonia, Norway, Russia and United Kingdom), North American (Panama) and South American (Brazil) countries. Taxa were described and illustrated based on morphological features coupled with phylogenetic analyses performed by maximum likelihood, maximum parsimony and Bayesian inference criteria. Colour codes followed the Methuen Handbook of Colour (Kornerup and Wanscher 1978). The new taxa are justified based on guidelines of Jeewon and Hyde (2016).
Ascomycota R.H. Whittaker
Notes: We follow the latest treatments and updated accounts of Ascomycota in Wijayawardene et al. (2017a, 2018a).
Subphylum Pezizomycotina O.E. Erikss. & Winka
Class Dothideomycetes O.E. Erikss. & Winka
Notes: We follow the latest treatments and updated accounts of Dothideomycetes in Hyde et al. (2013) and Liu et al. (2017a)
Subclass Dothideomycetidae P.M. Kirk et al.
Capnodiales Woron.
Notes: We follow the latest treatments and updated accounts of Capnodiales in Chomnunti et al. (2011, 2014) and Hyde et al. (2013). The evolution of Capnodiales with other fungal epiphytes using molecular clock dating was discussed in Hongsanan et al. (2016).
Dissoconiaceae Crous & de Hoog
Notes: Dissoconiaceae was introduced by Crous et al. (2009) to accommodate Dissoconium de Hoog, Oorschot & Hijwegen and Ramichloridium Stahel ex de Hoog. Species of Dissoconiaceae is characterised by immersed, globose, pseudothecial ascomata, bitunicate asci, ellipsoid-fusoid, 1-septate, hyaline ascospores, subcylindrical, subulate or lageniform to cylindrical conidiophores and, ellipsoid to obclavate or globose, 0–1-septate, olivaceous-brown conidia (Crous et al. 2009).
Dissoconium de Hoog, Oorschot & Hijwegen
Notes: Dissoconium was established based on D. aciculare de Hoog et al. as the type (de Hoog et al. 1983). The genus is characterised by medium brown, subcylindrical conidiophores and solitary, pale olivaceous-brown, smooth, ellipsoid to obclavate or globose, 0–1-septate conidia. There are no sexual morphs reported for the genus (Crous et al. 2007b, 2009). In this study, we report the sexual morph of D. eucalypti on Laurus nobilis from Italy for the first time with an updated phylogenetic tree for Dissoconium (Fig. 2).
Dissoconium eucalypti Crous & Carnegie, in Crous et al., Fungal Divers 26(1): 157 (2007)
Facesoffungi number: FoF 06961; Fig. 1
Holotype: AUSTRALIA, New South Wales, Morpeth Park, Plantation, Bonalbo, 152º 36′ 47″ E, 28º 46′ 3″, on leaves of Eucalyptus tereticornis, 8 February 2006, A. Carnegie, CBS-H 19770, cultures ex-type CPC 13004 = CBS 120039, CPC 13005–13006.
Associated with dead branches of Laurus nobilis. Appearing as black raised, spots, each surrounded by a thin, yellow border. Sexual morphAscomata up to 180–315 µm high, 240–340 µm wide, pseudothecial, single, globose, immersed becoming erumpent, dark brown, ostiolate. Ostiole 63–90 µm high, papillate, periphysate. Peridium 22–36 µm wide, composed of 3–4 layers of thick-walled, medium brown cells of textura angularis, with yellow vacuoles, inner layer of flattened, hyaline cells of textura angularis. Pseudoparaphyses lacking. Asci 90–105 × 15–28 µm (\( \bar{x} \) = 96 × 19.8 μm, n = 15), 8-spored, bitunicate, fasciculate, subsessile, cylindrical-clavate to broadly clavate, straight or slightly incurved. Ascospores 16.3–28.8 × 7.1– 10.6 µm (\( \bar{x} \) = 24.3 × 8.6 μm, n = 35), bi-seriate above, uni-seriate below, filling the ascus almost completely, ellipsoid-fusoid, with subobtuse ends, 1-septate, equilateral or inequilateral, constricted at the septum, with 2 larger guttules and many small guttules, straight to slightly curved, hyaline, with a mucoid sheath. Asexual morph See Crous et al. (2007b).
Material examined: ITALY, Province of Forlì-Cesena, Via Lombardini - Forlì, dead aerial branch of Laurus nobilis (Lauraceae), 24 November 2016, E. Camporesi, IT3170 (MFLU 16-2906).
Known host and distribution: Eucalyptus tereticornis (Australia), Malus domestica (United States) (Farr and Rossman 2020).
GenBank numbers: ITS = MN699134, LSU = MN699129.
Notes: Our new collection groups with ex-type strain of Dissoconium eucalypti (CBS 120039) which was isolated from Eucalyptus tereticornis in Australia (Crous et al. 2007b), with high statistical support (96% ML, 1.00 BYPP; Fig. 2). However, there were no sexual morphs have been reported for D. eucalypti or Dissoconium (Crous et al. 2007b). Our new fungus resembles Dissoconiaceae in having pseudothecial, immersed, globose, unilocular, papillate ascomata and ellipsoid-fusoid, 1-septate, hyaline ascospores (Crous et al. 2009). DNA sequences of D. eucalypti strains (MFLU 16-2906 and CBS 120039) differ in 2 nucleotides of the ITS region (0.4%, no gaps), while LSU sequences were identical. However, molecular data does not provide evidence for delimiting the new collection from D. eucalypti (Jeewon and Hyde 2016). Hence, it is reported here as the sexual morph of D. eucalypti.
Phylogram generated from RAxML analysis based on combined ITS and LSU sequence data of Dissoconium isolates. Related sequences were obtained from GenBank. Sixteen taxa are included in the analyses, which comprise 1290 characters including gaps. Single gene analyses were carried out and tree topologies of the tree and clade stability were compared. Tree is rooted to Uwebraunia musae CBS 122453, U. dekkeri CBS 567.89, U. commune CBS 114238 and U. australiensis CBS 120729. Tree topology of the ML analysis was similar to the BI. The best scoring RAxML tree with a final likelihood value of − 2658.535271 is presented. The matrix had 115 distinct alignment patterns, with 25.48% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.228902, C = 0.267138, G = 0.292715, T = 0.211244; substitution rates AC = 1.455206, AG = 1.069034, AT = 0.782623, CG = 1.123134, CT = 4.226060, GT = 1.000000; gamma distribution shape parameter α = 958.405199. RAxML bootstrap support values ≥ 70% (BT) and Bayesian posterior probabilities ≥ 0.99 (BYPP) are given at the nodes. The scale bar indicates 0.02 changes. The isolates obtained in this study are in blue and ex-types are in black bold
Mycosphaerellaceae Lindau
Notes: Mycosphaerellaceae is one of the largest ascomycetous families representing more than 5900 known species (Crous et al. 2009; Hongsanan et al. in press). Members of this family are commonly referred to as cercosporoid fungi and comprise dematiaceous, holoblastic asexual morphs and mycosphaerella-like sexual morphs (Braun et al. 2016; Videira et al. 2017). See Braun et al. (2014, 2015, 2016), Videira et al. (2017) and Hongsanan et al. (in press) for more details.
Pseudocercospora Speg.
Notes: Pseudocercospora was introduced based on P. vitis (Lév.) Speg. (type species) which was recognized as a foliar pathogen of grapevines by Spegazzini (1911). Pseudocercospora is a diverse genus which are mostly reported as plant pathogens associated with leaf and fruit spots as well as blights on a wide range of plant hosts (Crous et al. 2013; Videira et al. 2017; Wanasinghe et al. 2018). Species in this genus can occur in arid as well as wet environments and in a wide range of climates including cool temperate, subtropical and tropical regions (Crous et al. 2013a; Farr and Rossman 2020). Based on phylogenetic analyses of a combined ITS and LSU sequence dataset (Fig. 4), P. maetaengensis is introduced from unidentified fallen dead leaves.
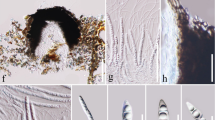
Pseudocercospora maetaengensis J.F. Li & Phookamsak, sp. nov.
Index Fungorum number: IF556890; Facesoffungi number: FoF 07054; Fig. 3
Pseudocercospora maetaengensis (MFLU 14-0206, holotype). a, b Appearance of fungal colonies on host substrate. c, e–g Close up conidiophores and conidiogenous cells. d Conidiophores. h–o Conidia. p Germination of conidia. q–r Culture. Scale bars: a = 1 cm, b = 50 µm, c–d, e–g = 10 µm, h–p = 5 µm, q–r = 0.5 cm
Etymology: Name reflects the location from which it was collected, Mae Taeng, Chiang Mai, Thailand.
Holotype: MFLU 14-0206.
Saprobic on fallen dead leaves. Sexual morph Undetermined. Asexual morphSynnemata 100–130 μm high, 5.9–8 μm wide, erect, simple, unbranched, dark brown to black, comprising various conidiophores twisted together in synnemata, with bubble-like, tightly interwoven, branched hyphae, compacted into an elongate bundle. Conidiophores 100–130 μm long × 1.8–3.5 μm diam. (\( \bar{x} \) = 124 × 2.6 μm, n = 10). Conidiogenous cells 2.5–3 μm long × 1.6–2 μm diam. (\( \bar{x} \) = 2.8 × 1.8 μm, n = 20), monoblastic, integrated, terminal, determinate or percurrent, cylindrical, doliiform, hyaline, smooth, thin-walled. Conidia 13–28 μm long × (2.5–)2.7–3.6(–4) μm diam. (\( \bar{x} \) = 21 × 3.1 μm, n = 20), acrogenous, solitary, hyaline, ellipsoidal, clavate, 2–4-septate, dry, simple, straight, curved, fusiform, smooth- and thin-walled.
Culture characteristics: Conidia germinating on PDA within 14 h and germ tubes produced from top cells. Colonies growing on PDA, hairy or cottony, grey to dark grey, reaching 5 mm in 20 days at 25 °C, mycelium superficial, effuse, grey to dark grey hyphae; Asexual spores and sexual spores were not formed within 60 days.
Material examined: THAILAND, Chiang Mai Province, Mae Teang District, Mushroom Research Center (M.R.C.), on unidentified fallen dead leaves, 25 November 2013, X. Zeng (MFLU 14-0206, holotype), ex-type living culture MFLUCC 14-0411.
GenBank numbers: ITS = MN648323, LSU = MN648328, SSU = MN648320, TEF1-α = MN821071.
Notes: Morphologically, Pseudocercospora maetaengensis resembles P. vitis in having septate, straight or slightly curved conidia, with a rounded apex and conidiophores grouped in synnema (Ellis 1971). However, P. maetaengensis is unique in its short, dark synnemata and smaller, less septate, subhyaline, fusiform conidia. Phylogeny based on a combined ITS and LSU sequence dataset reveals P. maetaengensis as an independent lineage distinct from P. vitis and further differs from other species in Pseudocercospora with significant support (77% ML, 0.98 BYPP; Fig. 4). Therefore, a new species, P. maetaengensis is established.
Phylogram generated from the best scoring of the RAxML tree based on combined ITS and LSU sequence data of taxa in Pseudocercospora. Pallidocercospora heimii (CPC 11716) is selected as the outgroup taxon. The best RAxML tree with a final likelihood value of − 10821.117889 is presented. RAxML analysis yielded 609 distinct alignment patterns and 19.20% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.248806, C = 0.241445, G = 0.281063, T = 0.228686, with substitution rates AC = 1.648825, AG = 3.373595, AT = 1.202983, CG = 0.838400, CT = 6.421832, GT = 1.000000. The gamma distribution shape parameter alpha = 0.165747. Bayesian posterior probabilities (BYPP) from MCMC were evaluated with final average standard deviation of split frequencies = 0.008723. Bootstrap support values for maximum likelihood (ML) equal to or greater than 70%; BYPP equal to or greater than 0.95 are given above or below the nodes as ML/BYPP. Type sequences are in black bold and newly generated sequences are indicated in blue bold
Subclass Pleosporomycetidae C.L. Schoch et al.
Hysteriales Lindau
Notes: The order Hysteriales was introduced by Engler and Prantl (1897). This order includes single family Hysteriaceae that is characterised by hysteriform apothecia. They are mostly saprobic and widely distributed (Jayasiri et al. 2018). We follow the latest treatment and updated accounts of Hysteriales in Hyde et al. (2013) and Jayasiri et al. (2018).
Hysteriaceae Chevall.
Notes: Hysteriaceae was introduced by Chevallier (1826). Currently this family includes 14 genera (Wijayawardene et al. 2018a). Taxa in this family are mostly found as saprobes on dead plant material and are characterised by erumpent or superficial, hysterothecial or navicular ascomata, sometimes branched or discoid. The exciple is composed of cells of textura angularis and is carbonaceous, 2–4-layered, thick, stout, rarely thin. Hamathecium composed of pseudoparaphyses with tips sometimes darkened or branched above the asci. Asci are clavate to cylindrical, fissitunicate, with a distinct apical chamber. Ascospores are ellipsoid, fusoid or clavate, hyaline or brown, variously septate, smooth or ornamented, sometimes with a sheath. Asexual morphs are pycnidial or hyphomycetous (Jaklitsch et al. 2016; Jayasiri et al. 2018). Based on phylogenetic analyses coupled with morphological characteristics, we introduce two novel species, Rhytidhysteron camporesii Ekanayaka & K.D. Hyde and R. erioi Ekanayaka & K.D. Hyde in this study. Furthermore, Hysterobrevium constrictum (N. Amano) E. Boehm & C.L. Schoch is also reported from Yunnan, China for the first time.
Hysterobrevium E. Boehm & C.L. Schoch
Notes: Hysterobrevium was introduced by Boehm et al. (2009). The genus is characterised by navicular hysterothecia with a prominent longitudinal slit, bitunicate, cylindrical to clavate asci and pigmented or hyaline, septate ascospores (Boehm et al. 2009). Currently, six species are accommodated in this genus (Index Fungorum 2020).
Hysterobrevium constrictum (N. Amano) E. Boehm & C.L. Schoch, in Boehm et al., Stud. Mycol. 64: 64 (2009)
Facesoffungi number: FoF 06461; Fig. 5
≡ Gloniopsis constricta N. Amano, Trans. Mycol. Soc. Japan 24(3): 289 (1983)
Holotype: JAPAN, Yunohana Onsen, Tateiwa-mura, Minaimiaizu-gun, Fukushima Pref., Y. Doi, 18 September 1982, F-237162.
Saprobic on dead stems. Sexual morphAscomata 350–450 µm high, 700–1000 µm long, apothecial, arising singly or in small groups, sessile, slightly erumpent from the substrate. Receptacle cupulate, black, hysteriform. Disc concave, black when fresh. Margins black. Excipulum 35–50 µm wide, ectal excipulum carbonaceous, thick-walled, black cells of textura globulosa to angularis, medullary excipulum composed of narrow, long, thin-walled, hyaline to brownish cells of textura porrecta. Hymenium hyaline. Pseudoparaphyses numerous, branched, septate. Asci 115–130 × 25–32 µm, 8-spored, short pedicellate, rounded at the apex, J-, croziers abscent at the asci base. Ascospores 27–35 × 10–12 µm, multi-seriate, hyaline, smooth-walled, ellipsoid to fusoid, muriform. Asexual morph Undetermined.
Material examined: CHINA, Yunnan Province, Kunming City, Kunming Institute of Botany, Botanical garden, on dead stems, 24 May 2018, A.H. Ekanayaka, HC13 (KUN-HKAS102101).
Known host and distribution: Lonicera xylosteum, Populus tremula, Quercus robur (Sweden), on decaying wood (Thailand, Japan, China and New Zealand) (Boehm et al. 2009; Farr and Rossman 2020).
GenBank numbers: ITS = MN429070, LSU = MN429073, SSU = MN420986, TEF1-α = MN442088.
Notes: This species is characterised by sessile, erumpent, naviculate apothecia, cylindrical asci and hyaline, muriform ascospores. In the comparison of ITS sequence, our new strain shows 93% (443/478 bp) similarity to Hysterobrevium mori (Schwein.) E. Boehm & C.L. Schoch (MFLUCC 14-0520) but differs from 35 base pairs including eight gaps. Phylogenetic analyses of the combined LSU, SSU, ITS and TEF1-α sequence dataset showed that our strain forms a robust clade with H. constrictum (GKM426N) with high support (96% ML, 0.99 BYPP; Fig. 8). Moreover, our strain is also similar to the description of H. constrictum provided by Boehm et al. (2009) except in having slightly larger ascospores. We therefore, identify our taxon as H. constrictum from Yunnan, China.
Rhytidhysteron Speg.
Notes: The genus was introduced by Spegazzini (1881). Taxa are characterised by hysterothecial ascomata, which are sometimes branched or discoid and erumpent or superficial. The exciple is composed of carbonaceous cells of textura angularis and hamathecium is composed of pseudoparaphyses and their tips are sometimes darkened or branched above asci. Asci are clavate to cylindrical, fissitunicate, with a distinct apical chamber. Ascospores are ellipsoid to fusoid, light brown to dark brown, septate and smooth or ornamented (Thambugala et al. 2016; Jayasiri et al. 2018). Twenty-one species are listed in the genus in Index Fungorum (2020).
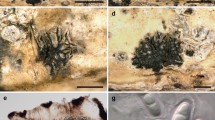
Rhytidhysteron camporesii Ekanayaka & K.D. Hyde, sp. nov.
Index Fungorum number: IF556783; Facesoffungi number: FoF 06459; Fig. 6
Rhytidhysteron camporesii (KUN-HKAS 104277, holotype). a Substrate. b Ascomata on wood. c Cross section of an ascoma. d Vertical section of the ascoma at margin. e Apically swollen paraphyses. f, g Cylindrical asci. h–j Ellipsoid ascospores. Scale bars: b = 500 µm, c = 400 µm, d = 100 µm, e–g = 50 µm, h–j = 10 µm
Etymology: Referring to the significant contribution Erio Camporesi made to mycology
Holotype: KUN-HKAS 104277
Saprobic on dead stems. Sexual morphAscomata 500–650 µm high, 800–1100 µm long (\( \bar{x} \) = 570.1 × 1002.4 µm, n = 10), apothecial, arising singly or in small groups, short stipitate, erumpent from the substrate. Receptacle cupulate, black, hysteriform. Disc concave covered by margins. Margins black, slightly dentate. Ectal excipulum 65–95 µm (\( \bar{x} \) = 71.9 µm, n = 10) carbonaceous, composed of blackish cells of textura globulosa to angularis. Medullary excipulum 19–22 µm (\( \bar{x} \) = 20.4 µm, n = 10), composed of narrow, long, thin-walled, hyaline to brownish cells of textura porrecta. Hymenium hyaline. Paraphyses 3–5 µm wide (\( \bar{x} \) = 4.4 µm, n = 20) at the apices, numerous, septate, branched at the base, exceed asci in length, apically swollen, slightly branched, slightly granulated and pigmented, pigments are brownish in water, magenta in KOH, apices glued together with gelatinous material to form pseudo-epithecium. Asci 165–175 × 13–15 µm (\( \bar{x} \) = 171.8 × 13.5 µm, n = 30) 8-spored, short pedicellate, long, cylindrical, rounded at the apex, J-, croziers abscent at the asci base. Ascospores 25–28 × 9–11 µm (\( \bar{x} \) = 26.1 × 10.4 µm, n = 40), 1-seriate, hyaline to light brown, 1–2-septate when immature, dark brown, 3-septate at maturity, smooth-walled, ellipsoidal to fusiform, slightly rounded or pointed at both ends, guttulate. Asexual morph Undetermined.
Material examined: CHINA, Yunnan Province, Kunming City, Kunming Institute of Botany, Botanical garden, on dead stems of unidentified woody plant, 14 April 2016, A.H. Ekanayaka, HC 005 (KUN-KHAS 104277, holotype).
GenBank numbers: ITS = MN429069, LSU = MN429072, TEF1-α = MN442087.
Notes: The new taxon is characterised by short stipitate apothecia, apically swollen, granulated and pigmented paraphyses, ascospores with slightly rounded and pointed ends. In the comparison of ITS sequences, our new strain shows 94% (486/517 bp) similarity to Rhytidhysteron rufulum (Spreng.) Speg. (MFLUCC 12-0568) and 94% (480/510 bp) to R. thailandicum Thambug. & K.D. Hyde (MFLUCC 14-0503) but differs from R. rufulum in 31 base pairs including six gaps and from R. thailandicum in 30 base pairs including five gaps. Phylogenetically this new species formed an independent lineage with strong statistical support (86% ML, 1.00 BYPP; Fig. 8). Rhytidhysteron camporesii is morphologically similar to R. neorufulum Thambug. & K.D. Hyde, but they are different in having sessile apothecia, and longer and highly guttulate ascospores in R. neorufulum (Thambugala et al. 2016). Rhytidhysteron tectonae Doilom & K.D. Hyde is also similar to R. camporesii, but differs in having apically swollen, granulated and pigmented paraphyses (Doilom et al. 2017).
Rhytidhysteron erioi Ekanayaka & K.D. Hyde, sp. nov.
Index Fungorum number: IF556788; Facesoffungi number: FoF 06460; Fig. 7
Rhytidhysteron erioi (MFLU 16-0584, holotype). a Substrate. b, c Ascomata on wood. d Cross section of an ascoma (mounted in KOH). e Close up of a vertical section of the ascoma at margin. f Septate paraphyses. g–j Cylindric-clavate asci. k–n Ellipsoid ascospores. Scale bars: b, c = 500 µm, d = 200 µm, e = 100 µm, f = 75 µm, g–j = 40 µm, k–n = 10 µm
Etymology: Referring to the significant contribution Erio Camporesi made to mycology
Holotype: MFLU 16-0584
Saprobic on dead stems. Sexual morphAscomata 270–360 µm high, 600–1200 µm long, apothecial, arising singly, substipitate, superficial or slightly erumpent from the substrate. Receptacle cupulate, convex or flat, black when fresh. Disc orange when fresh, become magenta with the presence of KOH. Margins black when fresh, dentate. Ectal excipulum 55–75 µm (\( \bar{x} \) = 63.3 µm, n = 10) composed of large, thin-walled, dark brown cells of textura angularis to textura globulosa. Medullary excipulum 14–20 µm (\( \bar{x} \) = 17.3 µm, n = 10) composed of hyaline cells of textura porrecta. Hymenium hyaline to brownish, enclosed in a thick gelatinous matrix. Paraphyses 2–2.5 µm wide (\( \bar{x} \) = 2.3 µm, n = 2 0) wide at the apices, numerous, filiform, septate, slightly branched at the base, slightly swollen at the apices, apices glued together to develop pseudo-epithecium, gelatinous material include yellowish to brownish pigments which turns magenta in KOH. Asci 140–200 × 9–16 µm (\( \bar{x} \) = 184.3 × 14.5 µm, n = 30) 8-spored, bitunicate, short pedicellate, cylindrical, rounded at the apex, J-. Ascospores 22–28 × 9–11 µm (\( \bar{x} \) = 27.1 × 10.3 µm, n = 40), 1-seriate, ellipsoid, hyaline to light brown, aseptate, with wrinkled walls when young, dark brown, 3-septate, smooth- and thick-walled, with guttulate. Asexual morph Undetermined (Fig. 8).
Phylogram generated from a maximum likelihood analysis of sequences of Hysteriaceae Based on LSU, SSU, ITS and TEF1-α sequence dataset. The newly generated nucleotide sequences were compared against the GenBank (http://www.ncbi.nlm.nih.gov/) database using the Mega BLAST program. Related sequences were obtained from GenBank. Thirty-eight strains were included in the sequence analyses, which comprised 3194 characters including gaps (LSU: 1–937, ITS: 938–1443, SSU: 1444–2464, TEF1-α: 2465–3194). Glonium circumserpens (CBS 123343) was used as the outgroup taxon. The best scoring RAxML tree with a final likelihood value of − 11291.754630 is presented. The matrix had 782 distinct alignment patterns, with 43.58% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.248, C = 0.235, G = 0.283, T = 0.233; substitution rates AC = 1.339408, AG = 2.211853, AT = 1.176124, CG = 0.711220, CT = 7.949676, GT = 1.000000; gamma distribution shape parameter α = 0.140525. Bayesian posterior probabilities equal or greater than 0.90 BYPP are given as the first set of numbers above the nodes and Bootstrap support values for ML equal or greater than 50% are given as the second set of numbers above the nodes. Newly generated sequences are in blue. Ex-type strains are indicated in bold
Material examined: THAILAND, Chiang Rai Province, Doi Pui, on unidentified decaying wood, 19 June 2015, A.H. Ekanayaka, HD 022 (MFLU 16-0584, holotype).
GenBank numbers: ITS = MN429068, LSU = MN429071, TEF1-α = MN442086.
Notes: Rhytidhysteron erioi is characterised by substipitate apothecia, paraphyses with slightly swollen apices without pigments and 3-septate ascospores. In the comparison of ITS data, our new strain shows 95% (466/492 bp) similarity to R. rufulum (MFLUCC 12-0568) and R. mangrovei (MFLUCC 18-1113) and 94% (461/492 bp) similarity to R. neorufulum (MFLUCC 13-0221), but differs in 26 base pairs including four gaps, 22 base pairs including five gaps and 31 base pairs including five gaps. Our strain is phylogenetically close to R. hysterinum (Dufour) Samuels & E. Müll. and R. thailandicum. However, R. hysterinum differs from our species in having 1-septate ascospores (Samuels and Müller 1979). Rhytidhysteron thailandicum differs in having yellowish to brown ascospores with smooth walls when both mature and immature (Thambugala et al. 2016). Rhytidhysteron mangrovei differs in having smaller asci (110–150 × 9.4–10 μm) and not having wrinkled ascospores at a young stage (Kumar et al. 2019).
Pleosporales Luttrell ex M.E. Barr
Notes: Among the Dothideomycetes, Pleosporales is the largest and most diverse order. This comprises over 4700 species (Ariyawansa et al. 2018) classified in more than 400 genera and 75 families (Wijayawardene et al. 2018a). The taxonomic circumscription of Pleosporales has changed frequently in recent years due to the addition of large numbers of families, genera and species.
Amorosiaceae Thambug. & K.D. Hyde
Notes: Amorosiaceae was established by Thambugala et al. (2015) to accommodate two genera namely Amorosia Mantle & D. Hawksw. and Angustimassarina Thambug., Kaz. Tanaka & K.D. Hyde. The family can easily be recognized by its solitary or gregarious ascomata, 8-spored, bitunicate cylindrical to cylindric-clavate, pedicellate asci and fusiform to cylindrical, or ellipsoidal-fusiform, 1(–3)-septate hyaline ascospores which sometimes appear as light-brown when mature (Thambugala et al. 2015) The asexual morph of Amorosia is hyphomycetous. Species of Amorosiaceae resemble species of Massariaceae Nitschke, Lophiostomataceae Sacc., Floricolaceae Thambug., Kaz. Tanaka & K.D. Hyde and Sporormiaceae Munk in having cylindrical to cylindric-clavate asci as well as ellipsoidal-fusiform ascospores. However, they differ from these families in having hyphomyceteous asexual morphs and appear to grow within ascomata of other ascomycetes and may be mycoparasites. We follow the latest treatment of Amorosiaceae in Thambugala et al. (2015) and updated accounts of taxa in Amorosiaceae in Wijayawardene et al. (2018a) and Hyde et al. (2019a). In this paper, we introduce a new species, Angustimassarina camporesii in Amorosiaceae.
Angustimassarina Thambug., Kaz. Tanaka & K.D. Hyde
Notes: Angustimassarina was introduced by Thambugala et al. (2015) with Angustimassarina populi Thambug. & K.D. Hyde as the type species. The morphological characters that define the genus are solitary or gregarious, immersed to semi-immersed ascomata, central, cylindrical, papillate ostioles usually composed of pseudoparenchymatous cells, peridium composed of several layers of dark brown to lightly pigmented cells of textura angularis, 8-spored cylindrical to cylindric-clavate asci and hyaline to brown fusiform to cylindrical or ellipsoidal-fusiform ascospores (Thambugala et al. 2015). The asexual morph is undetermined (Wijayawardene et al. 2017b). Eleven species epithets are registered under Angustimassarina (Index Fungorum 2020). Species of Angustimassarina can be found worldwide especially in Germany and Italy.
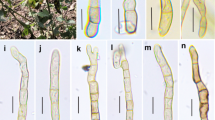
Angustimassarina camporesii D. Pem, Doilom & K.D. Hyde, sp. nov.
Index Fungorum number: IF556796; Facesoffungi number: FoF 06465; Fig. 9
Angustimassarina camporesii (MFLU 18-0057, holotype). a, b Appearance of ascomata on host surface. c Vertical section through the ascoma. d Peridium. e Hamathecium. f–i Asci. j–m Ascospores. n Ascospore in Indian ink, showing sheath. Scale bars: a = 2000 μm, b = 1000 μm, c, d = 50 μm, e = 5 μm, f–i = 25 μm, j–n = 10 μm
Etymology: The epithet honours Mr. Erio Camporesi who collected this fungus.
Holotype: MFLU 18-0057
Saprobic on dead aerial stem of Galium sp. Sexual morphAscomata 130–240 × 130–190 μm (\( \bar{x} \) = 177.8 × 170.7 µm, n = 20), semi-immersed to immersed with flat at the base, solitary or in small groups, globose to subglobose, visible as black dots on the host surface, conspicuous at the surface, ostiolate, papillate, black. Ostiole crest-like, papillate, immersed in ascomata, with a pore-like opening. Peridium 15–21 µm wide, thin comprised of dark brown to pale brown 3–5 layer of cells of textura angularis. Hamathecium comprising 1–2 µm wide, numerous, septate, hyaline, smooth, pseudoparaphyses, attached to the base, longer than asci. Asci 62–88 × 10–13 μm (\( \bar{x} \) = 77.1 × 11.2 µm, n = 20), 8-spored, bitunicate, fissitunicate, cylindric-clavate, slightly curved, long with a club-shaped pedicel, with apex rounded with a minute ocular chamber, smooth-walled. Ascospores 15–18 × 4–5 μm (\( \bar{x} \) = 17.1 × 4.5 µm, n = 20), overlapping 2–3-seriate at the centre and apex,1-seriate at the base, fusiform, hyaline, 1-septate at the centre, with 4–5 large guttules, enlarged cell near the central septum, constricted at the septum, conical at both ends, smooth-walled, surrounded by a mucilaginous sheath. Asexual morph Undetermined.
Material examined: ITALY, Province of Forlì-Cesena [FC], Borgo Paglia – Cesena, on dead aerial stem of Galium sp. (Rubiaceae), 26 March 2018, E. Camporesi, IT 3603 (MFLU 18-0057, holotype).
GenBank numbers: ITS = MN244197, LSU = MN244167, SSU = MN244173, TEF1-α = MN593307.
Notes: Our new collection is morphologically and phylogenetically (Fig. 10) related to Angustimassarina premilcurensis Tibpromma, Camporesi & K.D. Hyde, but differs in having smaller ascomata (138–241 × 138–190 μm versus 231–238 × 290–311 μm) and smaller ascospores (15–18 × 4–5 μm versus 19–23 × 4–7 μm). A comparison of ITS and TEF1-α nucleotide shows 7.95% and 2.5% differences between A. camporesii and A. premilcurensis. We, therefore, introduce A. camporesii as a new species in Angustimassarina.
Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, ITS and TEF1-α sequence data representing Amorosiaceae and the outgroup. Related sequences are taken from Hyde et al. (2019). Seventeen strains are included in the combined analyses which comprise 2701 characters (733 characters for LSU, 825 characters for SSU, 463 characters for ITS, 677 characters for TEF1-α) after alignment. Eremodothis angulata (CBS 610.74) in Sporormiaceae (Pleosporales) is used as the outgroup taxon. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian inference analysis. The best RaxML tree with a final likelihood values of − 4904.871734 is presented. The matrix had 162 distinct alignment patterns, with 27.94% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.241624, C = 0.242160, G = 0.272716, T = 0.243500; substitution rates AC = 0.606042, AG = 1.263408, AT = 1.243145, CG = 0.610875, CT = 5.612203, GT = 1.000000; gamma distribution shape parameter α = 3.242368. Bootstrap values for maximum likelihood (ML) equal to or greater than 50% and clade credibility values greater than 0.90 (the rounding of values to 2 decimal proportions) from Bayesian inference analysis labeled on the nodes. Ex-type strains are in bold and black, the new isolate is indicated in bold and blue
Camarosporidiellaceae Wanas., Wijayaw., Crous & K.D. Hyde
Notes: The family Camarosporidiellaceae was erected by Wanasinghe et al. (2017a) based on both morphology and multi-gene analysis, with Camarosporidiella Wanas., Wijayaw., K.D. Hyde as the type genus of this family and belongs to the order Pleosporales. This species can be saprobic, endophytic or pathogenic on leaves and wood (Wanasinghe et al. 2017a). The sexual morph is characterised by cylindrical asci with (2–)4–8-spored, ellipsoidal, muriform ascospores with 3–8 transverse septa and 1–2 longitudinal septa with a coelomycetous asexual morph (Wanasinghe et al. 2017a). Twenty-two epithets are listed in Index Fungorum (2020). We introduce a new species of Camarosporidiella isolated from Coronilla emerus based on morphology and phylogeny.
Camarosporidiella Wanas., Wijayaw. & K.D. Hyde
Notes: The genus Camarosporidiella was erected by Wanasinghe et al. (2017a) to accommodate C. caraganicola (Phukhams. et al.) Phukhams., Wanas. & K.D. Hyde. The members can be saprobic, endophytic or pathogenic on leaves and wood in terrestrial habitats (Wanasinghe et al. 2017a). The features of Camarosporidiella are cucurbitaria-like, fissitunicate, cylindrical asci containing eight ascospores with muriform, mostly ellipsoidal, 3–8-transverse septa, and without a mucilaginous sheath; the asexual morph has brown to dark brown, phragmosporous to muriform macroconidia and microconidia when present are oblong or ellipsoidal and hyaline (Wanasinghe et al. 2017a).
Camarosporidiella camporesii Tibpromma & K.D. Hyde, sp. nov.
Index Fungorum number: IF556781; Facesoffungi number: FoF 06383; Fig. 11
Camarosporidiella camporesii (MFLU 19-0296, holotype). a Appearance of ascomata on host substrate. b Section of ascoma. c Peridium. d Close up of ostiole. e Pseudoparaphyses. f, g Asci. h–j Ascospores. k Germinating ascospore. Scale bars: b = 100 μm, c, d = 20 μm, e = 5 μm, f, g = 20 μm, h–j = 5 μm, k = 10 μm
Etymology: Named in honour of Erio Camporesi, who is the best fungi collector from Italy and in recognition of his immense contribution to mycology.
Holotype: MFLU 19-0296
Saprobic on dead branch of Coronilla emerus. Sexual morphAscomata 370–500 × 340–430 μm (\( \bar{x} \) = 411 × 379 μm, n = 5), superficial to semi-immersed, globose to subglobose, solitary, scattered, conspicuous at the surface, uniloculate, black, ostioles, without papilla. Peridium 35–80 μm wide, thick-walled, comprising subhyaline to yellow–brown cells of textura angularis. Hamathecium 1.5–3 μm wide, comprising numerous, filamentous, branched, septate, pseudoparaphyses. Asci 100–190 × 10–15 μm (\( \bar{x} \) = 158 × 13 μm, n = 20), 4(6)–8-spored, mostly 8-spored, bitunicate, fissitunicate, cylindrical, pedicellate, with minute ocular chamber. Ascospores 19–25 × 8–13 μm (\( \bar{x} \) = 21 × 10 μm, n = 20), 1-seriate, sometimes overlapping, muriform, ellipsoidal to subfusiform, slightly curved, upper part wider than the lower part, 3–5 transversely septate, with 1–3 vertical septa, constricted at the central septum, slightly constricted at the septa, initially subhyaline, becoming yellow–brown to brown at maturity, with narrowly rounded ends, with guttules, thick and smooth-walled, lacking a mucilaginous sheath. Asexual morph Undetermined.
Culture characteristics: Colonies on MEA at room temperature reaching 9 cm in 6–8 weeks, circular with curved edges, white mycelium raised from the medium surface, not sporulating in culture within two months.
Material examined: ITALY, Province of Forlì-Cesena [FC], Monte Colombo-Predappio, on dead aerial branch of Coronilla emerus (L.) Lassen (Fabaceae), 25 November 2018, E. Camporesi, IT4136 (MFLU 19-0296, holotype), ex-type living cultures, KUMCC 19-0204, KUMCC 19-0205.
GenBank numbers: ITS = MN654369, LSU = MN417515, SSU = MN654373, TEF1-α = MN735983 (KUMCC 19-0204); ITS = MN654369, LSU = MN417516, SSU = MN654373, TEF1-α = MN735984 (KUMCC 19-0205).
Notes: Our new species is placed in Camarosporidiella (Camarosporidiellaceae), and well-separated from other species in Camarosporidiella (Fig. 12). Camarosporidiella camporesii is similar to C. caraganicola (Phukhams.) Phukhams., Wanas. & K.D. Hyde in having ellipsoidal ascospores with conical or narrow at the ends but differs in its ascomata and vertical septa. Camarosporidiella camporesii has ascomata without papilla and 1–3 vertically septate ascospores, while C. caraganicola has rough or hairy ascomata with 2–4 vertically septate ascospores (Wanasinghe et al. 2017a).
Phylogram generated from maximum likelihood analysis based on combined ITS, TEF1-α, LSU and SSU sequence dataset. Related sequences were obtained from Wanasinghe et al. (2017a). One hundred and two strains are included in the combined sequence analysis, which comprise 3466 characters with gaps. Pleospora herbarum (CBS 191.86) is used as the outgroup taxon. Tree topology of the ML analysis was similar to the BI analysis. The best scoring RAxML tree with a final likelihood value of − 7863.756111 is presented. The matrix had 433 distinct alignment patterns, with 12.36% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.241851, C = 0.243745, G = 0.267231, T = 0.247173; substitution rates AC = 1.388341, AG = 3.299126, AT = 2.098924, CG = 0.473406, CT = 7.522186, GT = 1.000000; gamma distribution shape parameter α = 0.020000. Bootstrap support values for ML equal to or greater than 60% and Bayesian posterior probabilities equal to or greater than 0.95 BYPP are given above the nodes. Newly generated sequences are in blue
In a BLASTn search on NCBI GenBank, the closest matches of ITS sequence of KUMCC 19-0204 and KUMCC 19-0205 are 99.65% identical to Camarosporidiella eufemiana Wanas., Camporesi & K.D. Hyde strain MFLUCC 17-0207 (MF434145), while the closest matches with the TEF1-α sequence were with 99.89% similarity with C. mirabellensis Wanas., Camporesi & K.D. Hyde strain MFLU 17-228 (MF434426). We also compared ITS and TEF1-α nucleotides and found that they are different 6 bp (1.09%) in 563 ITS (+5.8S) nucleotides and 26 bp (2.73%) in 951 TEF1-α nucleotides.
Coniothyriaceae W.B. Cooke
Notes: The family Coniothyriaceae was introduced by Cooke (1983) to accommodate Coniothyrium spp. Kirk et al. (2008) synonymized Coniothyriaceae with Leptosphaeriaceae M.E. Barr. De Gruyter et al. (2013) showed that C. palmarum Corda is distinct from Leptosphaeriaceae and they reinstated the family Coniothyriaceae in Pleosporales. Some Phoma species were also transferred to Coniothyrium Corda as they group in Coniothyriaceae. Three genera were accommodated in this family viz. Coniothyrium, Hazslinszkyomyces Crous & R.K. Schumach and Ochrocladosporium Crous & U. Braun (Wijayawardene et al. 2018a). Foliophoma Crous was added to the family by Crous and Groenewald (2017).
Foliophoma Crous
Notes: The monotypic genus Foliophoma was introduced by Crous and Groenewald (2017) with Foliophoma fallens (Sacc.) Crous as the type species. The latter was obtained from culture. The genus is characterised by eustromatic conidiomata, uni- to multi-loculate with 1–3 ostioles and conidiogenous cells with periclinal thickening or percurrent proliferation at the apex (Crous and Groenewald 2017). We introduce a new species Foliophoma camporesii isolated from dead branch of Maclura pomifera in Italy.
Foliophoma camporesii D. Pem, Doilom & K.D. Hyde, sp. nov.
Index Fungorum number: IF556797; Facesoffungi number: FoF 06466; Fig. 13
Etymology: Named in honour of Erio Camporesi, a prolific collector of fungi from Italy and in recognition of his immense contribution to mycology.
Holotype: MFLU 17-1006
Saprobic or pathogenic on dead aerial stem of Maclura pomifera. Sexual morph Undetermined. Asexual morphConidiomata 40–47 × 40–69 μm (\( \bar{x} \) = 43.3 × 48 µm, n = 20), pycnidial, globose to subglobose, ellipsoidal, or irregular, immersed to semi-immersed, carbonaceous. Pycnidial walls 15–40 μm, comprising 1–2-layered of cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 2–4 × 2–3 μm (\( \bar{x} \) = 2.6 × 2.2 µm, n = 20), phialidic with periclinal thickening or percurrent proliferation at apex, hyaline, smooth, globose to short cylindrical. Conidia 2–6 × 3–5 μm (\( \bar{x} \) = 4.9 × 3.9 µm, n = 20), ovoid to ellipsoidal, hyaline when immature, brown at maturity, aseptate, smooth- and thin-walled.
Culture characteristics: Colonies on MEA, 20–25 mm diam. after 7 days at 16 °C, margin regular, circular, aerial mycelia thinly hairy, white and flat; reverse grey and white at the margin.
Material examined: ITALY, Province of Forlì-Cesena [FC], Predappio Alta-Predappio, on dead aerial stem of Maclura pomifera (Moraceae), 5 May 2017, E. Camporesi, IT 3345 (MFLU 17-1006, holotype), ex-type living cultures, MFLUCC 18-1129.
GenBank numbers: ITS = MN244200, LSU = MN244170, SSU = MN244176.
Notes: Strain MFLUCC 18-1129 was isolated from dead stem of Maclura pomifera in Italy. Foliophoma camporesii has a close phylogenetic affinity to F. fallens, the type species of Foliophoma (Fig. 14). It differs from F. fallens in having smaller conidiomata (40–47 × 40–69 μm versus 120–250 μm) and brown conidia. ITS nucleotide comparisons show a difference of 12 base pairs or 2% difference between our new taxon and F. fallens. We, therefore, introduce F. camporesii as the second species in Foliophoma.
Phylogram generated from maximum likelihood analysis based on combined LSU, SSU and ITS sequence data representing Coniothyriaceae and the closely related families in Pleosporales. Related sequences are taken from Crous and Groenewald (2017). Thirty-four strains are included in the combined analyses which comprise 2573 characters (885 characters for LSU, 1023 characters for SSU and 665 characters for ITS) after alignment. Didymella exigua (CBS 183.55) and Ascochyta pisi (CBS 126.54) in Didymellaceae (Pleosporales) are used as the outgroup taxa. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian Inference analysis. The best RaxML tree with a final likelihood values of − 8723.938065 is presented. The matrix had 590 distinct alignment patterns, with 27.94% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.251588, C = 0.216344, G = 0.270099, T = 0.261970; substitution rates AC = 1.615107, AG = 3.847488, AT = 2.932232, CG = 0.678880, CT = 7.305403, GT = 1.000000; gamma distribution shape parameter α = 0.512104. Bootstrap values for maximum likelihood (ML) equal to or greater than 50% and clade credibility values greater than 0.90 (the rounding of values to 2 decimal proportions) from Bayesian inference analysis labeled on the nodes. Ex-type strains are in bold and black, the new isolate is indicated in bold and blue
Dictyosporiaceae Boonmee & K.D. Hyde
Notes: Boonmee et al. (2016) introduced this family and its members usually occur on decaying wood and plant debris in terrestrial and freshwater habitats (Boonmee et al. 2016; Liu et al. 2017c; Hyde et al. 2018b). The family currently comprises 12 genera. The new species Dictyosporium muriformis is introduced from decaying wood from a small freshwater stream in Guizhou, China and a new record of Dendryphiella phitsanulokensis is also reported from Brachiaria mutica in Thailand.
Dendryphiella Bubák & Ranoj
Notes: Ranojevic (1914) established Dendryphiella and designated D. interseminata (Berk. & Ravenel) Bubák as the type species. Dendryphiella is a hyphomycetous genus included in the family Dictyosporiaceae (Liu et al. 2017c; Hyde et al. 2018b) and 17 species are currently recognized in the genus (Liu et al. 2017c; Hyde et al. 2018b; Iturrieta-González et al. 2018; Index Fungorum 2020).
Dendryphiella phitsanulokensis N.G. Liu & K.D. Hyde, Mycosphere 9 (2): 287 (2018)
Facesoffungi number: FoF 03897; Fig. 15
Holotype: THAILAND, Phitsanulok Province, on decaying wood, 10 October 2016, N. Liu, J4 (MFLU 17-2651), ex-type living culture, MFLUCC 17-2513.
Saprobic on Brachiaria mutica. Sexual morph Undetermined. Asexual morphColonies on natural substrate superficial, effuse, dark brown to black. Conidiophores 150–300 μm long, 4–6 μm wide, macronematous, mononematous, fasciculate, dark brown at base, slightly paler towards the apex, thick-walled, erect, straight or slightly flexuous, finely verruculose, septate, unbranched or rarely branched. Conidiogenous cells 22–38 × 3.5–8 μm (\( \bar{x} \) = 31 × 6 μm, n = 20), polytretic, terminal, later becoming subterminal, proliferating asymmetrically, integrated, brown, finely verrucose, enlarged at apex. Conidia (10 –)16–31 × 5–9 μm (\( \bar{x} \) = 24 × 7 μm, n = 60), solitary to catenate, when catenate in acropetal chain, fusiform to ellipsoidal, rounded at apex, truncate at base, pale brown to brown or dark brown, 3-septate, constricted at the medium septum, slightly constricted at other septa, thick-walled, verrucose.
Culture characteristics: Colonies growing on MEA reaching 25 mm diam. after 10 days at 25 °C, circular, umbonate to flat, moderately dense, surface white, grayish white at the margin, reverse dark yellowish to blackish brown at the middle, white at the margin, smooth surface with edge entire to slightly curled.
Material examined: THAILAND, Chiang Rai Province, Mueang District, on dead culms of Brachiaria mutica (Forssk.) Stapf (Poaceae), 28 January 2017, N. Huanraluek, Gkk1 (MFLU 18-0757; HKAS 97496), living culture MFLUCC 17-2242.
Known host and distribution: On decaying wood and dead culms (Thailand) (Liu et al. 2017c).
GenBank numbers: ITS = MH118115, LSU = MH109525.
Notes: Dendryphiella phitsanulokensis was described in Thailand on decaying wood (Hyde et al. 2018b). This is the first report of D. phitsanulokensis on Brachiaria mutica. The new collection (MFLU 18-0757) and the type material (MFLU 17-2651; Hyde et al. 2018b) are morphologically very similar. In addition, there is also no phylogenetic divergence in the strain MFLUCC 17-2242 compared to the ex-type strain of D. phitsanulokensis (Fig. 17). Two Dendryphiella species have been so far recorded from Thailand (Liu et al. 2017c; Hyde et al. 2018b).
Dictyosporium Corda
Notes: Corda (1836) introduced Dictyosporium based on the type D. elegans Corda. The sexual morph of Dictyosporium species is characterised by subglobose ascomata, bitunicate cylindrical asci, and hyaline, fusiform, uniseptate ascospores with or without a sheath, while the asexual morph is characterised by micronematous to macronematous conidiophores and cheiroid, digitate complanate conidia with several parallel rows of cells (Goh et al. 1999; Boonmee et al. 2016; Yang et al. 2018a). Dictyosporium species have a worldwide distribution and have been reported from both terrestrial and aquatic habitats (Goh et al. 1999; Tanaka et al. 2015; Boonmee et al. 2016; Yang et al. 2018a).
Dictyosporium muriformis N.G. Liu, K.D. Hyde & J.K. Liu, sp. nov.
Index Fungorum number: IF557096; Facesoffungi number: FoF 06707; Fig. 16
Etymology: Name reflects the muriform conidia.
Holotype: MFLU 19-2853
Saprobic on decaying wood. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate sporodochial, scattered, black, glistening. Conidiophores semi-micronematous, mononematous, subhyaline to pale brown, aseptate, cylindrical, smooth-walled, thin-walled. Conidiogenous cells 3–3.7 μm wide, monoblastic, integrated, terminal, subhyaline to pale brown, enlarge at apex. Conidia 20–30 × 11–14.5 μm (\( \bar{x} \) = 24.5 × 14.5 μm, n = 20), solitary, acrogenous, cheiroid, median brown, not complanate, guttulate, consisting of 14–27 cells arranged in (3–)4(–5) closely compact rows, 2–7-euseptate in each column.
Culture characteristics: Conidia germinating on water agar within 24 h. Germ tubes produced from the base of conidia. Mycelia superficial, dense, circular, with entire edge, greyish brown from above and below.
Material examined: CHINA, Guizhou Province, Dushan, on decaying wood in a bank of a small freshwater, 6 July 2018, N.G. Liu, DS024 (MFLU 19-2853, holotype), ex-type living culture, GZCC 20-0006.
GenBank numbers: ITS = MT002304, LSU = MN897834, SSU = MN901117, TEF1-α = MT023011, RPB2 = MT023014.
Notes: Phylogenetic analyses (Fig. 17) show that Dictyosporium muriformis is basal to D. meiosporum Boonmee & K.D. Hyde, and D. tetrasporum L. Cai & K.D. Hyde and constitute an independent lineage within Dictyosporium. The conidia of D. muriformis resemble those of other species which consist mainly of four compact arms. However, D. nigroapice and D. tubulatum have conidial appendages which are absent in D. muriformis. Moreover, the conidia of D. muriformis are thinner than those of D. tetrasporum (11–14.5 versus 16–21.5 μm), and wider than those of D. meiosporum (11–14.5 versus 6–8.5 μm).
Phylogram generated from maximum likelihood analysis based on combined LSU, ITS and TEF1-α sequence data representing Dictyosporiaceae (Dothideomycetes). Sixty strains are included in the combined analyses which comprise 2364 characters (862 characters for LSU, 580 characters for ITS, 922 characters for TEF1-α) after alignment. Periconia igniaria (CBS 379.86) in Periconiaceae (Pleosporales) is used as the outgroup taxon. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood values of − 13815.246477 is presented. The matrix had 844 distinct alignment patterns, with 31.91% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.234018, C = 0.251157, G = 0.271122, T = 0.243703; substitution rates AC = 1.698556, AG = 3.692510, AT = 2.823016, CG = 0.799026, CT = 9.194311, GT = 1.000000; gamma distribution shape parameter α = 0.186936. Bootstrap values for maximum likelihood (ML) equal to or greater than 75% and clade credibility values greater than 0.95 from Bayesian-inference analysis labeled on the nodes. The new isolates are indicated in bold and blue
Didymellaceae Gruyter et al.
Notes: The family Didymellaceae comprises economically important plant pathogens, such as the causal agents of blackleg and ascochyta blight and various endophytic, fungicolous, lichenicolous and saprobic taxa (McDonald and Peck 2009; Aveskamp et al. 2010; de Gruyter et al. 2013; Chen et al. 2015, 2017; Doilom et al. 2018). The family Didymellaceae was established by de Gruyter et al. (2009). The type genus is Didymella Sacc. The family was established in order to accommodate the type species Didymella exigua (Niessl) Sacc., together with some Phoma Fr. or phoma-like genera which formed a strong familial clade in the phylogenetic tree (Hyde et al. 2013; Chen et al. 2017). Species of the family Didymellaceae are characterised by immersed, seldom superficial, separate or gregarious, globose to flattened, ostiolate, ascomata, with 2–5(–8) layers of pseudoparenchymatous cells. Asci are bitunicate, fissitunicate, cylindrical to clavate or saccate, 8-spored and arise from a broad hymenium among pseudoparaphyses. Ascospores are frequently hyaline or brownish and 1-septate to multi-septate (Jayasiri et al. 2017). Correct species identification in this family is difficult, mainly relying on morphology and plant host association (Aveskamp et al. 2010; Chen et al. 2015, 2017). Chen et al. (2017) provided a revision of Didymellaceae, considering all genera and proposed a new classification based on an initial phylogeny of LSU, ITS, RPB2 and TUB2 sequence data of 108 didymellaceous taxa, containing total 19 recognized genera. Consequently, 27 genera are included in the family Didymellaceae. The asexual morphs are coelomycetous or hyphomycetous which are formed on natural substrates or culture (Woudenberg et al. 2009; Chen et al. 2015).
Ascochyta Lib.
Notes: Ascochyta was introduced by Libert (1830) with A. pisi Lib. as type species. The peculiar morphological characters are the globose locules with perithecial protuberances immersed in the stroma. Species are mostly endophytes, pathogens and saprobes and associated with a number of hosts worldwide (Wijayawardene et al. 2017a; Farr and Rossman 2020). Ascochyta was accommodated in Didymellaceae (Hyde et al. 2013; Kirk et al. 2013). The asexual morphs of Ascochyta are coelomycetes (Chen et al. 2015). There are around 400 species epithets in the genus Ascochyta, however, the genus needs revision as more than 1000 records are available. We report Ascochyta medicaginicola from Medicago sp. in Italy and A. pisi is also reported as a new host record from Dioscorea communis (Dioscoreaceae) in Italy.
Ascochyta medicaginicola Qian Chen & L. Cai, Stud. Mycol. 82: 187 (2015)
Facesoffungi number: FoF 03938; Fig. 18
Ascochyta medicaginicola (MFLU 17- 0826). a Appearance of fruiting bodies on host substrate. b Close up view of conidiomata. c Section through conidioma. d Close up of ostiole. e Section through pycnidial wall. f Immature and mature conidia attached to conidiogenous cells. g Conidium. h Germinated conidium. Scale bars: a = 1000 µm, b = 200 µm, c = 50 µm, d–h = 10 µm
Replaced synonym: Phoma medicaginis Malbr. & Roum., in Roumeguère, Fungi Selecti Galliaei Exs., Cent. 37: no. 3675 (1886)
Type: USA, Minnesota, from Medicago sativa, September 1953, M.F. Kernkamp, CBS H-16487, ex-type culture CBS 112.53.
Saprobic on dead aerial stem of Medicago sp. Conidiomata visible as round to oval dark brown to black dots on the host surface. Sexual morph Undetermined. Asexual morph Coelomycetous. Conidiomata 100–200 μm high, 80–200 μm diam., pycnidial, solitary, scattered or gregarious, globose to subglobose, semi-immersed to immersed, sometimes erumpent, unilocular, thin-walled, with a single papillate, centrally located ostiole. Pycnidial walls composed 3–4 layers, similarly dense at the apex and base, with ostiole thicker than other parts, outer 1–2 layers dark brown to black, inner 1–2 layers hyaline, with thin-walled cells of textura angularis. Conidiogenous cells 4–7 × 2–4 μm (\( \bar{x} \) = 6 × 3 µm, n = 20), hyaline, phialidic, globose. Conidia 5–10 × 1–4 μm (\( \bar{x} \) = 8 × 3 µm, n = 20), ellipsoidal to cylindrical, rounded at both ends, hyaline, straight or slightly bent at the middle, aseptate, thin- and smooth-walled, with guttulate.
Culture characteristics: Colonies on PDA reaching 80 mm diam. after 7 days at 25 °C, circular colony, olivaceous to gray greenish, concentrically zonate, effuse mycelium, entire edge, reverse colony black in middle, grey olivaceous at the edge, without any diffusible pigments. Pycnidia globose to subglobose, without a distinct ostiole, glabrous, conidial matrix off whitish. Conidia sub cylindrical, aseptate.
Material examined: ITALY, Province of Forlì-Cesena [FC], Forlì-Via Pietro Nenni, on dead aerial stem of Medicago sp. (Fabaceae), 9 March 2017, E. Camporesi, IT 3281 (MFLU 17-0826), living culture, MFLUCC 18-0095, KUMCC 17-0326
Known host and distribution: Medicago sativa (Canada Czech Republic, France, USA), Melilotus albus (Russia), Trichosanthes dioica (India) (Chen et al. 2015; Jayasiri et al. 2017; Farr and Rossman 2020).
GenBank numbers: ITS = MN944408, LSU = MT020375.
Notes: We have collected the fungal isolate from dead aerial stem of Medicago sp. and it was identified as Ascochyta medicaginicola with the support from both morphology and phylogeny (Fig. 27). Our isolate clustered with A. medicaginicola (CBS 404.65, BRIP450.51, CBS 112.53) and another strain of A. medicaginicola var. medicaginicola Q. Chen & L. Cai (CBS 316.90) in the combined LSU, ITS, RPB2 and TUB2 sequence phylogeny with 98% ML and 0.98 BYPP statistical support. This is the first record of A. medicaginicola from Medicago sp. in Italy.
Ascochyta pisi Lib., Pl. crypt. Arduenna, fasc. (Liège) 1(nos 1-100): no. 59 (1830)
Facesoffungi number: FoF 06808; Fig. 19
Ascochyta pisi (MFLU 18-0904). a Ascomata on dead host surface of Dioscorea communis. b Longitudinal section of ascoma. c Longitudinal section of ostiole. d Longitudinal section of peridial wall. e Pseudoparaphyses. f–h Immature to mature asci. i Arrangement of the ascospores. j–l Ascospores. m Germinating ascospore. n, o Colonies on PDA (n = above part, o = lower part). Scale bars: a–c = 100 μm, d, f–i, n = 50 μm, e, j–m = 20 μm
≡ Septoria leguminum var. pisorum (Lib.) Desm., Ann. Sci. Nat. Bot., ser. 2, 19: 344. 1843.
= Didymella pisi Chilvers et al., Mycol. Res. 113: 396. 2009.
Epitype: NETHERLANDS, Venlo, from Pisum sativum, M.M.J. Dorenbosch, HMAS 246705, MBT202502, ex-epitype culture CBS 122785 = PD 78/517; isotype: BELGIUM, from pods of Pisum sativum, BR 5020059493320.
Saprobic on dead stem of Dioscorea communis. Sexual morphAscomata 100–150 × 300–350 µm (\( \bar{x} \) = 120 × 250 µm, n = 5), semi-immersed, solitary, globose to subglobose with long neck, uni-loculate, conspicuous and appear as black dots on host surface, shiny. Ostiole central. Ostiolar neck 100–130 µm long, 90–95 µm wide, papillate, filled with dark brown to hyaline cells. Peridium 45–60 µm wide (\( \bar{x} \) = 54 µm, n = 5) at the upper-most region of vertical wall, comprising of 3–5 cell layers, outermost layer composed dark brown cells of textura globulosa, inner layer composed light brown to hyaline cells of textura angularis. Hamathecium comprising numerous, 2.7–4.4 µm wide (\( \bar{x} \) = 3.35 µm, n = 20), filamentous, septate, pseudoparaphyses, branching and anastomosing between and above the asci. Asci 120–240 × 17–24 µm (\( \bar{x} \) = 149.7 × 21.4 µm, n = 15), 8-spored, bitunicate, cylindric-clavate, short pedicellate, rounded at the apex, with a conspicuous ocular chamber. Ascospores 21–49 × 8–12 µm (\( \bar{x} \) = 29.8 × 9.4 µm, n = 20) 1-seriate, hyaline, fusiform, 1-septate, constricted at the septum, acute at the apices, smooth, thin-walled, guttulate, lower cell is slightly wider than the upper cell. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on PDA within 24 h from single spore isolation. Colonies on PDA reaching 25–30 mm diam. after one week at 16 °C, circular, entire edge, white, with dense, convex with papillate surface, brown in reverse.
Material examined: ITALY, Province of Forlì-Cesena [FC], Pianetto-Galeata, on a dead hanging stem of Dioscorea communis (L.) Caddick & Wilkin (Dioscoreaceae), 10 March 2018, E. Camporesi, IT3777 (MFLU 18- 0904), living culture MFLUCC 18-1351.
Known host and distribution: Pisum sativum (Belgium, Canada, Netherlands, USA, Washington), Dioscorea communis (Italy, Province of Forlì-Cesena), Juglans regia (Netherlands) as well as various hosts worldwide (Chilvers et al. 2009; Chen et al. 2015; Farr and Rossman 2020; this study).
GenBank numbers: ITS = MN640976, LSU = MN640975, TUB2 = MN650642.
Notes: Ascochyta pisi is a species complex which causes ascochyta blight disease mainly on Pisum sativum (Chilvers et al. 2009). The sexual morph of Ascochyta has been referred to Didymella and Mycosphaerella (Corlett 1981; Peever et al. 2007; Tibpromma et al. 2017). The holotype (WSP 71448) and isotype (BPI 878440, K 157110) material of A. pisi are available (Chilvers et al. 2009). The phylogeny based on a combined LSU, ITS, RPB2 and TUB2 sequence dataset (Fig. 27) showed that our strain is sister to the strain CBS 122750 and clusters with other representative strains (CBS 122751, CBS 126.54, CBS 108.49) and the ex-type strain of A. pisi (CBS 122785). These isolates were collected from Pisum sativum in different localities, except the strain CBS 108.49 which was isolated from Juglans regia in Netherlands. Our isolate shares similar morphological characters to the type of A. pisi (Chen et al. 2015). Both isolates are characterised by scattered groups or individually scattered ascomata, 8-spored, cylindrical asci and hyaline, fusiform, 1-septate ascospores. However, the ascospores of our new collection are longer and slightly wider than the type (HMAS 246705) (Chen et al. 2015). This could be due to environmental factors. In addition, the hamathecium of our collection comprises numerous filamentous, septate, pseudoparaphyses, ostiole is conspicuous with a long neck and asci show a deep ocular chamber (Fig. 19) while in HMAS 246705, the hamathecial elements are lacking, the ostiole is inconspicuous and the ocular chamber in the asci is shallow (Chen et al. 2015). As per the guidelines of Jeewon and Hyde (2016) we have analyzed nucleotide differences within the rRNA gene region for further clarification. In comparison of ITS regions (ITS1-5.8S-ITS2) from 422 nucleotides there is 1 bp (0.23%) difference between MFLUCC 18-1351 and CBS 122750 strains. LSU rDNA region has 2 bp (0.24%) differences from 832 bp. In considering morpho-molecular data analysis, we conclude that our new collection is another record of Ascochyta pisi and also a new host record on Dioscorea communis (Dioscoreaceae) in Italy.
Didymella Sacc. ex Sacc.
Notes: Didymella was emended by Chen et al. (2015) to accommodate Peyronellaea and several other associated phoma-like species. They are pathogenic and saprobic on many crops and plants (Chen et al. 2015). We introduce a new species belonging to genus Didymella and a new host record.
Didymella camporesii Manawasinghe & K.D. Hyde, sp. nov.
Index Fungorum number: IF556895; Facesoffungi number: FoF 06802; Fig. 20
Didymella camporesii (JZB380040, holotype). a, b Appearance of conidiomata on the host surface. c–e Cross section through conidiomata on host. f, g Conidia from natural substrate. h Conidioma on PDA. i Pycnidial wall. j Conidia sporulated on PDA. k Upper view of colony on PDA. l Reverse view of colony on PDA. Scale bars: a, b = 100 µm, c–j = 10 μm
Etymology: Name reflects the collector of the specimens, Mr Erio Camporesi
Holotype: JZBH380040
Saprobic or necrotrophic on Amorpha fruticosa. Sexual morph Undetermined. Asexual morphConidiomata 125–149 μm diam. (\( \bar{x} \) = 74.5 μm, n = 20) on host submerged, pycnidial, solitary or confluent, pycnidial solitary or confluent, subglobose, glabrous or with some hyphal outgrows, produced on the agar surface or immersed. Pycnidial walls 11–16.8 μm (\( \bar{x} \) = 13.4 μm, n = 10) textura angularis, 5–8 layered, thick, outer walls pigmented. Conidiophores reduced to conidiogenous cells. Conidiogenous cells phialidic, hyaline, smooth, ampulliform. Conidia 8–4 × 1.5–2.6 μm (\( \bar{x} \) = 1.9 × 5.2 μm, n = 40), on host cylindrical, smooth- and thin-walled, hyaline, aseptate. On culture allantoid to fabiform, smooth- and thin-walled, hyaline, aseptate.
Culture characteristics: Colonies on PDA, reach 40 mm diam. after 7 days of incubation at 25 °C, margin regular, densely covered by floccose, and yellowish-green mycelia, immersed to submersed thin mycelial mat. Reverse initially yellowish, with age it becomes black, yellowish at margins.
Material examined: ITALY, Province of Forlì-Cesena, Fiumana di Predappio, on the dead aerial branch of Amorpha fruticosa L. (Fabaceae), 15 December 2015, E. Camporesi, IT 2743 (ZB380040, holotype), ex-type living culture, JZB380040.
GenBank numbers: ITS = MN648211, LSU = MN640406, RPB2 = MN984253, TUB2 = MN537432.
Notes: Didymella camporesii was isolated from dead and dying twigs and branches of Amorpha fruticosa in Forlì- Cesena, Italy. The characters of conidia and colony (Fig. 20) match the species concept of Didymella (Chen et al. 2015). The species identified in the present study is sister to Didymella macrophylla Qian Chen, Crous & L. Cai (CGMCC 318357) with 99% ML and 1.00 BYPP (Fig. 27). Didymella macrophylla has larger conidia (3.5–5.5 × 1.5–2.5 μm) than the current taxon (Chen et al. 2017). In pairwise nucleotide comparisons of type species of Didymella macrophylla and current taxon (JZB380040), there are 1.25% nucleotide differences in LSU (882 nucleotides) and 1.64% nucleotide difference in ITS (486 nucleotides). However, in the protein-coding genes, 2.12% nucleotide difference was observed for TUB2 across 330 nucleotides and 2.34% nucleotide difference was observed in RPB2 across 597 nucleotides. This is the first report of Didymella on Amorpha fruticosa (Farr and Rossman 2020).
Didymella macrostoma (Mont.) Qian Chen & L. Cai, in Chen et al., Stud. Mycol. 82: 177 (2015)
Facesoffungi number: FoF 02502; Fig. 21
≡ Phoma macrostoma Mont., Annls Sci. Nat., Bot., sér. 3 11: 52 (1849)
Saprobic on dead aerial stem of Origanum vulgare. Sexual morph Undetermined. Asexual morph Coelomycetous. Conidiomata 120–160 μm diam., black initially immersed, becoming erumpent through the host epidermis. Pycnidial walls composed of dark 3– 4 layers, thick-walled cells of textura angularis, outer 1–2 layers dark brown to black, inner 1–2 layers hyaline. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 4–10 μm long × 3–8 μm wide, hyaline, phialidic, ampulliform to cylindrical, smooth. Conidia 5–8 × 3.5–5 μm wide, aseptate, ellipsoidal, with several small, scattered guttules.
Material examined: ITALY, Province of Forlì-Cesena [FC], San Savino di Predappio, on dead aerial stem of Origanum vulgare L. (Lamiaceae), 22 April 2018, E. Camporesi, IT 3845 (JZB380045).
Known host and distribution: Acer pseudoplatanus (Switzerland), Ailanthus altissima (Italy), Larix decidua (Germany), Malus sylvestris (Netherlands) (Farr and Rossman 2020).
GenBank numbers: ITS = MN944409, LSU = MT020376.
Notes: The isolate obtained from dead aerial stem of Origanum vulgare was identified as Didymella macrostoma with support from both morphology and phylogeny. Our strain clusters with the reference strain of D. macrostoma (CBS 223.69), in the combined LSU, ITS, RPB2 and TUB2 sequence phylogeny with 100% ML and 1.00 BYPP statistical support (Fig. 27). This is the first record of Didymella macrostoma from Origanum vulgare in Italy.
Neodidymelliopsis Qian Chen & L. Cai
Notes: Neodidymelliopsis belongs to the family Didymellaceae (de Gruyter et al. 2009) and was introduced by Chen et al. (2015) to accommodate a small number of species formerly included in Ascochyta, Didymella and Phoma (Chen et al. 2015). The type species is Neodidymelliopsis cannabis (G. Winter) Qian Chen & L. Cai which was found on dead stem tip of Urtica dioica L. in the Netherlands. The genus is characterised by pycnidial, solitary conidiomata, hyaline, smooth conidiogenous cells, oblong to cylindrical hyaline conidia, sometimes with guttules. The sexual morph is undetermined. Ten species are accommodated in Neodidymelliopsis: N. achlydis L.W. Hou, Crous & L. Cai, N. cannabis (G. Winter) Q. Chen & L. Cai, N. farokhinejadii Ahmadp. & Mehr.-Koushk., N. longicolla L.W. Hou, Crous & L. Cai, N. moricola Chethana, Bulgakov & K.D. Hyde, N. negundinis Manawasinghe, Camporesi & K.D. Hyde, N. polemonii (Cooke) Q. Chen & L. Cai, N. ranunculi W.J. Li, Camporesi & K.D. Hyde N. sambuci Manawas., Camporesi & K.D. Hyde and N. xanthina (Sacc.) Q. Chen & L. Cai (Index Fungorum 2020).
Neodidymelliopsis camporesii D. Pem, Doilom & K.D. Hyde, sp. nov.
Index Fungorum number: IF556798; Facesoffungi number: FoF 06468; Fig. 22
Neodidymelliopsis camporesii (MFLU 18-0929, holotype). a, b Appearance of conidiomata on host surface. c Vertical section through the conidioma. d Pycnidial wall. e Conidiogenesis. f–j Conidia. k Germinating conidia. l, m Culture characteristics on MEA (l = above view, m = reverse view). Scale bars: a = 500 μm, b = 100 μm, c = 50 μm, d, k = 15 μm, e, f = 10 μm, g–j = 5 μm
Etymology: Name reflects the collector of the specimens, Mr Erio Camporesi.
Holotype: MFLU 18-0929
Saprobic or pathogenic on dead aerial stem of Galium sp. Sexual morph Undetermined. Asexual morphConidiomata 93–117 × 68–227 μm (\( \bar{x} \) = 100 × 150.8 µm, n = 20) pycnidial, globose to subglobose, ellipsoidal, or irregular, superficial to immersed solitary or confluent, ostiolate with an elongated neck. Pycnidial walls 11–15 μm, pseudoparenchymatous, comprising 1–2-layer of cells of textura angularis. Conidiophores indistinct. Conidiogenous cells 1–3 × 1.2–3.1 μm (\( \bar{x} \) = 2.4 × 2.1 µm, n = 20), annelidic, hyaline, smooth, flask-shaped, ampulliform to short cylindrical. Conidia 8–12 × 2–4 μm (\( \bar{x} \) = 10.8 × 3.5 µm, n = 20), smooth- and thin-walled, oblong, with rounded ends, sometime base obtuse, hyaline 1-septate, slightly constricted at the septum, slightly guttulate.
Culture characteristics: Colonies on MEA, 30–35 mm diam. after 7 days at 16 °C, margin regular, aerial mycelia white and woolly, greenish olivaceous, floccose; reverse dark brown, saffron near the margin.
Material examined: ITALY, Province of Forlì-Cesena [FC], Borgo Paglia- Cesena, on dead aerial stem of Galium sp. (Rubiaceae), 26 March 2018, E. Camporesi, IT3786 (MFLU 18-0929, holotype), ex-type living culture, MFLUCC 18-1136.
GenBank numbers: ITS = MN244199, LSU = MN244169, SSU = MN244175, TUB2 = MN879321.
Notes: Isolate MFLU 18-0929 obtained from dead aerial stem of Galium sp. was identified as a new species of Neodidymelliopsis, with support from both morphology and phylogeny (Figs. 22, 27). This species forms an independent lineage, clustered with the type strain of N. longicolla (CBS 382.96), N. farokhinejadii (SCUA4, SCUA6EZ) and N. ranunculi (JZB380046, MFLUCC 13-0490, MFLU 16-1870) in the combined LSU, ITS, RPB2 and TUB2 phylogeny (100% ML, 1.00BYPP; Fig. 27). Neodidymelliopsis camporesii differs from the closely related species, N. longicolla, in having larger conidiomata (200–490 × 150–360 μm versus 93–117 × 68–227 μm), narrower conidiophores (1–3 × 1.2–3.1 μm versus 4.5–6.5 × 4.5–6 μm) and shorter, hyaline ascospores (8–12 × 2–4 μm versus 12–15 × 4–7 μm) compared to pale brown ascospores of N. longicolla. We, therefore, introduce N. camporesii as a new species in Neodidymelliopsis.
Neodidymelliopsis ranunculi W.J. Li & K.D. Hyde, in Hyde et al., Fungal Diversity: https://doi.org/10.1007/s13225-016-0373-x, [41] (2016)
Facesoffungi number: FoF 02349; Fig. 23
Holotype: ITALY, Province of Forlì-Cesena [FC], Castrocaro Terme e Terra del Sole, near Converselle, on dead stem of Ranunculus sp. (Ranunculaceae), 2 December 2012, E.Camporesi, IT-936 (MFLU 16-1870), ex-type living culture, MFLUCC 13-0490.
Saprobic on dead aerial branches of Bougainvillea spectabilis. Conidiomata visible as round to oval small black dots on the host surface. Sexual morph Undetermined. Asexual morph Coelomycetous. Conidiomata 120–180 μm high, 30–80 μm diam., pycnidial, solitary, scattered or gregarious, globose to subglobose, semi-immersed to immersed, sometimes erumpent, unilocular, thin-walled, with a single papillate, centrally located ostiole. Pycnidial walls composed 3–4 layers, similarly dense at the apex and base, with ostiole thicker than other parts, outer 1–2 layers dark brown to black, inner 1–2 layers hyaline, with thin-walled cells of textura angularis. Conidiogenous cells 2–4 μm long × 1–2 μm wide, hyaline, phialidic, globose. Conidia 7–9 × 1–3 μm wide (\( \bar{x} \) = 8 × 2.5 µm, n = 20), ellipsoidal to cylindrical, rounded at both ends, hyaline, straight or slightly bent at the middle, aseptate, thin- and smooth-walled, sometimes guttulate.
Culture characteristics: Colonies on PDA reaching 30 mm diam. after 7 days at 25 °C, circular colony, white, concentrically zonate, effuse mycelium, entire edge, reverse colony color black, without any diffusible pigments. Pycnidia globose to subglobose, without a distinct ostiole, glabrous, conidial matrix off whitish. Conidia subcylindrical, aseptate.
Material examined: ITALY, Province of Forlì-Cesena [FC], Forli-Via Pietro Nenni, on dead aerial branches of Bougainvillea spectabilis Willd. (Nyctaginaceae), 9 March 2017, E. Camporesii, IT 3329 (MFLU 17-0980), living culture, JZB 380046.
Known host and distribution: Bougainvillea spectabilis, Ranunculus sp. (Italy) (Hyde et al. 2016; this study).
GenBank numbers: ITS = MN944410, LSU = MT020377.
Notes: The isolate JZB 380046 obtained from dead aerial branches of Bougainvillea spectabilis was identified as Neodidymelliopsis ranunculi with support from both morphology and phylogeny. This isolate clustered with the other type and reference isolates of N. ranunculi (MFLUCC 13-0490 and MFLU 16-1870), in the combined LSU, ITS, RPB2 and TUB2 phylogeny (Fig. 27). Further our isolate has 0.98% and 0.93% base pair differences with Neodidymelliopsis ranunculi (MFLUCC 13-0490) in ITS and LSU gene regions. Neodidymelliopsis species have been reported in many economically important host plants like Citrus, Acer, Pinus worldwide (Farr and Rossman 2020). This is the first record of Neodidymelliopsis ranunculi on Bougainvillea spectabilis from Italy.
Nothophoma Qian Chen & L. Cai
Notes: Nothophoma was introduced by Chen et al. (2015) with N. infossa (Ellis & Everh.) Qian Chen & L. Cai as the type species. The genus has been studied phylogenetically using ITS, LSU and TUB2 sequence data. Species belonging to this genus are characterised by large multi-loculate pycnidia, that are globose to elongated, or irregular, superficial on or immersed into the agar, solitary or confluent, ostiolate and sometimes with a short neck.
Nothophoma quercina (Syd. & P. Syd.) Qian Chen & L. Cai, Stud. Mycol. 82: 213 (2015)
Facesoffungi number: FoF 04918; Fig. 24
Nothophoma quercina (MFLU 17-2128). a, b Appearance of conidiomata on host surface. c, d Section of conidiomata. e Section through pycnidial wall (stained with congo red). f–h Conidiogenous cells (h = stained with congo red). i–l Conidia stained in congo red. Scale bars: d = 100 µm, e = 30 µm, c, f–l = 10 µm
≡ Cicinobolus quercinus Syd. & P. Syd. [as ‘Cicinnobolus’], Annls mycol. 13(1): 42 (1915)
Saprobic or weak pathogenic on Prunus mahaleb. Sexual morph Undetermined. Asexual morphConidiomata 220–265 μm high × 260–380 μm diam. (\( \bar{x} \) = 245 × 340 μm, n = 10), pycnidial, stromatic, solitary or clustered, semi-immersed, erumpent at maturity, dark brown to black, globose, without an ostiole. Pycnidial walls 25–35 μm wide, composed of two layers of hyaline to dark brown pseudoparenchymatous cells of textura angularis. Conidiogenous cells 10–22 μm high × 4–6 μm wide, hyaline, thin-walled, holoblastic, smooth, cylindrical, swollen at the base, discrete, producing a single conidium at the apex. Conidia 11–16 × 6–10 μm (\( \bar{x} \) = 14 × 9 μm, n = 50), aseptate, globose to subglobose, widest in the centre, with rounded apex, initially hyaline, becoming dark brown before release from the pycnidia, wall moderately thick, externally smooth, internally roughened. Spermatia 3–5 × 1.5–2 μm, rod-shaped with obtuse ends, hyaline, thin-walled, smooth.
Culture characteristics: Conidia germinating on water agar and germ tubes produced from conidia within 12 h from lower end. Colonies growing on PDA, circular, with flat surface, edge entire, reaching 28 mm in 3 weeks at 25 °C, white to pale brown in PDA medium on top. Mycelium superficial and partially immersed, branched, septate, hyaline grey to black, smooth.
Material examined: RUSSIA, Rostov region, Shakhty City, artificial ravine forest belt, on dead and dying branches of Prunus mahaleb L. (Rosaceae), 11 May 2017, T.S. Bulgakov, T-1826 (MFLU 17-2128), living culture, MFLUCC 18-0455.
Known host and distribution: Saprobic and pathogenic on various hosts from temperate regions (Aveskamp et al. 2010; Chen et al. 2015; Bai et al. 2016; Jiao et al. 2017; Liu et al. 2018b; Moral et al. 2018; Chethana et al. 2019; Farr and Rossman 2020).
GenBank numbers: ITS = MN909056, LSU = MN909057.
Notes: The culture on PDA of our isolate is similar to Nothophoma variabilis Valenz.-Lopez, Cano, Guarro & Stchigel strain CBS 142457 (Valenzuela-Lopez et al. 2018). However, based on phylogenetic analyses our strain clusters with N. quercina (CBS 633.92, UTHSC DI16-270) and morphologically, it is most similar to N. quercina (CBS 633.92) (Chen et al. 2015). This is the first report of N. quercina on Prunus mahaleb in Russia.
Xenodidymella Qian Chen & L. Cai.
Notes: Xenodidymella was introduced by Chen et al. (2015) to accommodate didymella-like taxa with X. applanata (Niessl) Q. Chen & L. Cai (≡ Didymella applanata (Niessl) Sacc.) as the type species. Xenodidymella is characterised by the pseudoparenchymatous wall comprising isodiametric cells and broadly fusiform hyaline ascospores (Chen et al. 2015). Species are mostly endophytes, pathogens and saprobes associated with a number of hosts from European countries (Farr and Rossman 2020). The asexual morph of Xenodidymella is coelomycetous having pycnidial, globose to subglobose conidiomata, phialidic, hyaline, globose to ampulliform conidiogenous cells and hyaline, smooth- and thin-walled conidia which are variable in shape (Chen et al. 2015). There are five epithets reported for the genus (Index Fungorum 2020). We introduce a new species, X. camporesii which was isolated from the stem of Dipsacus sp. in Italy.
Xenodidymella camporesii D. Pem, Doilom & K.D. Hyde, sp. nov.
Index Fungorum number: IF557076; Facesoffungi number: FoF 06467; Fig. 25
Xenodidymella camporesii (MFLU 17-1082, holotype). a, b Appearance of ascomata on host surface. c Vertical section through the ascoma. d Peridium. e Hamathecium. f–h Asci. i–k Ascospores. l Germinating ascospores. m, n Culture characteristics on MEA (m = above view, n = reverse view). Scale bars: a = 500 μm, b = 1000 μm, c = 100 μm, d, f = 30 μm, g, h = 50 μm, e, i–k = 10 μm, l = 20 μm
Etymology: The epithet honours Mr Erio Camporesi who collected this fungus.
Holotype: MFLU 17-1082
Saprobic on dead aerial stem of Dipsacus sp. Sexual morphAscomata 185–308 μm high, 185–330 μm wide, pseudothecial, separate or gregarious, or scattered, immersed to semi-immersed, dark brown to black, with central ostiole. Peridium 25–33 μm wide, comprising 1–2 layers of cells of textura angularis. Hamathecium lacking pseudoparaphyses. Asci 55–85 × 9–13 μm (\( \bar{x} \) = 66.6 × 12.7 µm, n = 20), 8-spored, bitunicate, fissitunicate, cylindric-clavate to clavate, short-pedicellate or apedicellate, apically rounded, with an ocular chamber. Ascospores 15–18 × 5–6 μm (\( \bar{x} \) = 14.5 × 5.6 µm, n = 20), overlapping 1–2-seriate, hyaline, ovoid to ellipsoidal, asymmetrical, upper cells wider than lower cells, 1-septate, slightly constricted at the septum, conical at both ends, smooth-walled. Asexual morph Undetermined.
Culture characteristics: Colonies on MEA, 35–45 mm diam. after 7 days at 16 °C, margin regular, aerial mycelia white and woolly, greenish olivaceous, floccose; reverse dark brown, saffron near the margin.
Material examined: ITALY, Province of Forlì-Cesena [FC], near Verghereto, on dead aerial stem of Dipsacus sp. (Scrophulariaceae), 5 June 2017, E. Camporesi, IT3374 (MFLU 17-1082, holotype), ex-type living culture, MFLUCC 17-2309.
GenBank numbers: ITS = MN244198, LSU = MN244168, SSU = MN244174, TUB2 = MN871955.
Notes: Our strain MFLUCC 17-2309 has a close phylogenetic affinity to Xenodidymella applanata (CBS 205.63) in our phylogeny analyses (Fig. 26). Morphologically, X. camporesii differs from X. applanata in having longer asci (55–85 × 9–13 μm versus 50–60 × 10.5–14.5 μm) and longer ascospores (15–18 × 5–6 μm versus 11.5–15.5 × 5.5–7.5 μm). Comparison of TUB2 nucleotides of X. applanata and X. camporesii reveals 24 (10.43%) nucleotide differences. We, therefore, introduce X. camporesii as a new species (Fig. 27).
Phylogram generated from maximum likelihood analysis based on combined LSU, ITS, and TUB2 sequence data. Twenty-seven strains are included in the combined gene analyses comprising 1577 characters after alignment (854 characters for LSU, 491 characters for ITS, 232 characters for TUB2). Microsphaeropsis olivacea (CBS 432.71, CBS 233.71) is used as the outgroup taxon. The tree topology of the Bayesian analysis was similar to the maximum likelihood analysis. The best RaxML tree with a final likelihood value of − 3643.265848 is presented. The matrix had 147 distinct alignment patterns, with 1.22% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.240330, C = 0.233630, G = 0.275019, T = 0.251022; substitution rates AC = 4.369019, AG = 9.812360, AT = 6.383275, CG = 0.483743, CT = 25.398844, GT = 1.000000; gamma distribution shape parameter α = 0.618184. Bootstrap values for maximum likelihood equal to or greater than 50% and Bayesian posterior probabilities equal or greater than 0.90 BYPP are placed above or below the branches. Ex-type strains are in bold and black. The newly generated sequence is indicated in blue
Phylogram generated from maximum likelihood analysis based on combined LSU, ITS, RPB2 and TUB2 sequence data representing genus Ascochyta, Didymella, Neodidymelliopsis, Nothophoma and Phomatodes in the family Didymellaceae. Reference sequences were downloaded from GenBank based on published data (Chen et al. 2015; Chethana et al. 2019). One hundred and twenty-eight strains are included in the combined gene analyses and Phoma herbarum (CBS 377.92 and CBS 502. 91) is used as the outgroup taxon. The tree topology of the Bayesian analysis was similar to the maximum likelihood analysis. The best RaxML tree with a final likelihood value of − 15328.506166 is presented. The matrix had 533 distinct alignment patterns, with 10.44% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.237260, C = 0.240627, G = 0.279223, T = 0.242890; substitution rates AC = 1.196224, AG = 5.897645, AT = 1.950521, CG = 0.765245, CT = 13.348334, GT = 1.000000; gamma distribution shape parameter α = 0.813932. Bootstrap values for maximum likelihood equal to or greater than 60% and Bayesian posterior probabilities equal or greater than 0.90 BYPP are placed near the branches as ML/BYPP. Ex-type strains indicated in T. The newly generated sequences are indicated in red
Didymosphaeriaceae Munk
Notes: We follow the treatment and classification of Didymosphaeriaceae in Ariyawansa et al. (2014) and Phookamsak et al. (2019). Twenty-seven genera were accepted in Didymosphaeriaceae (Wijayawardene et al. 2018a; Phookamsak et al. 2019). We provide an updated phylogenetic tree for Didymosphaeriaceae (Fig. 34).
Kalmusia Niessl
Notes: Kalmusia was introduced by Niessl (1872) and is typified by K. ebuli Niessl. The genus is characterised by septate pseudoparaphyses, narrowly ovoid to clavate asci, 3-septate ascospores in their sexual morph and coniothyrium-like asexual morphs (Ariyawansa et al. 2014; Zhang et al. 2014). Ariyawansa et al. (2014) revisited the genus and provided multi-gene phylogeny while synonymizing Dendrothyrium under Kalmusia and two new combinations. There are 44 Kalmusia species recorded in Index Fungorum (2020). However, only two new species have been introduced to the genus during the last few years viz. K. italica Thambugala et al. and K. spartii Wanasinghe et al. collected from Italy (Liu et al. 2015). We introduce a new Kalmusia species collected from northern Thailand based on morphological traits and multi-gene phylogeny.
Kalmusia erioi Samarak., Thambugala & K.D. Hyde, sp. nov.
Index Fungorum number: IF556802; Facesoffungi number: FoF 06462; Fig. 28
Etymology: In honour of Mr. Erio Camporesi, for his immense contribution to mycology.
Holotype: MFLU 18-0832
Saprobic on dead branches of unidentified dicotyledonous tree. Sexual morphAscomata 140–170 µm high, 145–240 µm diam. (\( \bar{x} \) = 157 × 182 µm, n = 10), immersed, slightly erumpent through the host tissue, scattered or gregarious, dark brown to black, uni- to bi-loculate, globose to subglobose, with a flattened base. Ostiole blackish-brown, smooth, ostiolar canal filled with hyaline cells. Peridium 10–18 µm wide, 4–5 layers, comprising 4.4–13.4 × 2–4.5 µm (\( \bar{x} \) = 8.5 × 3.2 µm, n = 20), brown to lightly pigmented, slightly flattened cells of textura angularis. Hamathecium comprising numerous, 2–3.5 µm wide (\( \bar{x} \) = 2.7 µm, n = 20), cellular, branched, septate pseudoparaphyses. Asci 76–113 µm × 13–18 µm (\( \bar{x} \) = 89.7 × 15 µm, n = 20), 8-spored, bitunicate fissitunicate, clavate, short-pedicellate, thick-walled at the apex, with an ocular chamber. Ascospores 15.5–20 × 7.7–8.5 µm (\( \bar{x} \) = 17.6 × 7.5 µm, n = 30), overlapping 1–2-seriate, mostly ellipsoidal, muriform, 3-transversely septate, with 1–2 vertical septa, deeply constricted at the middle septum, slightly constricted at the remaining septa, initially hyaline to pale yellow, becoming brown at maturity, rounded and pointed at ends remain in light brown, guttulate and smooth-walled. Asexual morph Undetermined.
Material examined: THAILAND, Chiang Rai Province, Mae Fah Luang University, on dead branch of a dicotyledonous tree, 9 July 2017, M.C. Samarakoon, SAMC075 (MFLU 18-0832, holotype); ibid., KUN-HKAS 102374 (isotype).
GenBank numbers: ITS = MN473058, LSU = MN473052, SSU = MN473046, TEF1-α = MN481599, TUB2 = MN481603.
Notes: The morphological characters of Kalmusia erioi fit into the generic concept of Kalmusia in having immersed to erumpent ascomata, filiform, delicate, septate pseudoparaphyses, bitunicate, clavate asci with narrowly ovoid to clavate, pale brown, 3-septate ascospores. Ascospores of Kalmusia erioi are characterised with 1–2 vertical septa, deeply constricted at the middle septum and rounded and pointed at ends remaining in light brown which are different from other Kalmusia species. The combined LSU, SSU, ITS and TEF1-α phylogenetic analyses show that our strain (MFLU 18-0832) clusters as basal to Kalmusia clade (97% ML, 1.00 BYPP; Fig. 34). BLASTn search of LSU is similar to Kalmusia variispora (Verkley, Göker & Stielow) Ariyaw. & K.D. Hyde strain M5 (MK138781, 99.5% similarity). Based on clear morphological differences and available molecular data, here we introduce Kalmusia erioi as a new species following guidelines by Jeewon and Hyde (2016). This is the first Kalmusia species reported from Thailand.
Montagnula Berl.
Montagnula was introduced by Berlese (1896) for saprobic taxa that occurred on dead wood and leaves in terrestrial habitats. Montagnula is characterised by its cylindric-clavate to clavate asci with long pedicel and brown ascospores. The genus was placed in the family Didymosphaeriaceae by Ariyawansa et al. (2014) with peculiar phragmosporous and didymosporous characters. There are 36 epithets of Montagnula listed in Species Fungurum (2020), however, only 11 species were confirmed by molecular data. We introduce a new species Montagnula camporesii which was collected from Italy.
Montagnula camporesii Phukhams. & K.D. Hyde, sp. nov.
Index Fungorum number: IF556760; Facesoffungi number: FoF 06343; Fig. 29
Montagnula camporesii (MFLU 16-2290, holotype). a Appearance of ascomata on Dipsacus sp. b Vertical section through ascoma. c Ostiolar canal. d Section of partial peridium layer. e Pseudoparaphyses. f–h Developmental state of asci. i–l Developmental state of ascospores. m Culture characteristic on MEA. Scale bars: b = 200 µm, c, d, f–h = 50 µm, e = 20 µm, i–l = 10 µm
Etymology: The species epithet is honouring Erio Camporesi.
Holotype: MFLU 16-2290
Saprobic on dead stems of Dipsacus L. Sexual morphAscomata 200–250 × 300–350 μm (\( \bar{x} \) = 220 × 330 μm, n = 5), covered by host peridium, superficial on the cortex of host, superficial, solitary, scattered, uni-loculate, subglobose to depressed globose, flatten base, ligth brown to brown, smooth-walled, papillate. Ostiole central, 60 × 150 μm, brown to light brown, papillate, opened pore, ostiolate filled with periphyses. Peridium 13–40 μm wide, thin, uniform, comprising of 4–6 layers of light brown to brown cells of textura angularis, inner layers comprising thin, hyaline cells. Hamathecium of dense, 1.5–3.0 μm wide (\( \bar{x} \) = 2.5 μm, n = 50), filiform branches, anastomosing above asci, reaching the ostiole part, transverse septate, cellular pseudoparaphyses. Asci 80–120 × 10–15 μm (\( \bar{x} \) = 95 × 15 μm, n = 20), 8-spored, bitunicate, fisitunicate, thick-walled, clavate, apically rounded, with long, furcated pedicel, ocular chamber clearly visible when immature. Ascospores 18–25 × 5–8 μm (\( \bar{x} \) = 20 × 6.5 μm, n = 50), 2-seriate, partial overlapping, hyaline when immature, reddish brown to brown at maturity, cell above median septum slightly wider than below, tapering towards the ends, slightly acute at both ends, with (1–)3 transversely septa, constricted at the septa, indentation present, without mucilaginous sheath. Asexual morph Undetermined.
Culture characteristics: Colonies on MEA, reaching 30 mm diam. after 4 weeks of incubation at 16 °C. Culture above cream with orange brown at the edge, medium dense mycelium, colonies circular, flat, umbonate, dull, covered with white aerial mycelium; reverse dark brown, dense, irregular, fimbriate margin.
Material examined: ITALY, Province of Forlì-Cesena [FC], Riofreddo-Bagno di Romagna, on dead aerial stem of Dipsacus sp. (Caprifoliaceae), 5 August 2016, E. Camporesi, IT 3059 (MFLU 16-2290, holotype), ex-type living culture, MFLUCC 16-1369.
GenBank numbers: ITS = MN401746, LSU = MN401742, SSU = MN401744, RPB2 = MN397909, TEF1-α = MN397908.
Notes: In the phylogenetic analysis (Fig. 34), Montagnula camporesii (strain MFLUCC 16-1369) forms a close relationship with M. bellevaliae Wanas., Camporesi, E.B.G. Jones & K.D. Hyde (strain MFLUCC 14-0924), M. cirsii Qing Tian, Camporesi & K.D. Hyde (strain MFLUCC 13-0680) and M. scabiosae (strain MFLUCC 14-0954) with strong statistical support (87% ML, 0.99 BYPP; Fig. 34). The Montagnula species in this clade are morphologically similar in having uniform peridium walls, reddish brown, broad fusiform ascospores, with (1–)3 transverse septa (Hongsanan et al. 2015; Hyde et al. 2016). Montagnula camporesii is reported from Dipsacus sp. and can be distinguished by its superficial ascomata located on the host surface and different sizes of asci. Montagnula camporesii has depressed globose ascomata with rather short pedicellate (Fig. 29).
Neokalmusia Ariyaw. & K.D. Hyde
Notes: Neokalmusia was introduced to accommodate N. brevispora (Nagas. & Y. Otani) Kaz. Tanaka, Ariyaw. & K.D. Hyde and N. scabrispora (Teng) Kaz. Tanaka, Ariyaw. & K.D. Hyde based on phylogeny and morphology (Ariyawansa et al. 2014). Later three other Neokalmusia species were discovered viz. N. didymospora D.Q. Dai & K.D. Hyde (Dai et al. 2016a), N. arundinis Thambugala & K.D. Hyde and N. thailandica Phukhams. & K.D. Hyde (Thambugala et al. 2017). The genus is characterised with subglobose to oblong ascostromata including several perithecia in a row, a clypeus-like structure composed of thin-walled cells and verrucose ascospores. In this study, another new species in the genus is introduced from China. Until now, the species of the genus are known from Italy, Japan and Thailand (Ariyawansa et al. 2014; Dai et al. 2016a; Thambugala et al. 2017).
Neokalmusia kunmingensis H.B. Jiang, Phookamsak & K.D. Hyde, sp. nov.
Index Fungorum number: IF555422; Facesoffungi number: FoF 05081; Fig. 30
Neokalmusia kunmingensis (KUN-HKAS 101765, holotype). a–c Ascostromata on dead bamboo surface (immersed within epidermis). d Vertical section of ascostroma. e Peridium. f–i Asci. j Pseudoparaphyses. k Germinating ascospore. l, n Ascospores. m Ascospore stained by Indian ink. o Culture frontage and back. Scale bars: d = 150 μm, e = 30 μm, j = 20 μm, f–i = 15 μm, k–n = 10 μm
Etymology: The epithet “kunmingensis” refers to the city where the taxon was collected.
Holotype: KUN-HKAS 101765
Saprobic on dead bamboo culms in terrestrial habitats. Sexual morphAscostromata 0.5–5 mm diam., 170–300 μm high, scattered or in groups, immersed under a black clypeus-like structure, dark brown to black, multi-loculate. Locules 280–370 μm diam., 175–290 μm high, immersed within ascostromata, dark brown to black, subglobose to ampulliform, ostiolate. Peridium 13–23 μm broad, comprising several layers; outer layers dark brown to brown, compressed cells of textura angularis; inner layers hyaline compressed pseudoparenchymatous cells, arranged in textura angularis. Hamathecium composed of 1.2–2 μm broad, septate, branched, anastomosing, filiform, hyaline, pseudoparaphyses. Asci 63–77 × 9.6–11.4 μm (\( \bar{x} \) = 70 × 10.5 μm, n = 20), 8-spored, bitunicate, cylindrical to clavate, curved, short pedicel with slightly furcate ends, apically rounded. Ascospores 13–15 × 4–5 μm (\( \bar{x} \) = 14 × 4.5 μm, n = 20), 1–2-seriate, partially overlapping, fusiform, dark brown to brown, 3-septate, constricted at the septa, straight to slightly curved, smooth-walled, guttulate, without sheath. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on PDA within 24 h. Colonies on PDA fast growing, 30–35 mm diam. after 4 weeks at 20–25 °C, colonies circulate, slightly raised to umbonate in the centre, floccose to woolly, slightly radiating, pale black at the middle, pale white at the margin from the above, pale black at the middle part, white at the margin from the below, not produced pigment in agar.
Material examined: CHINA, Yunnan Province, Kunming, Golden Temple, on dead bamboo culms, 5 December 2017, H.B. Jiang & R. Phookamsak, GT002 (KUN-HKAS 101765, holotype), ex-type living culture KUMCC 18-0120.
GenBank numbers: ITS = MK079886, LSU = MK079889, SSU = MK079887, TEF1-α = MK070172.
Notes: Neokalmusia kunmingensis is morphologically similar to N. brevispora in having multi-loculate, subglobose to ampulliform, immersed ascostromata under clypeus-like structure with several perithecia in a row, 8-spored, bitunicate, cylindrical to clavate, short-pedicellate asci and 3-septate, brown to dark brown, fusiform ascospores (Ariyawansa et al. 2014). However, N. kunmingensis differs from N. brevispora in having longer ascostromata (0.5–5 mm long versus 1–3.5 mm long), smaller asci (63–77 × 9.6–11.4 μm versus 80–118 × 10.5–15 μm), and having 4-guttulate, smaller ascospores without a mucilagenous sheath (13–15 × 4–5 μm versus 18–24 × 4–7 μm). Multi-gene phylogenetic analyses (Fig. 34) show that N. kunmingensis forms a sister clade to N. brevispora with high support (100% ML, 1.00 BYPP). A comparison of the ITS and TEF1-α nucleotides of N. kunmingensis and the type strain of N. brevispora (KT 2313) reveals nucleotide differences ≥ 1.5%, which indicates that N. kunmingensis is a new species in Neokalmusia.
Pseudocamarosporium Wijayaw. & K.D. Hyde
Notes: Pseudocamarosporium was introduced by Wijayawardene et al. (2014), and is typified by P. propinquum (Sacc.) Wijayaw., Camporesi & K.D. Hyde on Salix vitellina. The genus is closely related to Paracamarosporium phylogenetically, but it can easily be distinguished. We follow the latest treatment and updated accounts of Pseudocamarosporium in Wanasinghe et al. (2018). Thirteen species are known in this genus (Index Fungorum 2020). Pseudocamarosporium species are asexual morphs with brown, oblong to ellipsoidal, septate or muriform conidia and there is no sexual morph reported (Wijayawardene et al. 2014, 2016b; Crous et al. 2015b; Liu et al. 2015; Li et al. 2016a; Wanasinghe et al. 2018). We introduce a new species P. camporesii based on morphological characters and phylogenetic analysis.
Pseudocamarosporium camporesii Q. Tian & K.D. Hyde, sp. nov.
Index Fungorum number: IF556933; Facesoffungi number: FoF 06858; Fig. 31
Pseudocamarosporium camporesii (MFLU 16-2134, holotype). a Herbarium material, stem of Quercus cerris. b–d Appearance of black conidiomata semi-immersed to erumpent through the host. e, f Vertical section through conidioma. g Papillate ostiole. h Vertical section of conidioma wall. i, j Conidiogenous cells and developing conidia. k–p Conidia. Scale bars: b = 1000 µm, c = 500 µm, d = 200 µm, e, f = 50 µm, g, h = 25 µm, i, j = 10 µm, k–p = 5 µm
Etymology: Named in honor of Mr. Erio Camporesi who made an immense contribution to fungal investigation and collection.
Holotype: MFLU 16-2134
Saprobic on a dead stem of Quercus cerris. Sexual morph Undetermined. Asexual morph Coelomycetous. Conidiomata 300–400 μm diam., 180–250 μm high, pycnidial, solitary to gregarious, immersed to semi-immersed, erumpent to superficial at maturity, unilocular, globose to subglobose, sometimes depressed, ostiolate. Ostiole single, centrally located, papillate. Pycnidial walls 18–30 μm, multi-layered, with 3–5 outer layers of brown-walled cells of textura angularis, with innermost layer thin, hyaline. Conidiophores reduced to conidiogenous cells. Conidiogenous cells blastic, phialidic, hyaline, cylindrical, smooth, from the innermost layer of conidioma wall. Conidia 15–20(–30) × 8–12(–14) μm (\( \bar{x} \) = 18.5 × 10.3 μm, n = 20), oblong to ellipsoidal, initially hyaline, brown to dark brown at maturity, muriform, with 1–3 transverse septa, with 1–3 longitudinal septa, constricted at the septa, straight or infrequently slightly curved, narrowly rounded at both ends, smooth-walled, without mucilaginous sheath. Conidial secession schizolytic.
Material examined: ITALY, Province of Arezzo [AR], Montemezzano-Stia, on dead aerial branch of Quercus cerris L. (Fagaceae), 7 July 2016, E. Camporesi, IT 3028 (MFLU 16-2134, holotype).
GenBank numbers: ITS = MN653128, LSU = MN653129, SSU = MN653127.
Notes: A new camarosporium-like taxon, Pseudocamarosporium camporesii was collected from Italy on a dead stem of Quercus cerris during an investigation on the diversity of fungi in Italy. It is characterised by pycnidial, ostiolate conidiomata, blastic, phialidic conidiogenous cells and brown, muriform conidia that fit well within the species concept of Pseudocamarosporium in Didymosphaeriaceae. Based on a megablast search using the ITS sequence, the closest matches in NCBI’s GenBank nucleotide database was P. brabeji (Marinc., M.J. Wingf. & Crous) Crous (GenBank MG098280; Identities 596/596 (100%), no gap). The highest similarity using the LSU sequence was P. propinquum (GenBank MG8112621; Identities 808/808 (100%), no gaps). The highest similarities using the SSU sequence were P. propinquum (GenBank KJ819949; Identities 986/986 (100%), no gaps), P. corni Wijayaw., Camporesi & K.D. Hyde (GenBank NG_062183; Identities 984/986 (99%), no gaps), P. corni (GenBank KJ819946; Identities 984/986 (99%), no gaps). We analysed combined ITS, LSU, SSU and TEF1-α sequence data for Didymosphaeriaceae, but Pseudocamarosporium species do not have available protein (TEF1-α) sequence data in GenBank. Based on ITS, LSU and SSU sequences analyses from the ribosomal genes for Pseudocamarosporium our new taxon belongs to Pseudocamarosporium and shares close phylogenetic affinities to P. quercinum (Fig. 34). However, our new collection differs from P. quercinum in having larger conidia (\( \bar{x} \) = 18.5 × 10.3 μm versus \( \bar{x} \) = 16.4 × 6.8 μm) with 1–3 longitudinal septa and without a centrally located ostiole, while P. quercinum has smaller conidia with 1–2 longitudinal septa, and with a papillate ostiole. Pseudocamarosporium quercinum was discovered on dead branches of Quercus pubescens while our new collection occurred on a dead stem of Q. cerris (Wijayawardene et al. 2016b). Thus, Pseudocamarosporium camporesii is proposed as a new species.
Tremateia Kohlm., Volkm.-Kohlm. & O.E. Erikss.
Notes: Tremateia Kohlm. et al. was introduced by Kohlmeyer et al. (1995) to accommodate T. halophile Kohlm. et al. as a facultative marine genus which is characterised by depressed globose, immersed ascomata, numerous cellular pseudoparaphyses, fissitunicate and clavate asci, ellipsoid muriform ascospores, and a phoma-like asexual morph. Based on morphology and phylogeny, Ariyawansa et al. (2014) confirmed the familial status of Tremateia in Didymosphaeriaceae. The terrestrial distribution of Tremateia species was confirmed by Hyde et al. (2016) and Feng et al. (2019) while providing three novel taxa as T. arundicola Wanasinghe et al., T. guiyangensis J.F. Zhang et al. and T. murispora Y. Feng et al. from UK and China. We introduce two novel Tremateia species based on morphology and multi-gene phylogeny of a combined LSU, SSU, ITS and TEF1-α.
Tremateia camporesii Samarak. & K.D. Hyde, sp. nov.
Index Fungorum number: IF556787; Facesoffungi number: FoF 06464; Fig. 32
Tremateia camporesii (MFLU 19-2109, holotype). a–c Appearance of ascomata on the substrate. d Vertical section of ascoma. e Peridium. f Pseudoparaphyses. g–j Asci. k–q Ascospores (q in Indian Ink showing mucilaginous sheath). Scale bars: a = 1 cm, b = 500 µm, c, d = 200 µm, e, g–j = 20 µm, k–q = 10 µm, f = 5 µm
Etymology: In honour of Mr. Erio Camporesi, for his immense contribution to mycology.
Holotype: MFLU 19-2109
Saprobic on dead branch. Sexual morphAscomata 310–400 µm high, 230–320 µm diam. (\( \bar{x} \) = 282 × 155.7 µm, n = 5), immersed to erumpent, globose or subglobose, scattered, rarely gregarious, dark brown to black, coriaceous. Peridium 13–25 µm wide (\( \bar{x} \) = 19.6 µm, n = 10), comprising outer dark brown and inner hyaline cells of textura angularis. Hamathecium comprising numerous, 2–3.5 µm wide (\( \bar{x} \) = 2.8 µm, n = 20), filamentous, branched, septate, hyaline pseudoparaphyses. Asci 85–120 × 9–13.5 µm (\( \bar{x} \) = 101 × 11.5 µm, n = 20), 4–(2–6)-spored, bitunicate fissitunicate, clavate, pedicellate, thick-walled at the apex, with an ocular chamber. Ascospores 16–20 × 5–7 µm (\( \bar{x} \) = 17.4 × 6.3 µm, n = 30), overlapping 1–2-seriate, mostly ellipsoidal, muriform, 3–4-transversely septate, with 1–2 vertical septa, deeply constricted at the middle septum, slightly constricted at the remaining septa, initially hyaline to pale yellow, becoming brown at maturity, rounded at both ends, surrounded by a thick mucilaginous sheath. Asexual morph Undetermined.
Material examined: CHINA, Guizhou Province, Guiyang, Guizhou Academy of Agricultural Sciences, on dead branch, 22 July 2018, M.C. Samarakoon, SAMC170 (MFLU 19-2109, holotype); ibid., KUN-HKAS 102398 (isotype).
GenBank numbers: ITS = MN473061, LSU = MN473056, SSU = MN473050, TEF1-α = MN481602, TUB2 = MN481606.
Notes: Tremateia camporesii is similar to Tremateia species by immersed to erumpent, globose ascomata, numerous cellular pseudoparaphyses, fissitunicate and clavate asci and ellipsoid muriform ascospores. Tremateia camporesii, T. halophila Kohlm., Volkm.-Kohlm. & O.E. Erikss., T. lamiacearum Samarak. & K.D. Hyde and T. murispora Y. Feng, S.N. Zhang & Z.Y. Liu share mucilaginous ascospores. However, among all other Tremateia species, T. camporesii differs from all other Tremateia species by (2–)4(–6)-spored asci. Tremateia camporesii, T. guiyangensis and T. murispora have been collected from the same geographical region. Among them, T. camporesii and T. murispora cluster together in phylogenetic analyses (Fig. 34). However, T. murispora possesses ascospores with 3–7 transverse septa and 8-spored asci and is clearly distinguished from T. camporesii. In a BLASTn search of LSU the new species is similar to T. arunicola Wanas., E.B.G. Jones & K.D. Hyde (MFLUCC 16-1275; KX274248, 98% similarity), Bimuria novae-zelandiae D. Hawksw., Chea & Sheridan (CBS 107.79; MH872950, 97% similarity) and T. guiyangensis Jin F. Zhang et al. (GZAAS01; KX274247, 98% similarity). Phylogenetically, T. camporesii is sister to T. murispora with moderate statistical support (89% ML, 0.92 BYPP; Fig. 34). Based on clear morphological differences and available molecular data, we introduce Tremateia camporesii as a new species following the guidelines by Jeewon and Hyde (2016).
Tremateia lamiacearum Samarak. & K.D. Hyde, sp. nov.
Index Fungorum number: IF556786; Facesoffungi number: FoF 06463; Fig. 33
Etymology: Name reflects the host family Lamiaceae
Holotype: MFLU 18-0826
Saprobic on dead branch of Lamiaceae. Sexual morphAscomata 140–240 µm high, 110–190 µm diam. (\( \bar{x} \) = 200.5 × 155.7 µm, n = 5), immersed to erumpent, globose or subglobose, dark brown to black. Peridium 6–11.5 µm wide, comprising 3–4 layers, of dark brown cells of textura angularis. Hamathecium comprising 2–3 µm wide (\( \bar{x} \) = 2.6 µm, n = 20), numerous, cellular, branched, septate, hyaline pseudoparaphyses. Asci 82–105 × 13–17 µm (\( \bar{x} \) = 93.7 × 14.4 µm, n = 20), 8-spored, bitunicate fissitunicate, clavate, pedicellate, thick-walled at the apex, with ocular chamber. Ascospores 15–20 × 7–9 µm (\( \bar{x} \) = 17.2 × 7.3 µm, n = 30), overlapping 1–2-seriate, mostly ellipsoidal, muriform, 3–5-transversely septate, with 1–2 vertical septa, deeply constricted at the middle septum, slightly constricted at the remaining septa, initially hyaline to pale yellow, becoming brown at maturity, rounded at both ends, surrounded by a thick mucilaginous sheath. Asexual morph Undetermined.
Material examined: THAILAND, Phayao Province, Phachang Noi, Pong, 19.32 N 100.46E, 545 m, on dead branch of Lamiaceae sp., 11 September 2017, M.C. Samarakoon, SAMC012 (MFLU 18-0826, holotype); ibid., KUN-HKAS 102315 (isotype).
GenBank numbers: LSU = MN473055, SSU = MN473049, TEF1-α = MN481601, TUB2 = MN481605.
Notes: Tremateia lamiacearum has close affinity to Tremateia species by immersed to erumpent, globose ascomata, numerous cellular pseudoparaphyses, fissitunicate and clavate asci and ellipsoid muriform ascospores. Tremateia guiyangensis and T. lamiacearum share immersed to semi-immersed or erumpent, subglobose ascomata and muriform, normally overlapping 1–2-seriate, ellipsoid, with 3–5-transversely septate ascospores. However, T. lamiacearum differs from other Tremateia species in having comparatively small asci (93.7 × 14.4 µm) and ascospores (17.2 × 7.3 µm) with large mucilaginous sheath (Hyde et al. 2016). A BLASTn search of LSU in GenBank shows our new species is similar to T. arunicola (MFLUCC 16-1275; KX274248, 99% similarity) and T. guiyangensis (GZAAS01; KX274247, 99% similarity). Phylogenetically, T. lamiacearum shares a sister relationship to T. arunicola and T. guiyangensis with significant statistical support in the ML analysis (75% ML; Fig. 34). Based on clear morphological differences and available molecular data, we introduce T. lamiacearum as a new species following the guidelines by Jeewon and Hyde (2016).
Phylogenetic tree generated from maximum likelihood analysis (RAxML) based on a combined ITS, LSU, SSU and TEF1-α sequence dataset of genera in Didymosphaeriaceae. Eighty-nine strains are included in the combined analyses which comprise 4437 characters (1163 characters for ITS, 892 characters for LSU, 1459 characters for SSU, 923 characters for TEF1-α) after alignment. Pleospora herbarum (CBS 191.86) is used as the outgroup taxon. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian inference analysis. The best RaxML tree with a final likelihood values of − 25621.093114 is presented. The matrix had 1584 distinct alignment patterns, with 47.43% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.240096, C = 0.245090, G = 0.274017, T = 0.240797; substitution rates AC = 1.286291, AG = 2.381701, AT = 1.260781, CG = 1.047135, CT = 6.097921, GT = 1.000000; gamma distribution shape parameter α = 0.421449. Bootstrap values for maximum likelihood (ML) equal to or greater than 70% and clade credibility values greater than 0.90 (the rounding of values to 2 decimal proportions) from Bayesian inference analysis are labeled on the nodes. Ex-type strains are in bold. The new isolate is in blue
Fuscostagonosporaceae Jayasiri, Camporesi & K.D. Hyde
Notes: Fuscostagonosporaceae was introduced by Hyde et al. (2017) to accommodate a monophyletic genus Fuscostagonospora Kaz. Tanaka & K. Hiray. This family is characterised by immersed, globose to subglobose ascomata, branched trabeculate pseudoparaphyses (sensu Liew et al. 2000) and narrowly fusiform, hyaline ascospores with a sheath (Hyde et al. 2017). We introduce a new species with support from both morphology (ellipsoid to obovoid, brown ascospores without a mucilaginous sheath) and multi-gene phylogeny.
Fuscostagonospora Kaz. Tanaka & K. Hiray.
Notes: Fuscostagonospora was introduced by Tanaka et al. (2015), to accommodate a bambusicolous taxon, F. sasae Kaz. Tanaka & K. Hiray. Subsequently, Hyde et al. (2017) accommodated Fuscostagonospora sasae in Fuscostagonosporaceae with another species, F. cytisi Jayasiri, Camporesi & K.D. Hyde (MFLU 15-3607). Fuscostagonospora comprises only three species, viz. F. banksiae Crous & Carnegie, F. cytisi and F. sasae (Tanaka et al. 2015; Hyde et al. 2017; Crous et al. 2019b).
Fuscostagonospora camporesii Tennakoon & K.D. Hyde, sp. nov.
Index Fungorum number: IF557123; Facesoffungi number: FoF 06333; Fig. 35
Fuscostagonospora camporesii (MFLU 16-1362, holotype). a Ascomata visible as black dots on host surface. b, c Vertical sections of ascomata. d Section through peridium. e Pseudoparaphyses. f–i Asci. j–n Ascospores. o Germinated ascospore. p Colony from above. q Colony from below. Scale bars: b, c = 50 µm, d–i = 20 µm, j–n = 5 µm, o = 10 µm
Etymology: In honour of Mr. Erio Camporesi for his immense contribution to mycology
Holotype: MFLU 16-1362
Saprobic on dead branches of Cornus sanguinea. Sexual morphAscomata 100–130 μm high, 160–220 μm diam., solitary, scattered to clustered, immersed, visible as raised, black spots on host surface globose to subglobose, glabrous, uni-loculate, ostiole central with minute papilla. Peridium 22–33 μm wide, thin to thick-walled with equal thickness, composed of two type layers of pseudoparenchymatous cells, inner layer comprising 3–5 cell layers of flattened, hyaline cells, arranged in a textura prismatica, outer layer comprising several layers of dark brown to black cells, arranged in a textura angularis. Hamathecium composed of dense, broad, 1.8–2.5 μm wide, filamentous, cellular pseudoparaphyses, with indistinct septa, not constricted at the septa, anastomosing at the apex, embedded in a hyaline gelatinous matrix. Asci (69–)80–90(–110) × (7–)8–9(–9.5) μm (\( \bar{x} \) = 86 × 8.5 μm, n = 20), 8-spored, bitunicate, fissitunicate, cylindric-clavate, short pedicellate, with furcate to obtuse end, apically rounded with well-developed ocular chamber. Ascospores (11.8–)13–15(–15.7) × (5.5–)6–6.5(–7.3) μm (\( \bar{x} \) = 13.9 × 6.3 μm, n = 40), overlapping, 1-seriate, ellipsoid to obovoid, initially hyaline to slightly brown, becoming brown at maturity, aseptate when young, becoming 1-septate, straight to slightly curved, smooth-walled. Asexual morph Undetermined.
Culture characteristics: Colonies on PDA reaching 35–40 mm diam. after 5 weeks at 20–25 °C, colonies medium sparse, circular, convex, slightly rough surface with edge entire, well defined margin, cottony to fairly fluffy with sparse aspects, colony from above, white to cream at the margin, white to yellowish at the centre; from below, yellow to yellowish brown at the margin, light brown at the centre, mycelium white to cream with tufting, not producing pigmentation in PDA media agar.
Material examined: ITALY, Province of Forlì-Cesena [FC], Monte Maggiore-Predappio, on dead branches of Cornus sanguinea L. (Cornaceae), 9 March 2015, E. Camporesi, IT 2409 (MFLU 16-1362, holotype); ibid., KUN-HKAS 9370 (isotype), ex-type living culture, MFLUCC 16-0787, KUMCC 15-0554.
GenBank numbers: ITS = MN750611, LSU = MN750590, SSU = MN750605.
Notes: The morphological characteristics of Fuscostagonospora camporesii fit with F. cytisi Jayasiri, Camporesi & K.D. Hyde (MFLU 15-3607) in having immersed, subglobose to globose ascomata, cylindric-clavate, short pedicellate asci and 1-septate, ellipsoid to obovoid ascospores and trabeculate pseudoparaphyses (Hyde et al. 2017). But, F. camporesii differs from F. cytisi in having brown and smaller ascospores (13.9 × 6.3 µm versus 16 × 7.4 µm), whereas, F. cytisi has hyaline ascospores with a mucilaginous sheath (Hyde et al. 2017). We also compared the sequences of our newly-described taxon with F. cytisi.
According to the multi-gene phylogenetic analyses of a combined LSU, SSU, ITS and TEF1-α sequence dataset, Fuscostagonospora camporesii is closely related to other Fuscostagonospora species, in particularly with F. banksiae (CBS 144621) with significant support (89% ML, 79% MP; Fig. 36). The asexual morph of F. banksiae was isolated from Banksia sp. (Proteaceae) and introduced by Crous et al. (2019b). A comparison of the 665 nucleotides across the ITS (+5.8S) gene region of F. camporesii and F. banksiae shows 30 base pair differences (4.51%) which justifies the new species following the guidelines of Jeewon and Hyde (2016).
Phylogram generated from maximum likelihood analysis based on combined ITS, LSU, SSU and TEF1-α sequence dataset for Fuscostagonospora species and several closely related genera in Didymosphaeriaceae, Lentitheciaceae and Massarinaceae. Related sequences were taken from Hyde et al. (2017) and Crous et al. (2019b). Fifty-six strains are included in the combined analyses which comprise 3482 characters (543 characters for ITS, 933 characters for LSU, 1077 characters for SSU, 929 characters for TEF1-α) after alignment. Periconia byssoides (C292) and P. submersa (MFLUCC 16-1098) (Periconiaceae, Pleosporales) are used as the outgroup taxa. Single gene analyses are carried out and the topology of each tree had clade stability. Tree topology of the maximum likelihood analysis is similar to the maximum parsimony analysis and the Bayesian inference analysis. The best RaxML tree with a final likelihood value of − 18830.051624 is presented. The matrix had 1195 distinct alignment patterns, with 37.67% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.242753, C = 0.239241, G = 0.274146, T = 0.243860; substitution rates AC = 1.751566, AG = 2.491358, AT = 1.693843, CG = 1.162505, CT = 6.992228, GT = 1.000000; gamma distribution shape parameter α = 0.578052. Maximum parsimony analysis of 2543 constant characters and 591 informative characters (CI = 0.488, RI = 0.638, RC = 0.312, HI = 0.512). Bootstrap values for maximum likelihood (ML) and maximum parsimony (MP) equal to or greater than 60% and Bayesian posterior probabilities (BYPP) equal to or greater than 0.90 are placed above the branches. Newly generated sequences are indicated in blue
Halotthiaceae Y. Zhang, J. Fourn. & K.D. Hyde
Notes: Halotthiaceae was introduced by Zhang et al. (2013) and is typified by Halotthia Kohlm. with H. posidoniae (Durieu & Mont.) Kohlm. as the type species. The family was first introduced to accommodate freshwater and maritime Dothideomycetes genera Halotthia, Mauritiana Poonyth, Phaeoseptum Ying Zhang, J. Fourn. & K.D. Hyde and Pontoporeia Kohlm. (Hyde et al. 2013; Zhang et al. 2013). Subsequently, Ariyawansa et al. (2015a) introduced three new genera to Halotthiaceae viz. Brunneoclavispora Phook. & K.D. Hyde, Neolophiostoma Boonmee & K.D. Hyde and Sulcosporium Phook. & K.D. Hyde. Hyde et al. (2018b) introduced a new family Phaeoseptaceae Boonmee, Thambugala & K.D. Hyde to accommodate the genera Phaeoseptum, Lignosphaeria Boonmee et al. and Neolophiostoma Boonmee & K.D. Hyde based on phylogenetic analysis of LSU, SSU, RPB2 and TEF1-α sequence data. Based on present phylogenetic analyses in this study (Fig. 39), Neolophiostoma formed a basal lineage in Halotthiaceae. Hence, the phylogenetic status of this genus needs to be clarified with more taxon sampling. Currently, six genera are accepted in Halotthiaceae viz. Brunneoclavispora, Halotthia, Mauritiana, Neolophiostoma, Pontoporeia and Sulcosporium.
Brunneoclavispora Phookamsak & K.D. Hyde
Notes: Brunneoclavispora was introduced as a monotypic genus by Ariyawansa et al. (2015a) and is typified by B. bambusae Phookamsak & K.D. Hyde, which was collected from bamboo in Thailand in a terrestial habitat. Ariyawansa et al. (2015a) treated the genus in Halotthiaceae, while the genus formed an independent lineage basal to Sporormiaceae in Hyde et al. (2018b). Phukhamsakda et al. (2019) treated the genus in Pleosporales genera incertae sedis together with Neolophiostoma. The phylogenetic status of Brunneoclavispora is unresovled and it clusters in Halotthiaceae in this study. Thus, we tentatively accommodate Brunneoclavispora in Halotthiaceae until phylogenetic affinity of the genus is clarified with more taxon sampling. In this study, the second new species, B. camporesii, isolated from decaying wood in Thailand in terrestial environment is introduced.
Brunneoclavispora camporesii Boonmee & Phookamsak, sp. nov.
Index Fungorum number: IF557029; Facesoffungi number: FoF 06507; Figs. 37, 38
Brunneoclavispora camporesii (MFLU 11-0001, holotype). a Material and habit on wood. b Appearance of ascomata immersed in wood tissues (arrows). c Cross section of ascoma. d Peridium. e Pseudoparaphyses. f–i Asci. j–l Close up of apical asci with fissitunicate dehiscence. m–p Ascospores. Scale bars: b = 500 µm, c, m–p = 10 µm, d, f–i = 50 µm, e = 5 µm, j–l = 20 µm
Etymology: The specific epithet “camporesii” named after Mr. Erio Camporesi for his immense contribution to mycology
Holotype: MFLU 11-0001
Saprobic on decaying wood. Sexual morphAscomata 230–380 µm high, 210–290 µm diam. (\( \bar{x} \) = 310 × 252 µm, n = 5), present as dark spots on host surface, immersed, uni-loculate, scattered, subglobose, dark brown, with ostiole. Peridium 23–38.5 µm thick, composed of 4–5 layers of textura angularis, with dark brown cells. Hamathecium 2 µm wide, anatomosed, branched, septate, hyaline, pseudoparaphyses. Asci 109–193 × 21.5–28 µm (\( \bar{x} \) = 140 × 24.5 µm, n = 20), 8-spored, bitunicate, fissitunicate, cylindric-clavate to clavate, thickened at the apex, with 2–2.5 µm wide ocular chamber, short pedicellate (ca. 18–29.5 µm long), sometimes expanding up to 59 µm long. Ascospores 21–26.5 × 11–15 µm (\( \bar{x} \) = 25 × 12 µm, n = 20), overlapping 1–2-seriate, muriform, suboblong-ellipsoidal to clavate, transversely 6–7-septate, with longitudinally 1-septate in each cell, brown to dark brown, slightly curved, constricted at the septa, septa thickened, smooth-walled. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on MEA within 12 h. Colonies on MEA reaching 9 mm diam. in 7 days at 28 °C, colonies compressed, slightly umbonate, undulate edge, colour of colonies at initially white to yellowish and white at the margin with orange-brown pigmented in medium after 45 days. Mycelium superficial, slightly effuse, radially striate with lobate edge, septate, hyaline, smooth-walled.
Material examined: THAILAND, Chiang Mai Province, Chom Thong, N 18° 31′ E 98° 29′, on decaying wood, 16 November 2010, R. Phookamsak, ITN-03 (MFLU 11-0001, holotype), ex-type living culture, MFLUCC 11-0001.
GenBank numbers: ITS = MN809329, LSU = MN809328.
Notes: Brunneoclavispora camporesii is morphologically similar to B. bambusae in having brown, muriform ascospores and was collected from a terrestial environment. However, the species can be distinguished from B. bambusae in having subglobose ascomata, cylindric-clavate to clavate, short pedicellate asci and suboblong-ellipsoidal to clavate, transversely 6–7-septate, and longitudinally 1-septate in each cell of the ascospores. Whereas, B. bambusae has elongate conical ascomata with a flattened base, with slit-like openings ostiole. Asci of B. bambusae are clavate, with short to long pedicels and its ascospores are clavate to fusiform, 7–8 transverse septa, with 1–4 longitudinal septa, with a tail-like appendage at the basal cell (Ariyawansa et al. 2015a). Phylogenetic analyses of a combined LSU, SSU and ITS sequence dataset showed that B. camporesii forms a sister lineage with B. bambusae with high support (98% ML, 1.00 BYPP; Fig. 39). Therefore, we introduce B. camporesii as a second species in Brunneoclavispora with both species in this genus being saprobes collected from dead and decaying wood in Thailand.
Phylogram generated from maximum likelihood analysis based on combined LSU, SSU and ITS sequence dataset representing Halotthiaceae and related families. Related sequences are taken from Phukhamsakda et al. (2019). Thirty-nine strains are included in the combined analyses which comprise 2444 characters 858 characters for LSU, 1021 characters for SSU and 565 characters for ITS) after alignement. Aigialus grandis (BCC 20000) and Neoastrosphaeriella krabiensis (MFLUCC 11-0025) in Aigialaceae (Pleosporales) are used as the outgroup taxa. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood values of − 15614.164113 is presented. The matrix had 1062 distinct alignment patterns, with 29.90% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.249630, C = 0.230631, G = 0.282717, T = 0.237022; substitution rates AC = 1.426187, AG = 2.223185, AT = 1.439924, CG = 1.247317, CT = 6.150653, GT = 1.000000; gamma distribution shape parameter α = 0.266758. Bootstrap values for maximum likelihood (ML) equal to or greater than 50% and clade credibility values greater than 0.95 BYPP (the rounding of values to 2 decimal proportions) from Bayesian-inference analysis labeled on the nodes. The ex-type strains are indicated in bold and the newly generated sequence is indicated in bold and blue
Lentitheciaceae Y. Zhang ter et al.
Notes: Lentitheciaceae was introduced by Zhang et al. (2009) with L. fluviatile (Aptroot & Van Ryck.) K.D. Hyde, J. Fourn. & Y. Zhang as the type species. We follow the latest treatment and updated accounts of Lentitheciaceae in Phookamsak et al. (2019). In this study, two new Keissleriella species and a new genus, Pseudomurilentithecium are introduced, based on morphology and molecular data, together with description and illustrations. A phylogenetic tree based on combined LSU, ITS, SSU, TEF1-α and RPB2 sequence data is presented in Fig. 43.
Keissleriella Höhn.
Notes: Keissleriella was introduced by von Höhnel (1919) for wood associated fungal species. Keissleriella aesculi (Höhn.) Höhn. is the type species of the genus. Keisseleriella has unique characters of black setae on the papilla, hyaline, uni- to multi-septate ascospores with a mucilaginous sheath (Zhang et al. 2012; Liu et al. 2015; Tippromma et al. 2017; Wanasinghe et al. 2018). There are 41 epithets of Keissleriella species listed in Species Fungorum (2020). We introduce two additional species of Keissleriella camporesiana and K. camporesii which were collected from Italy (Figs. 40, 41).
Keissleriella camporesiana (MFLU 15-3288, holotype). a Ascomata on Centaurea. b Close up of ascomata. c Vertical section of ascoma. d Ostiolar canal contain of brown setae. e Peridium. f Pseudoparaphyses. g–i Developing state of asci. j–n Developing state of ascospores. o Ascospore stained with India ink showed mucilaginous sheath. Scale bar: b = 500 µm, c = 100 µm, d = 20 µm, e–i = 50 µm, j–o = 10 µm
Keissleriella camporesiana Phukhams. & K.D. Hyde, sp. nov.
Index Fungorum number: IF556759; Facesoffungi number: FoF 06344; Fig. 40
Etymology: The species epithet was honouring Erio Camporesi.
Holotype: MFLU 15-3288
Saprobic on dead stems of Centaurea L. Sexual morphAscomata 170–240 × 270–320 μm (\( \bar{x} \) = 165 × 240 μm, n = 5), on surface of the host, covered by a pseudoclypeus, immersed to superficial, solitary, scattered, uni-loculate, obpyriform to compressed globose, flatten base, brown to dark brown, rough-walled, papillate. Ostiole central, 55 × 49 μm, brown to dark brown, papillate, opened pore, easily open, ostiolate with black setae on the papilla. Peridium 16–40 μm wide, thick, multi-layer, comprising of 7–9 layers of dark brown to brown cells of textura angularis, inner layers comprising thin, hyaline cells. Hamathecium of dense, 1.15–2.3 μm wide (\( \bar{x} \) = 1.5 μm, n = 50), filiform branches, anastomosing above asci, reaching the ostiole part, transverse septate, trabeculate pseudoparaphyses. Asci 68–120 × 15–20 μm (\( \bar{x} \) = 95 × 20 μm, n = 40), 8-spored, bitunicate, fissitunicate, thick-walled, cylindric-clavate to clavate, apically rounded, with furcated pedicel, ocular chamber clearly visible when immature. Ascospores 20–30 × 7–8 μm (\( \bar{x} \) = 26 × 7.5 μm, n = 50), bi-seriate, partial overlapping, hyaline, broad fusiform, cell above median septum slightly wider than below, lower part longer than the upper part, tapering towards the ends, slightly acute at both ends, with (1–)4–5 transverse septa, constricted at the septa, large guttules each cell, deeply constricted at the median septum, smooth-walled, with mucilaginous sheath. Asexual morph Undetermined.
Culture characteristics: Colonies on MEA, reaching 30 mm diam. after 4 weeks of incubation at 16 °C. Culture grey brown, with orange brown mixed in the mycelium after 4 weeks, dense mycelia, colonies circular, flat, umbonate, dull, covered with aerial mycelium, white mycelium at the edges; reverse dark brown, dense, irregular, fimbriate margin.
Material examined: ITALY, Province of Forlì-Cesena [FC], Strada San Zeno-Galeata, on dead stems of Centaurea L. (Asteraceae), 22 December 2012, N. Camporesi, IT 971 (MFLU 15-3288, holotype), ex-type living culture, MFLUCC 15-0029.
GenBank numbers: ITS = MN401745, LSU = MN40174, SSU = MN401743, TEF1-α = MN397907.
Notes: Keissleriella is characterised by ascomata with black setae on the papilla (Zhang et al. 2012) and trabeculate pseudoparaphyses (sensu Liew et al. 2000). In the phylogenetic analyses (Fig. 43), K. camporesiana (strain MFLUCC 15-0029) and K. dactylis Singtr., Camporesi & K.D. Hyde (strain MFLUCC 13-0751) form a separate subclade with moderate support (75% ML, 0.94 BYPP). Keissleriella camporesiana is morphologically distinguished from K. dactylis by the second cell of ascospores being enlarged and being 4–5-septate (Singtripop et al. 2015). Keissleriella dactylis was described from the host family Poaceae while K. camporesiana occurs on Asteraceae.
Keissleriella camporesii C.G. Lin & K.D. Hyde, sp. nov.
Index Fungorum number: IF557105, Facesoffungi number: FoF 06182; Fig. 41
Etymology: The specific is named in honour of the prolific fungus collector, Erio Camporesi.
Holotype: MFLU 15-1068
Saprobic on dried aerial spines of Rosa canina. Sexual morph Undetermined. Asexual morphConidiomata up to 140 μm diam., immersed, pycnidial, scattered, brown to dark brown, globlose to subglobose, uni-loculate, indistinct ostiolate. Pycnidial walls thin-walled of brown to dark brown pseudoparenchymatous cells, of textura angularis. Conidiophores 9–25 × 2.4–3.4 μm, hyaline, unbranched or branched, smooth, flexuous, formed from the inner cells of the pycnidial wall. Conidiogenous cells monophialidic, cylindrical, hyaline. Conidia 5–10 × 1.5–5 μm, solitary, dry, acropleurogenous, simple, hyaline to pale brown, ellipsoidal, fusiform.
Culture characteristics: Conidia germinating on PDA within 24 h. Colonies on PDA effuse, greyish white from above, reaching 0.7–1.5 cm diam. in 7 days at 25 °C.
Material examined: ITALY, Province of Forli-Cesena Province, on dried aerial spines of Rosa canina L. (Rosaceae), 2014, Erio Camporesi, IT 2246 (MFLU 15-1068, holotype), ex-type living culture MFLUCC 15-0117.
GenBank numbers: ITS = MN252879, LSU = MN252886, SSU = MN252907.
Notes: The asexual morph of Keissleriella was reported as Dendrophoma Sacc. (Sivanesan 1984). Keissleriella camporesii shares morphological characters with Dendrophoma species, such as branched, hyaline conidiophores, phialidic conidiogenous cells and hyaline to brown, aseptate or septate conidia. Keissleriella camporesii grouped with K. cladophila (Niessl) Corbaz (CBS 104.55), K. rosacearum Phukhams., Camporesi & K.D. Hyde (MFLUCC 15-0089) and K. spartiicola (MFLUCC 14-0196) as a monophyletic clade (Fig. 43). However, the asexual morph of K. rosacearum and K. spartiicola are undetermined (Liu et al. 2015; Wanasinghe et al. 2018), and thus we cannot compare the morphology. The asexual morph of K. cladophila was described as pycnidia up to 150 μm broad, globose, ostiolate, branched, with 10–40 × 2–2.5 μm conidiogenous cells and hyaline, one-celled, oblong, 5–7 × 2–2.5 μm conidia (Sivanesan 1984). Keissleriella camporesii differs from K. cladophila by its hyaline to brown, 0–1-septate conidia.
Pseudomurilentithecium Mapook & K.D. Hyde, gen.nov.
Index Fungorum number: IF556904; Facesoffungi number: FoF 06791
Etymology: Referring to its similarity with Murilentithecium.
Saprobic on dead aerial stem. Sexual morphAscomata immersed, solitary or scattered, coriaceous, subglobose to globose, dark brown to black, ostiolar neck protruding. Peridium 2–3 layers, comprising dark brown cells of textura angularis. Hamathecium with cylindrical, septate, branching pseudoparaphyses. Asci 8-spored, bitunicate, cylindric-clavate, straight or slightly curved, apically rounded, pedicellate. Ascospores overlapping, 1–2-seriate, initially hyaline to pale yellow, 1-septate when immature, becoming golden-brown to brown at maturity, ellipsoid to broadly fusiform, muriform, 3–7-transversely septate, with 1–2-longitudinal septate, slightly constricted at the central septum, straight or slightly curved, surrounded by hyaline gelatinous sheath observed clearly when mounted in Indian ink. Asexual morph Undetermined.
Type species: Pseudomurilentithecium camporesii Mapook & K.D. Hyde
Notes: A phylogenetic analysis based on combined dataset of LSU, ITS, SSU, TEF1-α and RPB2 sequence data show that Pseudomurilentithecium clusters with Poaceascoma and Setoseptoria (Fig. 43). However, Pseudomurilentithecium differs from Poaceascoma in having ellipsoid to broadly fusiform, muriform ascospores with 3–7 transverse septa, and 1–2 longitudinal septa, while Poaceascoma has hyaline, filiform, multi-septate ascospores with spirally arranged within the ascus (Phookamsak et al. 2015). Therefore, Pseudomurilentithecium is described as a new genus based on phylogeny and morphological comparison.
Pseudomurilentithecium camporesii Mapook & K.D. Hyde, sp. nov.
Index Fungorum number: IF556903; Facesoffungi number: FoF 06792; Fig. 42
Pseudomurilentithecium camporesii (MFLU 20-0047, holotype). a, b Appearance of ascomata on substrate. c Section through ascoma. d Peridium. e Pseudoparaphyses. f, g asci. h–j Ascospores. k Ascospores with gelatinous sheath in Indian ink. Scale bars: a, b = 200 µm, c, f, g = 50 µm, d = 20 µm, e, h–k = 10 µm
Etymology: Named in honour of the collector, Erio Camporesi.
Holotype: MFLU 20-0047
Saprobic on dead aerial stem of Vicia sp. Sexual morphAscomata (90–)130–145 µm high, 140–160 µm diam. (\( \bar{x} \) = 123 × 154 µm, n = 5), immersed, solitary or scattered, coriaceous, subglobose to globose, dark brown to black, ostiolar neck protruding. Peridium 14–20 µm wide, 2–3 layers, comprising dark brown cells of textura angularis. Hamathecium comprising 2.5–5(–7) µm wide, cylindrical, septate, branching pseudoparaphyses. Asci 90–115 × 16–22 µm (\( \bar{x} \) = 104 × 19 µm, n = 10), 8-spored, bitunicate, cylindric-clavate, straight or slightly curved, apically rounded, pedicellate. Ascospores (20–)27–32 × 8–12 µm (\( \bar{x} \) = 28.5 × 10 µm, n = 40), overlapping, 1–2 seriate, initially hyaline to pale yellow, 1-septate when immature, becoming golden-brown to brown at maturity, ellipsoid to broadly fusiform, muriform, 3–7-transversely septate, with 1–2 longitudinal septa, slightly constricted at the central septum, straight or slightly curved, surrounded by hyaline gelatinous sheath clearly observed when mounted in Indian ink. Asexual morph Undetermined.
Material examined: ITALY, Province of Forlì-Cesena [FC], Monte Coronaro-Verghereto, on dead aerial stem of Vicia sp. (Fabaceae), 26 July 2014, E. Camporesi, IT 2018 (MFLU 20-0047, holotype), ex-type living culture, MFLUCC 14-1118.
GenBank numbers: ITS = MN638861, LSU = MN638846, SSU = MN638850, TEF1-α = MN648730.
Notes: In a BLASTn search of NCBI GenBank, the closest matches of the ITS sequence of Pseudomurilentithecium camporesii (MFLUCC 14-1118, ex-holotype) with 90.38% similarity, was to Keissleriella cladophila (strain CBS 104.55, MH857391). The closest matches of the LSU sequence with 94.57% similarity, was to Parathyridaria ramulicola Jaklitsch, Fourn. & Voglmayr (strain MF4, KX650564), 94.65% similarity, was to Bimuria novae-zelandiae D. Hawksw., Chea & Sheridan (strain CBS 107.79, NG_058623), and 94.45% similarity, was to Keissleriella cladophila (strain CBS 104.55, GU205221). The closest matches with the SSU sequences with 90.98% similarity, was to Darksidea alpha D.G. Knapp et al. (strain CBS 135628, KP184054). The closest matches of the TEF1-α sequences with 91.35% similarity, was with Misturatosphaeria sp. (strain MFLUCC 10-0126, KR075170), while the closest matches of the RPB2 sequences with 78.79% similarity, was to Murilentithecium clematidis Wanas. et al. (strain MFLUCC 14-0562, KM454447). In the phylogenetic analyses (Fig. 43), P. camporesii forms a basal clade to Poaceascoma.
Phylogram generated from maximum likelihood analysis based on combined dataset of LSU, ITS, SSU, TEF1-α and RPB2 sequence data. Seventy-seven strains are included in the combined sequence analysis, which comprise 4008 characters with gaps. Tree topology of the ML analysis was similar to the BI. The best scoring RAxML tree with a final likelihood value of − 18684.534541 is presented. The matrix had 1199 distinct alignment patterns, with 37% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.241414, C = 0.246131, G = 0.273783, T = 0.238671; substitution rates: AC = 1.203261, AG = 1.960685, AT = 1.267413, CG = 1.440469, CT = 7.447474, GT = 1.000000; gamma distribution shape parameter α = 0.166789. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. Newly generated sequences are in blue bold and type strains are in black bold. Bambusicola bambusae (MFLUCC 11-0614) and B. irregulispora (MFLUCC 11-0437) are used as outgroup taxa
Leptosphaeriaceae M.E. Barr
Notes: We follow the latest treatment and updated accounts of Leptosphaeriaceae in Ariyawansa et al. (2015b), Hyde et al. (2016, 2017), Tennakoon et al. (2017), Tibpromma et al. (2017) and Phookamsak et al. (2019). Eleven genera are accepted in this family viz. Acicuseptoria Quaedvl. et al., Alloleptosphaeria Ariyaw. et al., Alternariaster E.G. Simmons, Chaetoplea (Sacc.) Clem., Heterosporicola Crous, Leptosphaeria Ces. & De Not., Paraleptosphaeria Gruyter et al., Plenodomus Preuss, Querciphoma Crous, Subplenodomus Gruyter et al. and Sphaerellopsis Cooke.
Plenodomus Preuss
Notes: Plenodomus introduced by Preuss (1851) was typified by P. rabenhorstii Preuss. Species of Plenodomus are saprobic or parasitic on plants in terrestrial habitats (Ariyawansa et al. 2015b). We follow the latest treatment and updated accounts of Plenodomus in Phookamsak et al. (2019). A novel species P. triseptatus is introduced from Russia.
Plenodomus triseptatus S.N.Wijesinghe, Bulgakov & K.D. Hyde, sp. nov.
Index Fungorum: IF556925; Facesoffungi number: FoF 06807; Fig. 44
Plenodomus triseptatus (MFLU 17-0789, holotype). a, b Ascomata on host. c Vertical section of ascoma. d Section of peridium. e Pseudoparaphyses. f–i Immature to mature asci. j–o Immature to mature ascospores. p, q Colony on PDA (p = from above, q = from below). Scale bars: a = 500 μm, b = 100 μm, c = 100 μm, d–i = 20 μm, j–o = 5 μm
Etymology: The specific epithet “triseptatus” refers to the number of septa in ascospores.
Holotype: MFLU 17-0789
Saprobic on decaying wood. Sexual morphAscomata 276–324 μm high, 217–250 μm diam., (\( \bar{x} \) = 298 × 228 μm, n = 15), solitary, scattered or small group, raised, erumpent on host, uni-loculate, black, globose to subglobose. Peridium 26–51 μm wide, thin-walled, composed of several layers of small, brown to dark brown cells of textura angularis. Hamathecium comprising numerous, 2–3 μm wide, septate, branched, cellular pseudoparaphyses, embedded in a gelatinous matrix. Asci 58–112 × 7–13 μm (\( \bar{x} \) = 85 × 9.5 μm, n = 30), 8-spored, bitunicate, fissitunicate, cylindrical, short pedicel, with furcate ends, apically rounded, with well-developed ocular chamber. Ascospores 19–25 × 5.7–7.2 μm (\( \bar{x} \) = 22 × 6.5 μm, n = 30), overlapping 1-seriate, hyaline to pale brown, fusiform, with pointed ends, 3-septate, slightly constricted at the septa, guttulate, thick and smooth-walled. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on PDA within 24 h. Germ tubes produced both sides of ascospore. Colonies on PDA reaching 0.5–1 cm diam. after 5 days in day light at 16 °C, colonies circular, medium dense, flat or effuse, from above, white in middle and dark brown at the margin, dark brown in reverse side.
Material examined: RUSSIA, Republic of Crimea, Bakhchisaraisky District, near Nauchny settlement, Selbukhra mountain, stony steppe on the slope, on dead stem of Daucus carota L. (Apiaceae), 4 July 2016, T.S. Bulgakov, CR-089 (MFLU 17-0789, holotype), ex-type living culture, MFLUCC 17-1345.
GenBank numbers: ITS = MN648452, LSU = MN648451, SSU = MN648453.
Notes: Plenodomus triseptatus (MFLUCC 17-1345) is similar to P. libanotidis (Fuckel) Gruyter, Aveskamp & Verkley (CBS 113795) in having a peridium with brown to dark brown cells of textura angularis, 8-spored asci with a well-developed ocular chamber and guttulate, 3-septate, hyaline to pale brown ascospores constricted at each septum. However, P. triseptatus differs from P. libanotidis by its smaller ascospores (17–21 μm versus 19–25 μm; de Gruyter et al. 2013). Phylogenetically, P. triseptatus shares a sister relationship to P. libanotidis with high support (96% ML, 1.00 BYPP; Fig. 45). Also, the phylogeny supports that they are distinct species. Therefore, P. triseptatus is introduced as a novel species in family Leptosphaeriaceae.
Phylogram generated from maximum likelihood analysis based on a combined LSU, SSU and ITS sequence dataset. Paraphoma radicinia (CBS 111.79), Phaeosphaeria oryzae (CBS 110110) and Phaeosphaeriopsis glaucopunctata (MFLUCC 13-0265) were selected as the outgroup taxa under Pleosporales. Related sequences are taken from Phookamsak et al. (2019) and GenBank. Seventy-five strains are included in the multy-gene phylogenetic analyses which comprise 2477 characters (882 characters for LSU, 998 characters for SSU and 597 characters for ITS) after alignment. Single gene analyses were also compared for the topology and clade stability with the combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood values of − 12964.530032 is presented. The matrix had 577 distinct alignment patterns, with 26.62% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.244203, C = 0.220508, G = 0.272622, T = 0.262667; substitution rates AC = 1.857458, AG = 3.628940, AT = 2.129510, CG = 0.931715, CT = 7.125511, GT = 1.000000; gamma distribution shape parameter α = 0.109630. Bootstrap values for maximum likelihood (ML) equal to or greater than 70% and clade credibility values greater than 0.95 BYPP (the rounding of values to 2 decimal proportions) from Bayesian-inference analysis demarcated above the nodes. The newly generated sequences are indicated in bold and blue and ex-type strains are in bold
Lophiostomataceae Sacc.
Notes: Lophiostomataceae was introduced by Saccardo (1883) with Lophiostoma macrostomum (Tode) Ces. & De Not. as the type species (Hashimoto et al. 2018). It is characterised by carbonaceous ascomata with a slit-like ostiolar neck, clavate asci and hyaline to dark brown ascospores with mucilaginous sheaths and/or appendages at each end (Thambugala et al. 2015; Hyde et al. 2016, 2019a; Hashimoto et al. 2018; Tennakoon et al. 2018b). Members of Lophiostomataceae are usually saprobic on herbaceous and woody plants in terrestrial, freshwater and marine habitats (Chesters and Bell 1970; Thambugala et al. 2015; Hyde et al. 2016, 2019a; Hashimoto et al. 2018).
Neovaginatispora A. Hashim., K. Hiray. & Kaz. Tanaka
Notes: The monotypic genus Neovaginatispora is typified by N. fuckelii (Sacc.) A. Hashim., K. Hiray. & Kaz. Tanaka. The genus was introduced by Hashimoto et al. (2018) based on phylogenetic distinction and its typical characters of a thinner peridium than Vaginatispora. In this study, N. fuckelii is reported from Prunus in Yunnan, China.
Neovaginatispora fuckelii (Sacc.) A. Hashim., K. Hiray. & Kaz. Tanaka, Stud Mycol 90: 188 (2018)
Facesoffungi number: FoF 06527; Fig. 46
Neovaginatispora fuckelii (KUN-HKAS 99593). a Material. b Appearance of ascomata on the host. c Crest-like ostiole of ascoma. d Section through ascoma. e, f Peridium. g–i Asci with pseudoparaphyses. j–m Ascospores. n Germinating ascospore. Notes: l, m stained in black ink reagent. Scale bars: c = 500 μm, d = 200 μm, e = 100 μm, f–i, n = 50 μm, j–m = 10 μm
Saprobic on decaying wood. Sexual morphAscomata 250–580 × 190–230 × 210–250 μm (\( \bar{x} \) = 400 × 220 × 235 μm, n = 5), perithecial, solitary and scattered, immersed to semi-immersed, unilocular, subglobose to ellipsoid, black, ostiolate. Ostiole rounded or crest-like, periphysate. Peridium 10–40 μm wide, outer layer composed of brown to dark brown, thick-walled cells of textura angularis; inner layer composed of hyaline to lightly pigmented cells of textura prismatica. Hamathecium comprising 1.5–2.5 μm wide, numerous, filiform, septate, branched, anastomosed pseudoparaphyses, embedded in a gelatinous matrix. Asci 45–75 × 6–10 μm (\( \bar{x} \) = 60 × 8 μm, n = 40), 8-spored, bitunicate, fissitunicate, cylindrical to clavate, short-pedicellate, rounded at the apex, with a distinct ocular chamber. Ascospores 10–15 × 4–6 μm (\( \bar{x} \) = 14 × 5 μm, n = 50), 1–2-seriate, partially overlapping, hyaline, broadly fusiform with obtuse ends, 1-septate at median, constricted, slightly inflated of the cell nearby septum, straight or slightly curved, smooth-walled, guttulate, with cap-shaped, globose appendages at each end. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on PDA within one week at 23 °C. Colony 1.5 cm diam. after 30 days. Mycelium superficial to semi-immersed, filamentous, branched, septate, hyaline to brown. Colonies on PDA, initially, pale brown, becoming dark brown, rough surface and raised elevation with concentric zonations, uneven margins; reverse pale brown to dark brown at the zonations.
Material examined: CHINA, Yunnan Province, Kunming, Songhuaba Lake, on decaying wood of Prunus sp. (Rosaceae), 3 September 2017, S.K. Huang, SHB04 (KUN-HKAS 99593), living culture (KUMCC 17-0319).
Known host and distribution: This species has been recorded from various dead herbaceous plants and leaves of Mangifera in Europe, Japan and China (Wang and Lin 2004; Thambugala et al. 2015; Hyde et al. 2016, 2019a; Tennakoon et al. 2018b).
GenBank numbers: ITS = MN473470, LSU = MN473513, RPB2 = MN482130.
Notes: Hashimoto et al. (2018) found that Vaginatispora fuckelii is phylogenetically distinct from other species of Vaginatispora based on their multi-gene phylogenetic analysis. They proposed a monotypic genus Neovaginatispora typified by N. fuckelii (Hashimoto et al. 2018). In this study, our new strain clusters with other N. fuckelii strains in the phylogenetic analysis (Fig. 47). The present collection shares similar morphological features with previously reported collections, but the ascomata and ascospore dimensions are slightly different (Thambugala et al. 2015; Hyde et al. 2016; Tennakoon et al. 2018b). Some morphological characters may be slightly different on different host species and locality (Thambugala et al. 2015; Hyde et al. 2016; Tennakoon et al. 2018b). In Tennakoon et al. (2018b), N. fuckelii was found on Mangifera indica and its ascospores have a thin mucilaginous sheath. This is the first record of Neovaginatispora species on dead branches of Prunus species.
Phylogram generated from maximum likelihood analysis based on combined ITS, LSU, SSU, TEF1-α and RPB2 sequence data in Lophiostomataceae. Related sequences are taken from Hyde et al. (2019a). Thirty-two strains are included in the combined analyses which comprise 4177 characters (505 characters for ITS, 822 characters for LSU, 937 characters for SSU, 893 characters for TEF1-α, 1020 characters for RPB2) after alignment. Angustimassarina populi (MFLUCC 13-0034) and A. acerina (MFLUCC 14-0505) in Amorosiaceae were used as outgroup taxa. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood values of − 17538.189018 is presented. The matrix had 1073 distinct alignment patterns, with 22.77% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.2484248, C = 0.2519696, G = 0.2665036, T = 0.2331016; substitution rates AC = 1.4621358, AG = 22.4568366, AT = 1.1916542, CG = 1.3290588, CT = 17.2712454, GT = 1.000000; gamma distribution shape parameter α = 1.003752. Bootstrap values for maximum likelihood (ML) equal to or greater than 70% and clade credibility values greater than 0.90 BYPP (the rounding of values to 2 decimal proportions) from Bayesian-inference analysis labeled on the nodes. Ex-type strains are in bold and black, the new isolate is indicated in blue
Macrodiplodiopsidaceae Voglmayr, Jaklitsch & Crous
Notes: Macrodiplodiopsidaceae was introduced by Crous et al. (2015a) to accommodate Macrodiplodiopsis Petr. and Pseudochaetosphaeronema Punith based on molecular phylogeny. Tanaka et al. (2015) accepted the asexual genera Camarographium Bubák, Macrodiplodiopsis and Pseudochaetosphaeronema in Macrodiplodiopsidaceae and treated the family in suborder Massarineae. Ariyawansa et al. (2015a) introduced the genus Pseudomonodictys Doilom et al. to this family. Wijayawardene et al. (2018a) accepted only two genera in Macrodiplodiopsidaceae viz. Macrodiplodiopsis and Pseudomonodictys. While, Pseudochaetosphaeronema and Camarographium were treated in Pleosporales genera incertae sedis (Wijayawardene et al. 2018a). In this study, Pseudochaetosphaeronema forms a clade with Macrodiplodiopsis and Pseudomonodictys in Macrodiplodiopsidaceae (Fig. 49). Therefore, we reinstate the genus in Macrodiplodiopsidaceae.
Pseudochaetosphaeronema Punith.
Notes: Pseudochaetosphaeronema was proposed by Punithalingam (1979) based on Pseudochaetosphaeronema larense (Borelli & R. Zamora) Punith. Later, four species P. martinelli S.A. Ahmed, Desbois, Miossec, Atoche, Bonifaz & de Hoog (Ahmed et al. 2015), P. ginkgonis X.Y. Deng, T. Yuan Zhang & Yi X. Zhang (Zhang et al. 2016), P. pandanicola Tibpromma & K.D. Hyde (Tibpromma et al. 2018) and P. siamensis Jayasiri, E.B.G. Jones & K.D. Hyde (Jayasiri et al. 2019) were added to this genus. Species of Pseudochaetosphaeronema can be human pathogens, endophytes and saprobes. The asexual morphs are characterised by dark brown to black, nearly globose, scattered or gregarious, surficial conidiomata, monophialidic, cylindrical conidiogenous cells, hyaline, subglobose to oval, aseptate conidia and grey colonies on PDA (Jayasiri et al. 2019). The sexual morphs are undetermined. The LSU, SSU, ITS, TEF1-α and RPB2 loci are commonly considered to be reliable phylogenetic markers in the classification of Macrodiplodiopsidaceae (Ahmed et al. 2015; Zhang et al. 2016; Tibpromma et al. 2018; Jayasiri et al. 2019).
Pseudochaetosphaeronema kunmingense D.P. Wei, Wanas. & K.D. Hyde, sp. nov.
Index Fungorum number: IF557019; Facesoffungi number: FoF 07078; Fig. 48
Pseudochaetosphaeronema kunmingense (KUN-HKAS 102564, holotype). a Substrate (twig). b, c Conidiomata on substrate. d Vertical section of conidioma. e Pycnidial wall. f–i Conidiogenous cells bearing conidia. j, k Colony on PDA from above and below. l–o Conidia. p Germinating conidium. Scale bars: b = 500 µm, c = 200 µm, d = 50 µm, e, f, p = 20 µm, l–o = 10 µm, g–i = 5 µm
Etymology: The specific epithet “kunmingense” refers to the collecting site, Kunming City, Yunnan, China.
Holotype: KUN-HKAS 102564
Saprobic on decaying twig. Sexual morph Undetermined. Asexual morph Coelomycetous. Conidiomata 180–250 μm diam. (\( \bar{x} \) = 221 μm, n = 5), pycnidial, erumpent to superficial, globose to subglobose scattered, black, glabrous, solitary, uni-loculate, without papilla. Pycnidial walls 15–50 μm (\( \bar{x} \) = 30 μm, n = 30), outer layers comprised of thick-walled, brown cells of textura angularis, inner layers comprised of hyaline, thin-walled cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 3–9 × 1–2.5 μm (\( \bar{x} \) = 6 × 1.3 μm, n = 20), produced from inner stromatic tissue, monophialidic, cylindrical or ampulliform, integrated, hyaline, smooth-walled. Conidia 10–15 × 4–6 μm (\( \bar{x} \) = 13 × 5 μm, n = 50), hyaline when immature, becoming brown at maturity, typically fusiform with round ends, occasionally oval, smooth-walled, 3-septate, slightly constricted at the septa.
Culture characteristics: Conidia germinating on WA and germ tubes produced from conidium within 24 h. Colonies growing on PDA, circular, with flat surface, edge entire, cotton, reaching 20 mm diam. in 10 days at 25 °C, gray upper and reverse view with pale gray margin on PDA medium.
Material examined: CHINA, Yunnan Province, Kunming City, Kunming Institute of Botany, on decaying twig of Cerasus pseudocerasus (Lindl.) Loudon (Rosaceae), 19 August 2018, D.P. Wei, KIB1902 (KUN-HKAS 102564, holotype), ex-type living culture, KUMCC 19-0215.
GenBank numbers: ITS = MN792812, LSU = MN792815, SSU = MN792814, TEF1-α = MN794017.
Notes: In the concatenated gene analyses, Pseudochaetosphaeronema kunmingense shows a close phylogenetic affinity to P. siamensis Jayasiri, E.B.G. Jones & K.D. Hyde (MFULCC 17-2287) with strong support (100% ML, 1.00 BYPP; Fig. 49). Morphologically, P. kunmingense is similar to P. siamensis in having dark brown, uni-loculate, globose to subglobose, glabrous perithecia, monophialidic, cylindrical conidiogenous cells and gray to brown colonies. But it differs from P. siamensis in conidial morphology with P. kunmingense having septate, fusiform, yellow-brown, conidia and P. siamensis in having hyaline to subhyaline, subglobose to oval, aseptate conidia (Jayasiri et al. 2019). The comparison of ITS sequences between P. kunmingense (KUN-HKAS 102564) and P. siamensis (MFULCC 17-2287) shows 37 bp differences (6%) within 510 bp. Thus, we introduce our isolates as a new species according to the guidelines of Jeewon and Hyde (2016).
Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, ITS and TEF1-α sequence data representing suborder Massarineae in Pleosporales. Related sequences are taken from Phukhamsakda et al. (2016). Sixty-four strains are included in the combined analyses which comprised 3085 characters (808 characters for LSU, 963 characters for SSU, 471 characters for ITS, 843 characters for TEF1-α) after alignment. The best scoring RAxML tree with a final likelihood value of − 17310.739106 is presented. The matrix had 992 distinct alignment patterns, with 34.19% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.241498, C = 0.240612, G = 0.272871, T = 0.245019; substitution rates: AC = 1.471176, AG = 2.947321, AT = 1.799343, CG = 1.117933, CT = 8.219803, GT = 1.000000; gamma distribution shape parameter α = 0.486274. Bootstrap support values for ML equal to or greater than 70% and BYPP equal to or greater than 0.95 are given above the nodes. Newly generated sequences are in blue bold and type species are in black bold. Alternaria alternata (CBS 916.96) and Leptosphaeria doliolum (CBS 505.75) in Pleosporineae (Pleosporales) were used as the outgroup taxa
Melanommataceae G. Winter
Notes: The family Melanommataceae was introduced by Winter (1885) with Melanomma Nitschke ex Fuckel as the type genus. The family is characterised by globose or depressed perithecial ascomata, trabeculate pseudoparaphyses, bitunicate and fissitunicate asci, pigmented and phragmosporous ascospores (Hyde et al. 2013; Tian et al. 2015; Li et al. 2016b, 2017a; Tennakoon et al. 2018a). Most species of this family are saprobic or hyperparasitic and occur mainly on twigs or bark of various woody plants from terrestrial, marine or freshwater habitats, and are widespread in temperate and subtropical regions (Zhang et al. 2012; Hyde et al. 2013; Tian et al. 2015). Tian et al. (2015) accepted 20 genera and provided the most detailed of morphological and multi-gene (LSU, SSU, TEF1-α and RPB2) phylogenetic data for this family. Many new species and genera have been added to this family (Li et al. 2016b, 2017a; Hyde et al. 2016; Almeida et al. 2017; Jaklitsch and Voglmayr 2017; Tibpromma et al. 2017; Tennakoon et al. 2018a; Pem et al. 2019c). The family includes 33 genera (Hongsanan et al. in prep.).
Camposporium Harkn.
Notes: Camposporium was introduced by Harkness (1884) with a single species Camposporium antennatum Harkn. The genus is characterised by dematiaceous, simple conidiophores, terminal, integrated, denticulate conidiogenous cells, and the conidia are typically cylindrical and elongate, multi-septate, rounded at one or both ends, often the cells at each end are paler in pigmentation than the central cells, the apex is either simple or has one or more cylindrical appendages, the base typically has a persistent portion of the denticle attached (Hughes 1951; Ellis 1971; Ichinoe 1971; Whitton et al. 2002). Species of this genus have been reported in many countries (Argentina, Australia, Brunei, India, Canada, Hawaii, Hungary and Japan) from freshwater and terrestrial habitats. (Rao and Rao 1964; Dudka 1966; Ichinoe 1971; Matsushima 1971, 1983; Shearer 1974; Ellis 1976; Abdullah 1980; Castañeda 1985; Mercado et al. 1995; Whitton et al. 2002).
Hughes (1951) reviewed this genus and accepted four species. Subsequently, 16 additional species have been described in this genus (Rao and Rao 1964; Dudka 1966; Ichinoe 1971; Matsushima 1971, 1983; Shearer 1974; Ellis 1976; Abdullah 1980; Castañeda 1985; Mercado et al. 1995; Whitton et al. 2002; Thakur et al. 2014; Adamčík et al. 2015), and 20 species are now accepted in this genus. Crous et al. (2018b) found that Camposporium antennatum and Fusiconidium mackenziei clustered together within Melanommataceae in their phylogenetic analyses. However, the placement of Camposporium is still doubtful. In this study, we added all the strains of Camposporium in our multi-gene phylogenetic analyses of Melanommataceae and the result showed that Camposporium and Fusiconidium grouped together within Melanommataceae. However, these two genera are morphological different in the conidiogenesis cells and conidial shape. We, therefore, treat these two genera as distinct until phylogenetic affinities between these two genera are well-resolved. We place Camposporium in Melanommataceae based on phylogeny. We added three new species in Camposporium and introduced a new record of C. pellucidum based on morphological characters and phylogenetic analyses. In addition, a new combination C. lycopodiellae is introduced for Fusiconidium lycopodiellae.
Camposporium appendiculatum D.F. Bao, Z.L. Luo, K.D. Hyde & H.Y. Su, sp. nov.
Index Fungorum number: IF557026; Facesoffungi number: FoF 07064; Fig. 50
Etymology: Referring to the apendages of this fungus.
Holotype: DLU 1234
Saprobic on submerged decaying wood in freshwater. Sexual morph Undetermined. Asexual morphColonies on substratum effuse, superficial, black. Mycelium immersed, composed of brown, branched, smooth hyphae. Conidiophores micronematous, mononematous, very short, simple, unbranched, flexuous, pale brown to subhyaline, smooth. Conidiogenous cells mono- or polyblastic, terminal, integrated, denticulate, subhyaline. Conidia 107–119 μm long (\( \bar{x} \) = 113.2 μm, SD = 6.1, n = 30), 9.5–11.5 μm wide (\( \bar{x} \) = 10.4 μm, SD = 1, n = 30), solitary, cylindrical, elongate, 10–13-septate, pale brown at middle cells, hyaline at both the end cells, truncate at both of ends, the apex with a filiform, septate, hyaline apical appendage, 72–114 μm long (\( \bar{x} \) = 107.8 μm, SD = 35.9, n = 25), 3.5–4.8 μm wide (\( \bar{x} \) = 4.2 μm, SD = 0.6, n = 25), smooth.
Culture characteristics: Conidia germinating on PDA within 24 h. Colonies on PDA at room temperature reaching 2.5 cm diam. in 3 weeks, mycelium pale brown to greyish brown after 3 weeks, composed of pale brown to brown, septate, smooth hyphae.
Material examined: CHINA, Yunnan Province, saprobic on submerged decaying wood in Lancang River, December 2016, Z.L. Luo, 2L H 6–3–2, S-1234 (DLU 1234, holotype), ex-type living culture, DLUCC 1234.
GenBank numbers: ITS = MN758890, LSU = MN759021, SSU = MN758956, TEF1-α = MN784094.
Notes: Morphologically, Camposporium appendiculatum is similar to C. cambrense S. Hughes in having short, simple, unbranched conidiophores, mono- or polyblastic, terminal, integrated conidiogenous cells and cylindrical, elongate, septate conidia with a single appendage at apex. However, C. appendiculatum has 10–13-septate conidia with truncate at both ends, whereas, C. cambrense has 3–15-septate conidia, rounded at apex and truncate base. Phylogenetically, C. appendiculatum is not close to C. cambrense, rather it clusters with C. multiseptatum and is sister to C. lycopodiellae (≡ Fusiconidium lycopodiellae) with a strong statistical support (99% ML, 1.00 BYPP; Fig. 55).
Camposporium lycopodiellae (Crous & R.K. Schumach.) Tibpromma & K.D. Hyde, comb. nov.
Index Fungorum number: IF557244; Facesoffungi number: FoF 06524
≡ Fusiconidium lycopodiellae Crous & R.K. Schumach., Fungal Systematics and Evolution 1: 188 (2018)
Description and illustration: See Crous et al. (2018b)
Notes: Fusiconidium lycopodiellae was introduced by Crous et al. (2018b) and was collected from Germany on stems of Lycopodiella inundata. Crous et al. (2018b) mentioned that this species is reminiscent of Clasterosporium but it lacks hyphopodia. Based on LSU sequence data F. lycopodiellae allied with Fusiconidium, but it also lacks percurrent proliferation of the conidiogenous cells and fusoid to ellipsoid conidia. Therefore Crous et al. (2018b) tentatively named their collection as Fusiconidium.
In our phylogenetic analysis based on combined LSU, SSU, ITS and TEF1-α sequence, Fusiconidium lycopodiellae clustered with Camposporium with high support (99% ML, 1.00 BYPP; Fig. 55). Therefore, the description of the new combined Camposporium lycopodiellae (≡ Fusiconidium lycopodiellae) follows Crous et al. (2018b). Thus, we propose the transfer of Fusiconidium lycopodiellae under the new combination Camposporium lycopodiellae, based on morphological similarities and phylogenetic analysis.
Camposporium multiseptatum D.F. Bao, Z.L. Luo, K.D. Hyde & H.Y. Su, sp. nov.
Index Fungorum number: IF557025; Facesoffungi number: FoF 07063; Fig. 51
Etymology: Referring to multi-septate conidia of this species.
Holotype: DLU 792
Saprobic on submerged decaying wood in freshwater. Sexual morph Undetermined. Asexual morphColonies on substratum effuse, superficial, black. Mycelium immersed, composed of hyaline, branched, smooth hyphae. Conidiophores micronematous, mononematous, short, simple, unbranched, flexuous, pale brown to subhyaline, smooth. Conidiogenous cells mono- or polyblastic, terminal, determinate, subhyaline. Conidia 97–111 μm long (\( \bar{x} \) = 103.8 μm, SD = 7.2, n = 30), 9–11 μm wide (\( \bar{x} \) = 9.8 μm, SD = 0.8, n = 30), fusiform to cylindrical, truncate at both of the ends, 10–13-septate, dark brown at central cells, paler both of end cells, with a short, septate, subhyaline, single appendage at apex, 11–17 μm long (\( \bar{x} \) = 13.8 μm, SD = 3, n = 15), 3.5–4.7 μm wide (\( \bar{x} \) = 4.1 μm, SD = 0.6, n = 15), smooth.
Culture characteristics: Conidia germinating on PDA within 24 h. Colonies on MEA at room temperature reaching 3.3 cm diam. in 3 weeks, mycelium grayish brown to dark brown after 4 weeks, composed brown to dark brown, septate, smooth hyphae.
Material examined: CHINA, Yunnan Province, saprobic on submerged decaying wood in Dulong River, October 2016, Z.L. Lou, 2D H 1–4–2, S-792 (DLU 792, holotype), ex-type living culture, DLUCC 792.
GenBank numbers: ITS = MN758889, LSU = MN759020, SSU = MN758955, TEF1-α = MN784093.
Notes: Camposporium multiseptatum is phylogenetically close to C. appendiculatum (Fig. 55). Morphologically, C. multiseptatum is similar to C. appendiculatum in having short, simple conidiophores, mono- or polyblastic, terminal, determinate conidiogenous cells and cylindrical, septate conidia, truncate at both ends, with a single appendage at apex. However, C. multiseptatum differs from C. appendiculatum in having shorter appendage (11–17 μm versus 72–114 μm). Moreover, we compared the base pairs of TEF1-α gene regions, and there are 17 base pairs that were different in 875 nucleotides of the TEF1-α region. These results strongly support our isolate to be a new species.
Camposporium pellucidum (Grove) S. Hughes, Mycological Papers 36: 9 (1951)
Facesoffungi number: FoF 07065, Fig. 52
≡ Bactrodesmium caulincola var. pellucidum Grove, J. Bot., Lond. 24: 200 (1886)
Saprobic on submerged decaying wood in freshwater. Sexual morph Undetermined. Asexual morphColonies on substratum effuse, superficial, black. Mycelium mostly immersed, composed of hyaline, branched, smooth hyphae. Conidiophores micronematous, mononematous, solitary, short, unbranched, flexuous, septate, pale brown to subhyaline, smooth. Conidiogenous cells monoblastic, terminal, integrated into the apical region of the conidiophores, subhyaline, smooth. Conidia 112–157 μm long (\( \bar{x} \) = 134.5 μm, SD = 22.3, n = 30), 8.8–12.3 μm wide (\( \bar{x} \) = 10.6 μm, SD = 1.7, n = 30), solitary, cylindrical to elongate, rounded at apex, truncate at the ends, 10–16-septate, the middle cells are brown, both of end cells are pale brown to subhyaline, without appendage.
Culture characteristics: Conidia germinating on PDA within 24 h. Colonies on PDA at room temperature reaching 2 cm diam. in 2 weeks, mycelium pale brown to grayish brown after 3 weeks, composed pale brown to brown, septate, smooth hyphae.
Material examined: CHINA, Yunnan Province, saprobic on submerged decaying wood in Lancang River, December 2016, Z.L. Luo, 2L H 4–36–3, S-1239 (DLU 1239), living culture, DLUCC 1239.
Known host and distribution: Occurring on wood, bark, fruit of various trees, shrubs and dead herbaceous stems (Finland, Britain and Netherlands; Gönczöl and Révay 2004).
GenBank numbers: ITS = MN758891, LSU = MN759022, SSU = MN758957, TEF1-α = MN784095.
Notes: Camposporium pellucidum S. Hughes was introduced by Hughes (1951) based on the variety Bactrodesmium caulincola var. pellucidum Grove. It is characterized by macronematous, mononematous, unbranched conidiophores, cylindrical or more usually cylindric-fusoid conidia, up to 16-septate, truncated at base, rounded at apex or subulate elongated into a long filiform septate appendage. Morphologically, our new isolate fits well with the characters of C. pellucidum in having macronematous, mononematous, unbranched conidiophores and similar size (112–157 × 8.8–12.3 versus 80–140 × 7.5–12 μm), septation (10–17 versus 7–16) and shape of conidia (cylindrical to elongate, rounded at apex, truncate at the ends). However, in our specimen we did not observe the appendage. Phylogenetic analysis also showed that our isolate clustered with C. pellucidum with high statistical support (97% ML, 0.99 BYPP; Fig. 55). Therefore, we identified our isolate as C. pellucidum and it is a new record for China.
Camposporium septatum N.G. Liu, J.K. Liu & K.D. Hyde, sp. nov.
Index Fungorum number: IF557182; Facesoffungi number: FoF 06711; Fig. 53
Etymology: Name reflects the septate conidia.
Holotype: MFLU 19-2851
Saprobic on submerged decaying wood in a freshwater. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate, effuse, golden brown, velvety. Mycelium mostly immersed, composed of white, septate, branched and guttulate hyphae. Conidiophores macronematous, mononematous, often procumbent on substrate, pale brown to median brown, unbranched, irregularly cylindrical, flexuous, septate, thick-walled. Conidiogenous cells monoblastic, terminal, integrated, subcylindrical, pale brown. Conidia 98–125 μm (\( \bar{x} \) = 110 μm, n = 30) long, 7–11.5 μm (\( \bar{x} \) = 9 μm, n = 30) wide at middle, 3.5–6 μm (\( \bar{x} \) = 4.5 μm, n = 30) wide at base, solitary, dry, cylindrical, elongate, median brown, paler at base, finely verrucose, (8–)9(–11)-septate, not constricted or slightly constricted at the septa, apex rounded, basal cell truncate, the apical cell gives rise to (2–)3 simple appendages; appendage hyaline, aseptate, smooth, tapering from the base to the apex.
Culture characteristics: Conidia germinating on water agar within 24 h. Germ tubes produced from the basal cell of conidia. Mycelia superficial, irregularly circular, with entire edge, from above yellowish gray in the centre, gray at the edge.
Material examined: THAILAND, Chiang Rai Province, Muang, Ban Nang Lae Nai, on decaying wood submerged in a freshwater stream, 6 March 2018, N.G. Liu, CR089 (MFLU 19-2851, holotype), ex-type living culture, MFLUCC 19-0483.
GenBank numbers: ITS = MN758892, LSU = MN759023, SSU = MN758958, TEF1-α = MN784096.
Notes: Camposporium septatum resembles C. fusisporum Whitton, McKenzie & K.D. Hyde in conidial length (98–125 versus 86–115 μm) and number of septa ((8–)9(–11) versus 8–11). Moreover, these two species have 2–3 apical appendages. However, the conidia of C. fusisporum are much wider than those of C. septatum (13.5–19 versus 7–11.5 μm) (Whitton et al. 2002). Phylogenetic analyses of a combined LSU, SSU, ITS and TEF1-α sequence dataset showed that C. septatum forms a distinct lineage basal to Fusiconidium with high support (91% ML, 1.00 BYPP; Fig. 55). However, Camposporium septatum is morphologically different from Fusiconidium in conidiogenesis and conidial shape (Li et al. 2017a), but fits well to Camposporium. Therefore, we introduce our new species as C. septatum.
Uzbekistanica Wanas., Gafforov & K.D. Hyde
Notes: Uzbekistanica was introduced by Wanasinghe et al. (2018) with U. rosae-hissaricae Wanas., Gafforov & K.D. Hyde as the type species. The genus was introduced to accommodate taxa having dictyosporous ascospores with a coelomycetous asexual morph which was collected from Rosa in Uzbekistan. Two species are accommodated in this genus viz. U. rosae-hissaricae and U. yakutkhanika Wanas., Gafforov & K.D. Hyde (Wanasinghe et al. 2018; Index Fungorum 2020). We follow the latest treatment and updated account of Uzbekistanica in Wanasinghe et al. (2018). We introduce a new Uzbekistanica pruni based on phylogenetic and morphological evidence.
Uzbekistanica pruni Chaiwan, Wanas., Bulgakov & K.D. Hyde, sp. nov.
Index Fungorum number: IF557062; Facesoffungi number: FoF 02777; Fig. 54
Uzbekistanica pruni (MFLU 17-2136, holotype). a, b Appearance of conidiomata on host. c Section of conidioma. d Ostiole. e Pycnidial wall. f–h Conidiogenous cells (g, h = stained with congo red). i–m Conidia (j, l = stained with congo red). Scale bars: c = 200 µm, d = 50 µm, e = 20 µm, f–h = 15 µm, i–m = 10 µm
Etymology: The specific epithet “pruni” refers to the host genus, Prunus, from which the holotype was collected.
Holotype: MFLU 17-2136
Saprobic or weak pathogenic on dead twigs of Prunus armeniaca. Sexual morph Undetermined. Asexual morphConidiomata 220–265 μm high, 260–380 μm diam. (\( \bar{x} \) = 245 × 340 μm, n = 10), pycnidial, solitary, scattered, semi-immersed in the host, subglobose to ampulliform, indistinct ostiolate. Pycnidial walls 25–35 μm wide, outer and inner layers composed of dark brown cells, 3–4 layers and thick-walled, hyaline cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 10–22 μm × 4–6 μm, holoblastic, ampulliform, hyaline, smooth, swollen at the base, discrete, producing a single conidium at the apex. Conidia 11–16 × 6–10 μm (\( \bar{x} \) = 14 × 9 μm, n = 50), oblong to ellipsoidal, with rounded ends, occassionally truncate base, initially hyaline, unicellular, becoming dark brown before release from the pycnidia, 3–4-septate, not constricted at the septa, with thickenned, septal bands, slightly thick-walled.
Material examined: RUSSIA, Rostov region, Shakhty City, private garden, on dead and dying branches of Prunus armeniaca L. (Rosaceae), 11 May 2017, T.S. Bulgakov, T-1834 (MFLU 17-2136, holotype).
GenBank numbers: ITS = MN758893, LSU = MN759024, TEF1-α = MN784097.
Notes: Uzbekistanica pruni has brown, oblong to ellipsoidal, 3–4-septate conidia whereas U. yakutkhanika has dark brown, oval to ovoid and 1-septate conidia (Wanasinghe et al. 2018). Phylogenetically this species forms a distinct lineage basal to Uzbekistanica (0.98 BYPP; Fig. 55). Therefore, U. pruni is introduced as a novel species from Prunus armeniaca in Russia.
Phylogenetic tree generated from maximum likelihood (ML) analysis based on combined LSU, SSU, ITS and TEF1-α sequence dataset for the species from Melanommataceae. Cyclothyriella rubronotata (CBS 121892 and CBS 141486) were used as the outgroup taxon. The dataset comprised 3216 characters including gaps (LSU: 1–840; SSU: 841–1850; ITS: 1851–2341 and TEF1-α: 2342–3216). The RAxML analysis of the combined dataset yielded a best scoring tree with a final ML optimization likelihood value of − 15146.729361. The matrix had 882 distinct alignment patterns, with 26.56% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.246328, C = 0.238141, G = 0.275690, T = 0.239841; substitution rates AC = 2.336359, AG = 4.382849, AT = 2.267052, CG = 1.191360, CT = 17.215273, GT = 1.000000; gamma distribution shape parameter α = 0.180969. Support values for maximum likelihood (ML) higher than 75% and Bayesian posterior probabilities (BYPP) greater than 0.95 are given at the nodes
Occultibambusaceae D.Q. Dai & K.D. Hyde
Notes: We follow the latest treatment and updated account of Occultibambusaceae in Phookamsak et al. (2019).
Occultibambusa D.Q. Dai & K.D. Hyde
Notes: Occultibambusa was introduced to accommodate O. bambusae D.Q. Dai & K.D. Hyde. Dai et al. (2017) and Zhang et al. (2017) provided updated accounts for this genus.
Occultibambusa bambusae D.Q. Dai & K.D. Hyde, in Dai et al., Fungal Divers.: https://doi.org/10.1007/s13225-016-0367-8, [26] (2016)
Facesoffungi number: FoF 01975; Fig. 56
Occultibambusa bambusae (MFLU 19-2696) a, b Appearance of ascomata on the host. c Section through ascoma. d Section through peridium. e Mycelial structures on the ascomatal wall. f Pseudoparaphyses. g–i Different developing stages of the asci. j–m Ascospores. n Ascospore stained in Indian ink. o Germinating ascospore. Scale bars: c = 100 μm, d, g–i, o = 20 μm, e, f, j–n = 10 μm
Holotype: THAILAND, Chiang Rai Province, Mae Fah Luang Unversity, on dead culms of bamboo, 9 July 2013, D.Q. Dai, DDQ00262 (MFLU 15-1212), ex-type living cultures, MFLUCC 13-0855 = CBS 139960 = MUCL 55882.
Saprobic on dead culms of Miscanthus gigantius, forming dark, round spots on host surface, with ascostromata on raised areas, erumpent. Sexual morphAscostromata 151–196 μm high, 368–518 μm diam., solitary, scattered, immersed, subglobose, slightly conical in section, uni-loculate, black at the ostiolar regions, coriaceous, with a central, rounded, papillate ostiole, lined internally with periphyses. Peridium comprising host and fungal tissues, 26–42 μm thick, composed of brown to hyaline, thick- to thin-walled cells of textura angularis, intermingled with host cells, wedge of palisade-like cells at the periphery: 46–63 μm thick, composed of large, brown cells. Hamathecium composed of septate, cellular pseudoparaphyses branching above the asci. Asci 60–84 × 11–18 μm (\( \bar{x} \) = 72 × 14.5 μm, n = 20), 8-spored, bitunicate, broadly cylindrical, with a short furcate pedicel, with a shallow ocular apical chamber. Ascospores 20–28 × 3.8–7 μm (\( \bar{x} \) = 24 × 5.4 μm, n = 20), 1-seriate, slightly broad and fusiform, 1-septate, occasionally with larger upper cell, narrowly acute at both ends, dark brown, straight to curved, surrounded by a gelatinous sheath, each cell with 2–3 guttules. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on PDA within 24 h with germ tubes produced from both cells. Colonies growing slowly on PDA circular, entire, floccose, with even margin, dark green to brown above and below.
Material examined: TAIWAN, Chia Yi Province, Kwang Hwa, Miscanthus gigantius (Poaceae), 27 April 2018, A. Karunarathna, AKTW 65 (MFLU 19-2696), living culture, NCYUCC 19-0370.
Known host and distribution: On dead culms of bamboo (Thailand; Dai et al. 2017), on Miscanthus gigantius (Taiwan; this study).
GenBank numbers: ITS = MN937238, LSU = MN937220.
Notes: Our strain NCYUCC 19-0370 isolated from the collection MFLU 19-2696, shows many similarities with Occultibambusa bambusae in having broadly cylindrical asci, with a short furcate pedicel and a shallow ocular apical chamber, and brown, fusiform, 1-septate ascospores. Our collection also shares the size range of ascomata, asci and ascospores with O. bambusae (Dai et al. 2017). However, our collection shows some variations from O. bambusae in having setae on the upper surface of the ascomata. In the phylogenetic analysis (Fig. 57), our strain NCYUCC 19-0370 has a close relationship with O. chiangraiensis Phukhams. & K.D. Hyde (MFLUCC 16-0380) and O. bambusae (MFLUCC 11-0394, MFLUCC 13-0855) with high support (98% ML, 1.00 BYPP). However, our collection differs from O. chiangraiensis in having frequently 1-septate ascospores, whereas O. chiangraiensis has 3-septate ascospores (Hyde et al. 2016). The strain NCYUCC 19-0370 lacks protein coding genes, hence this can affect the phylogenetic placement of NCYUCC 19-0370 within the Occultibambusa clade. The sequence pairwise comparison of ITS region shows 1.04% dissimilarity between NCYUCC 19-0370 and O. bambusae (MFLUCC 13-0855). Based on the guidelines of Jeewon and Hyde (2016), we identify our taxon as Occultibambusa bambusae. The species is reported on a new host, Miscanthus gigantius and a new geographical record from Taiwan.
Phylogram generated from maximum likelihood analysis based on a combined LSU, SSU, RPB2 and TEF1-α sequence dataset of taxa in Occultibambusaceae. Twenty strains are included in the combined sequence analyses. Roussoella hysterioides (HH 26988) and Neoroussoella bambusae (MFLUCC 11-0124) were used as the outgroup taxa. Bootstrap support values for ML equal to or greater than 70% and Bayesian posterior probabilities equal to or greater than 0.95 BYPP are indicated at the nodes. Newly generated sequences are in red
Parabambusicolaceae Kaz. Tanaka & K. Hiray.
Notes: The family Parabambusicolaceae was introduced by Tanaka et al. (2015) to accommodate Aquastroma Kaz. Tanaka & K. Hiray., Multiseptospora Phookamsak & K.D. Hyde and Parabambusicola Kaz. Tanaka & K. Hiray. (type genus), together with two unnamed “Monodictys” species. Subsequently, another four genera, i.e. Lonicericola Phookamsak, Jayasiri & K.D. Hyde, Neoaquastroma Wanas., E.B.G. Jones & K.D. Hyde, Paratrimmatostroma Jayasiri, Phookamsak, D.J. Bhat & K.D. Hyde, Pseudomonodictys Doilom, Ariyaw., Bhat & K.D. Hyde, were described in Parabambusicolaceae based on both morphology and phylogeny (Ariyawansa et al. 2015a; Wanasinghe et al. 2017b; Phookamsak et al. 2019). We introduce a new genus Paramonodictys based on morphological and molecular evidence.
Paramonodictys N.G. Liu, K.D. Hyde & J.K. Liu, gen. nov.
Index Fungorum number: IF557092; Facesoffungi number: FoF 06709
Etymology: Referring to the genus similar to Monodictys
Saprobic on decaying wood. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate superficial, effuse, scattered, solitary, black. Mycelium partly immersed, partly superficial, composed of brown, branched, septate hyphae. Stroma present, erumpent, erect, subcylindrical or truncated-cone-form, pale brown to olivaceous brown. Conidiophores absent. Conidiogenous cells monoblastic. Conidia solitary, dictyosporous, muriform, globose or subglobose, olivaceous brown to dark brown, broadly rounded at apex, sometimes subtruncate at base.
Type species: Paramonodictys solitarius N.G. Liu, K.D. Hyde & J.K. Liu
Notes: Hughes (1958) introduced the polyphyletic genus Monodictys which presently reside in three ascomycete classes, Dothideomycetes, Leotiomycetes, and Sordariomycetes (Tanaka et al. 2015). Since the sequence data of type species M. putredinis (Wallr.) S. Hughes is unavailable, the phylogenetic placement of Monodictyssensu stricto could not be clarified. However, our collection differs from M. putredinis in having a stroma (Ellis 1971). Therefore, we introduce our collection as a new genus to avoid taxonomic confusion for Monodictys.
Paramonodictys solitarius N.G. Liu, K.D. Hyde & J.K. Liu, sp. nov.
Index Fungorum number: IF557093; Facesoffungi number: FoF 06710; Fig. 58
Etymology: Name reflects the solitary conidia.
Holotype: MFLU 19-2854
Saprobic on decaying wood. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate superficial, effuse, scattered, solitary, black. Mycelium partly immersed, partly superficial, composed of brown, branched, septate hyphae. Stroma present, erumpent, erect, subcylindrical or truncated-cone-form, pale brown to olivaceous brown, up to 20 μm high. Conidiophores absent. Conidiogenous cells monoblastic. Conidia 50–87 × 40–61 μm (\( \bar{x} \) = 64 × 52 μm, n = 30), solitary, dictyosporous, muriform, globose or subglobose, olivaceous brown to dark brown, broadly rounded at apex, sometimes subtruncate at base.
Culture characteristics: Conidia germinating on water agar within 24 h. Germ tubes produced peripherally. Mycelia superficial, irregularly circular, with entire edge, greyish white from above, dark brown at centre, pale brown at circumference from below.
Material examined: CHINA, Guizhou Province, Guiyang, Guizhou Academy of Agricultural Sciences, on decaying wood, 27 August 2018, N.G. Liu, NKY036 (MFLU 19-2854, holotype), ex-type living culture, GZCC 20-0007.
GenBank numbers: ITS = MN901152, LSU = MN897835, SSU = MN901118, RPB2 = MT023015, TEF1-α = MT023012.
Notes: Tanaka et al. (2015) provided sequence data for an unnamed Monodictys species without any morphological information. However, molecular comparison indicated our collection and their strain are the same species (Fig. 59). Paramonodictys solitarius, characterised by absence of conidiophores, differs from Pseudomonodictys tectonae Doilom et al. The latter has semi-macronematous to sometimes macronematous conidiophores. Moreover, the conidia of P. solitarius are produced directly from stroma.
Phylogram generated from maximum likelihood analysis based on combined LSU, ITS, SSU, TEF1-α and RPB2 sequence data representing Parabambusicolaceae and the closely related families in Pleosporales. Fifty strains are included in the combined analyses which comprise 4416 characters (872 characters for LSU, 543 characters for ITS, 1034 characters for SSU, 922 characters for TEF1-α, 1045 characters for RPB2) after alignment. Melanomma pulvis-pyrius (CBS 124080) in Melanommataceae (Pleosporales) is used as the outgroup taxon. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RAxML tree with a final likelihood values of − 28950.933465 is presented. The matrix had 1788 distinct alignment patterns, with 32.59% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.240257, C = 0.252649, G = 0.271819, T = 0.235275; substitution rates AC = 1.353719, AG = 3.137124, AT = 1.266860, CG = 1.010760, CT = 6.055808, GT = 1.000000; gamma distribution shape parameter α = 0.189550. Bootstrap values for maximum likelihood (ML) equal to or greater than 75% and clade credibility values greater than 0.95 from Bayesian-inference analysis labeled on the nodes. The ex-type and ex-epitype strains are indicated in bold. The new isolate is indicated in bold and blue
Periconiaceae (Sacc.) Nann.
Notes: Periconiaceae has long been placed as members of Massarinaceae until Tanaka et al. (2015) revised Massarineae and placed it as a distinct family based on phylogenetic analysis (Tanaka et al. 2015; Hyde et al. 2017, 2018b). We follow the latest treatments and updated accounts of Periconiaceae in Hyde et al. (2017, 2018b), Liu et al. (2017b), Thambugala et al. (2017) and Phookamsak et al. (2019). A novel species, Periconia palmicola, is introduced in this study based on molecular data coupled with morphological characteristics.
Periconia Tode
Notes: Periconia was first descripted by Tode (1791). This genus is morphologically unique in having macronematous, branched conidiophores with a stipe and spherical apex. Conidiogenous cells are monoblastic or polyblastic, discrete on stipe and branched. Conidia are verruculose or echinulate, pale to dark brown, unicellular which are catenate, usually sphaerical to subsphaerical (Ellis 1971).
Periconia palmicola J.F. Li & Phookamsak, sp. nov.
Index Fungorum number: IF556889, Facesoffungi number: FoF 07053; Fig. 60
Periconia palmicola (MFLU 14-0198, holotype). a Appearance of fungal colonies on host substrate. b Conidiophores. c–e Close up conidiophores attached with conidiogenous cells. f–h Conidiogenous cells. i–l Conidia. m Germinated conidia. Scale bars: a = 100 µm, c–e = 20 µm, b, m = 10 µm, f–h = 5 µm, i–l = 2 µm
Etymolgy: The specific epithet “palmicola” refers to the host palm, of which the fungus was living.
Holotype: MFLU 14-0198
Saprobic on dead, fallen leaves of unidentified palm. Sexual morph Undetermined. Asexual morphColonies on the substratum superficial, numerous, effuse, black, floccose. Mycelium immersed on the substrate, composed of septate, branched, smooth, dark hyphae. Conidiophores 151–188 μm long × 5.6–8 μm wide (\( \bar{x} \) = 175.4 × 6.8 μm, n = 10), macronematous, mononematous, septate, partly forming sphaerical heads and branched at apex, erect, mostly slightly flexuous to curved, dark brown to black. Conidiogenous cells 3–3.5 μm long × 3–4.8 μm wide (\( \bar{x} \) = 3.2 × 4 μm, n = 10), mono- to polyblastic, discrete, terminal, globose, hyaline to subhyaline. Conidia 5.1–7.4 μm long × 4.8–6.1 μm wide (\( \bar{x} \) = 6.2 × 5.5 μm, n = 20), solitary to catenate, in acropetal chains, subglobose to globose, light brown to brown, verruculose.
Culture characteristics: Conidia germinating on PDA within 14 h and germ tubes produced from top cells. Colonies growing on PDA, hairy or cottony, white to grey, reaching 5 mm in 20 days at 25 °C, mycelium superficial, effuse, radially striate, with regular edge, subhyaline to pale white hyphae; Asexual spores and sexual spores were not formed within two months.
Material examined: THAILAND, Chiang Rai Province, Mae Fah Luang University, Herb Garden, on dead fallen leaves of palm, 28 December 2013, J.F. Li, H-13 (MFLU 14-0198, holotype), ex-type living culture MFLUCC14-0400.
GenBank numbers: LSU = MN648327, SSU = MN648319.
Notes: Periconia palmicola is described herein as a new species. Periconia palmicola is unique in its colourless conidiogenous cells and dark conidiophores. In the phylogenetic analyses of combined ITS, LSU and TEF1-α, P. palmicola formed an independent subclade to P. digitata (77% ML, 0.98 BYPP; Fig. 61). Therefore, P. palmicola is established as a new species.
Phylogram generated from the best scoring of the RAxML tree based on combined ITS, LSU and TEF1-α sequence data of taxa in Periconiaceae and other related families (Massarinaceae, Didymellaceae and Lentitheciaceae). Taxa in Morosphaeriaceae, Morosphaeria ramunculicola (KH 220) and M. velatispora (KH 221) were selected as the outgroup taxa. The best RAxML tree with a final likelihood value of − 13065.728774 is presented. RAxML analysis yielded 982 distinct alignment patterns and 28.84% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.245718, C = 0.219687, G = 0.310546, T = 0.224049, with substitution rates AC = 1.522214, AG = 2.988589, AT = 1.846846, CG = 0.504980, CT = 12.777992, GT = 1.000000. The gamma distribution shape parameter alpha = 0.317094. Bayesian posterior probabilities (BYPP) from MCMC were evaluated with final average standard deviation of split frequencies = 0.008294. Bootstrap support values for maximum likelihood (ML) equal to or greater than 70% and Bayesian posterior probabilities (BYPP) equal to or greater than 0.95 are given above or below the nodes. Type strains are in black bold and newly generated sequences is indicated in blue bold
Phaeosphaeriaceae M.E. Barr
Notes: Barr (1979) established Phaeosphaeriaceae which is a large and important family of Pleosporales (Phookamsak et al. 2014). It includes economically important plant pathogens and some endophytes as well as saprobes on plant hosts of monocotyledons (e.g., Cannaceae, Cyperaceae, Juncaceae, Poaceae), while others are found on dicotyledonous (Phookamsak et al. 2014, 2017, 2019; Tennakoon et al. 2016; Tibpromma et al. 2018; Wanasinghe et al. 2018; Hyde et al. 2019a). Phaeosphaeriaceous taxa are morphologically diverse. The morphological characters of taxa in this family are often ambiguous having overlapping characters with other taxa in Leptosphaeriaceae and Pleosporaceae. However, genera in this family can be distinguished based on morphological characteristics of both sexual and asexual morph coupled with multi-gene phylogenetic analysis. (Phookamsak et al. 2014, 2017).
We follow the latest treatment and updated accounts of Pheaeosphaeriaceae in Phookamsak et al. (2014, 2019), Crous et al. (2019b) and Hyde et al. (2019a). Currently, 68 genera are accommodated in this family (Phookamsak et al. 2014, 2019; Wanasinghe et al.; 2018; Bakhshi et al. 2019; Crous et al. 2019b; Maharachchikumbura et al. 2019; Yang et al. 2019a, b). We introduce a monotypic genus Bhagirathimyces to accommodate a novel hyphomycetous species, B. himalayensis and a new genus Paraloratospora to accommodate Paraloratospora camporesii sp. nov. and Paraloratospora gahniae comb. nov. In addition, three other new species are introduced viz. Loratospora arezzoensis, Neosetophoma camporesii and Septoriella camporesii. Following various treatments and introduction of new taxa, the family has become somewhat confused and requires a detailed study with much more taxa and molecular data. We therefore refrain from synonymising species and genera in this study and wait a monograph of the family. The sexual morph of Phaeosphaeriopsis pseudoagavacearum is reported and new host records are provided for Phaeosphaeria chinensis and Wojnowiciella dactylidis.
Bhagirathimyces S.M. Singh & S.K. Singh, gen. nov.
MycoBank number: MB830751; Facesoffungi number: FoF 06121
Etymology: Generic epithet refers to Holy River “Bhagirathi” (a turbulent Himalayan River) originated from Indian Himalayas
Isolated from Cryconite of glacier. Sexual morph Undetermined. Asexual morphColonies slow growing, brown to black, reverse black. Mycelium partly immersed, partly superficial, single or interwoven, subhyaline to light olivaceous, brown, smooth to rough-walled. Chlamydospores produced in chain, intercalary, swollen, subhyaline to light olivaceous. Conidiophores semi-macronematous, mononematous, small to median large, arising from superficial hyphae, simple to branched, subhyaline to light olivaceous, smooth to minutely irregularly roughened, 1–3-septate. Conidia solitary or catenate, highly variable in shape and size, lobed, globose to subglobose, clavate, muriform, often with a pale protuberant basal stalk cell, persistent to sessile, smooth-walled to sometimes spinulose, dark brown to blackish brown, sometimes olivaceous brown.
Type species: Bhagirathimyces himalayensis S.M. Singh & S.K. Singh
Notes: Bhagirathimyces can be morphologically compared with genera like Dematiopleospora (Wanasinghe et al. 2014), Nodulosphaeria Rabenh (Mapook et al. 2016a), Chaetosphaeronema Moesz., Muriphaeosphaeria (Phukhamsakda et al. 2015), Ophiobolopsis, Paraophiobolus, and Pseudoophiobolus (Phookamsak et al. 2017). It is distinct from these genera in having a combination of morphological characters such as dimensions of conidia and conidiophores, shape and septation patterns in conidia. In addition, the proposed genus Bhagirathimyces, is distinct from all known genera in the Phaeosphaeriaceae based on the sequence and phylogenetic analysis. On megablast analysis, ITS sequence of Bhagirathimyces, showed less than 93.6% similarity with known taxa. Phylogenetically, it was found to form a separate distinct subclade with strong support.
Bhagirathimyces himalayensis S.M. Singh & S.K. Singh, sp. nov.
MycoBank number: MB830752; Facesoffungi number: FoF 06122; Fig. 62
Bhagirathimyces himalayensis (AMH 10127, holotype). a Landscape of the sampling location at Hamtah glacier. b Cryoconites with pH probe. c Colony morphology on PDA (front view). d Reverse view of colony. e Numerous conidia. f Lobbed conidia (muriform) attached with conidiophores. g Globose to subglobose and obclavate conidia. h Branched conidiophore with attached conidia. i Attached conidia and conidiophores. j Conidia with a pale basal stalk cell. k Branched conidiophore with muriform conidium. l Macronematous conidiophore with conidium. m Conidia in chain. Scale bars: e–m = 20 μm
Etymology: The specific epithet “himalayensis” refers to the Himalaya, the region of origin of the sample.
Holotype: AMH 10127
Isolated from Cryconite of glacier. Sexual morph Undetermined. Asexual morph Hyphomycetous. Vegetative hyphae smooth to rough walled, septate subhyaline to light olivaceous, constricted near septa, up to 6.2 μm wide. Conidiophores arising from superficial hyphae, small to median large, micronematous, mononematous, simple, rarely branched, straight, light olivaceous to olivaceous brown, smooth-walled, 1–3-septate, 5.9–44.1 × 4.2–19.4 μm. Conidiogenous cells monoblastic, integrated, terminal, determinate, cylindrical. Conidia solitary or catenate, very variable in shape and size, lobed, globose to subglobose, clavate, muriform, often with a pale protuberant basal stalk cell, persistent to sessile, smooth-walled, sometimes spinulose, dark brown to blackish brown, sometimes olivaceous brown, 15.4–50.4 × 12.8–46.2 μm (\( \bar{x} \) = 26.8 × 22.4 μm, n = 30).
Culture characteristics: On PDA greyish black (10F-1), reaching 2.2 cm diam. in 15 days at 25 °C, velvety, sulcate with successive growth rings, regular margin, reverse greyish black (10F-1).
Material examined: INDIA, Himachal Pradesh, Hamtah Glacier, Lahaul and Spiti District, from Cryconites, 16 September 2016, S.M. Singh, AMH 10127 (holotype), ex-type living culture, NFCCI 4580.
GenBank numbers: ITS = MK836021, LSU = MK836020, TEF1-α = MT091965.
Notes: Because of the presence of muriform conidia, Bhagirathimyces himalayensis is distinct from many allied genera. To some extent it resembles Trimmatostroma scutellare (Berk. & Br.) Ellis and Epicoccum andropogonis (Ces.) Schol-Schwarz (= Cerebella andropogonis Ces.), but it lacks sporodochial stromata. It differs from Tetraploa in having muriform conidia, while they are setiform in Tetraploa. Bhagirathimyces himalayensis shows no morphological resemblance to the asexual coelomycetous state of Muriphaerosphaeria. Unfortunately, B. himalayensis cannot be compared with the allied genus Dematiopleospora, due to lack of a sexual morph. Bhagirathimyces himalayensis is also similar to Paradictyoathrinium diffractum, but differs in having catenate muriform conidia with a cylindrical basal stalk. The colour of the conidia are jet black in the former taxon, while they are brown in latter taxon.
Interestingly, Bhagirathimyces himalayensis was isolated from an entirely different habitat/ecological niche, i.e. from cryoconites of glacier of Indian Himalayan region as against its allied taxa such as Nodulosphaeria multiseptata Tibpromma, Camporesi & K.D. Hyde, (MFLUCC 15-0078/IT1889, type) isolated from forest soil of Sambucus ebulus (from Italy), N. guttulatum Tibpromma, Camporesi & K.D. Hyde (MFLUCC 14-1111) and N. scabiosae Mapook, Camporesi & K.D. Hyde (MFLUCC 15-0069/IT1826) from forest soil of Scabiosa sp. (from Italy), Dematiopleospora cirsii Wanas. et al. isolated from Cirsium sp. (from Italy), and D. mariae Wanas. et al. (MFLUCC 13-0612/IT1293) isolated from dead and hanging branches of Ononis spinosa (from Forli-Cesena, Italy).
Bhagirathimyces himalayensis (NFCCI 4580), is also different from known taxa based on the molecular and phylogenetic analysis. On megablast analysis, ITS sequence of B. himalayensis, showed less than 93.6% similarity with known taxa, therefore, is new to science; 93.03% (401/431) similarity with 2 gaps with Nodulosphaeria multiseptata (MFLUCC 15-0078/IT1889, type), 93.27% (402/431) similarity with 2 gaps with N. guttulatum (MFLUCC 15-0069/IT1826, type), 93.49% (402/430) similarity with 1 gaps with N. scabiosae (MFLUCC 14-1111, type), 87.35% (511/585) similarity with 29 gaps (4.95%) with Dematiopleospora cirsii (MFLUCC 15-0615, type) and 85.97% (478/556) similarity with 25 gaps (4.49%) with D. mariae (MFLUCC 13-0612/IT1293, type). Phylogenetic analysis clearly establishes Bhagirathimyces as a novel genus, and B. himalayensis as the type species with strong bootstrap values.
Loratospora Kohlm. & Volkm.-Kohlm.
Notes: Loratospora was introduced for the single species L. aestuarii, isolated from both submerged and aerial parts of dead culms of Juncus roemerianus growing in a marine habitat (Kohlmeyer and Volkmann-Kohlmeyer 1993). The genus was placed in Dothidealesincertae sedis until Barr (1996) placed it in Planistromellaceae owing to its locule openings which cracked the surface of the host and presence of periphysate ostioles. Its placement was maintained in Planistromellaceae (Jones et al. 2009) when finally, it was found to belong in Phaeosphaeriaceae based on multi-locus molecular phylogeny (Schoch et al. 2009; Suetrong et al. 2009; Zhang et al. 2012; Hyde et al. 2013).
Phookamsak et al. (2014) reported that Loratospora comprised a mucilaginous hamathecium instead of pseudoparaphyses which differentiated it from other genera in Phaeosphaeriaceae. They also remarked that this character might not be significant to differentiate the genera at family level. In the present study, a new Loratospora species, L. arezzoensis is introduced and while the former contains a mucilaginous hamathecium, the latter comprises pseudoparaphyses (Fig. 63). Addition of species to the genus therefore provides insight into the morphology of Loratospora and this observation seconds the statement made by Phookamsak et al. (2014) about the centrum-type not being sufficient to distinguish among genera in Phaeosphaeriaceae.
Loratospora arezzoensis (MFLU 17-1942, holotype). a Ascomata on host substrate. b, c Sections of ascomata. d Section through peridium. e Asci embedded in pseudoparaphyses. f–h Asci (note: h in congo red). i–l Ascospores (note: l stained with Indian ink). Scale bars: b, e = 50 µm, c, f–h = 30 µm, d, i–l = 10 µm
The phylogram generated in this study (Fig. 70) revealed our strains (MFLU 17-1942, MFLU 18-0915) group with two strains of Loratospora aestuarii Kohlm. & Volkm.-Kohlm. (JK 5535B and CBS 117592), Phaeosphaeria juncophila Leuchtm. (CBS 575.86), P. eustoma (Fuckel) L. Holm (CBS 307.71), P. glyceriae-plicatae (Săvul. & Sandu) Shoemaker & C.E. Babc. (CBS 101261), P. parvula (Niessl) Leuchtm. (CBS 605.86), P. caricicola (Fautrey) Leuchtm. (CBS 603.86), P. gahniae Crous (CPC 32454), P. norfolcia (Cooke) Leuchtm. (CBS 593.86), Wingfieldomyces cyperi (Crous & M.J. Wingf.) Y. Marín & Crous (CBS 141450), ‘Phaeosphaeria pleurospora’ (Niessl) Leuchtm. (CBS 460.84) and Sulcispora supratumida Senan., Camporesi & K.D. Hyde (MFLUCC 14-0995) with strong support (97% ML, 1.00 BYPP; Fig. 70). None of the Phaeosphaeria taxa mentioned above clustered with the type species of Phaeosphaeria (P. oryzae) in the phylogenetic analyses. Based on morphology coupled with phylogenetic analyses, we introduce our novel species as Loratospora arezzoensis. The new genus Paraloratospora is also introduced to accommodate Paraloratospora camporesii sp. nov. and Paraloratospora gahniae comb. nov., these taxa clustered with Phaeosphaeriasensu lata (Fig. 70).
Loratospora arezzoensis Bundhun, Wanas., Jeewon & K.D. Hyde, sp. nov.
Index Fungorum number: IF556992; Facesoffungi number: FoF 06979; Fig. 63
Etymology: The specific epithet “arezzoensis” refers to the name of the Province (Arezzo) from where the fungus was collected.
Holotype: MFLU 17-1942
Saprobic on dead land stem of rush (Juncus sp.). Sexual morphAscomata 109–119 µm high, 104–123 µm diam. (\( \bar{x} \) = 114.4 × 116.3 µm, n = 7), solitary, scattered to aggregated, immersed to erumpent through host surface, noticeable as small black spots on host surface, uni- to bi-loculate, globose to subglobose, dark brown to black, glabrous, centrally ostiolate. Ostiole up to 27 µm and 31 µm wide, central, minutely papillate, dark brown, composed of light brown periphyses. Peridium of unequal thickness, thicker at the apex (19–25 µm) and thinner at the sides and base (11–16 µm), composed of 5–7 layers of thick-walled, dark brown to black pseudoparenchymatous cells of textura angularis to textura globulosa. Hamathecium composed of sparse, filamentous, indistinct, septate pseudoparaphyses, anastomosed above the asci, embebbed in mucilaginous matrix. Asci 66–75(–83) × 14–19 µm (\( \bar{x} \) = 71.4 × 16.3 µm, n = 11), 8-spored, bitunicate, fissitunicate, cylindric-clavate, sessile to short pedicellate; apex rounded, thick-walled, with well-developed ocular chamber. Ascospores 32–37 × 4–5 µm (\( \bar{x} \) = 34.5 × 4.3 µm, n = 20), overlapping or parallel, 3–5-seriate, phragmosporous, narrowly elongate fusiform with rounded ends, slightly curved, 3-transversely septate, faintly constricted at the central septum, initially hyaline, becoming pale yellow at maturity, rough-walled, surrounded by a thick, distinct sheath. Asexual morph Undetermined.
Material examined: ITALY, Province of Arezzo, near Eremo di Camaldoli, on dead aerial stem of Juncus sp., 2 October 2017, E. Camporesi, IT 775 (MFLU 17-1942, holotype).
GenBank numbers: ITS = MN756638, LSU = MN756636, SSU = MN756634.
Notes:Loratospora arezzoensis shares similar characteristics with the type species, L. aestuarii in its ostiole comprising brown periphyses, peridium made up of cells of textura angularis to textura globulosa and shape of the asci and ascospores. It also occurs on Juncus sp. similar to L. aestuarii. However, L. arezzoensis differs from L. aestuarii in its smaller ascomata (109–119 µm high, 104–123 µm diam. versus 130–250 μm high, 160–310 μm diam.; Phookamsak et al. 2014). Its asci are smaller and cylindric-clavate (\( \bar{x} \) = 71.4 × 16.3 µm versus \( \bar{x} \) = 80.9 × 28.2 μm; Phookamsak et al. 2014) as compared to the ovoid or ampulliform asci of L. aestuarii. The ocular chambers for the asci of L. arezzoensis are conspicuous as compared to the asci of L. aestuarii which have indistinct ocular chambers. The ascospores of L. arezzoensis are also smaller than L. aestuarii (\( \bar{x} \) = 34.5 × 4.3 µm versus \( \bar{x} \) = 46.7 × 6.7 μm; Phookamsak et al. 2014) with a slight constriction at the central septum and a thick surrounding mucilaginous sheath. Furthermore, while L. aestuarii occurs in a marine environment (Kohlmeyer and Volkmann-Kohlmeyer 1993; Monkai et al. 2013), L. arezzoensis occurs on dead terrestrial hosts.
Neosetophoma Gruyter, Aveskamp & Verkley
Notes: Neosetophoma was introduced by de Gruyter et al. (2010) to accommodate N. samarorum. It is closely related to Paraphoma, but differs based on morphological characters and molecular phylogeny. Neosetophoma was accepted as a distinct genus in the family Phaeosphaeriaceae in several recent studies (de Gruyter et al. 2010; Hyde et al. 2013, 2018b; Quaedvlieg et al. 2013; Tibpromma et al. 2017; Marin-Felix et al. 2017; Wanasinghe et al. 2018; Phookamsak et al. 2014, 2019). We follow the latest treatment and updated accounts of Neosetophoma in Phookamsak et al. (2019). Nineteen species are known in this genus (Index Fungorum 2020). Neosetophoma species are mostly asexual morphs with yellowish conidia that are attenuated at one end and are regarded as pathogens causing leaf spots of various hosts (Phookamsak et al. 2014). Tibpromma et al. (2017) introduced N. garethjonesii as the first report of the sexual morph of Neosetophoma and Hyde et al. (2018b) introduced three sexual morph species, viz. N. guiyangensis, N. shoemaker and N. xingrensis. We introduce another new sexual species N. camporesii based on morphological characters and phylogenetic analysis.
Neosetophoma camporesii Q. Tian & K.D. Hyde sp. nov.
Index Fungorum number: IF556934; Facesoffungi number: FoF 06859; Fig. 64
Neosetophoma camporesii (MFLU 15-3394, holotype). a Specimen, dead spines of Rosa canina. b Appearance of black ascomata on the host surface. c Pseudoparaphyses. d Vertical section of ascoma. e, f Vertical section of ascoma, note the reddish tinge at the pore. g–i Asci with ascospores. j Germinating ascospore. k–n Ascospores. Scale bars: b = 200 μm, d = 50 μm, e, f = 20 μm, g–j = 10 μm, c, k–n = 5 μm
Etymology: Named in honor of Mr. Erio Camporesi who made an immense contribution to fungal investigation and collection.
Holotype: MFLU 15-3394
Saprobic on leaves of Rosa sp. Sexual morphAscomata 235–360 μm high × 170–280 μm diam. (\( \bar{x} \) = 295 × 220 μm, n = 10), scattered or sometimes gregarious, globose, subglobose to turbinate, coriaceous, black, smooth, ostiolate. Ostiole flat, epapillate, with a reddish tinge at the pore. Peridium thick at apex, 15–26 μm thick (\( \bar{x} \) = 23 μm, n = 10) at the sides, 2-layered, outer layer composed of irregular, thick-walled, brown to dark brown cells of textura epidermoidea, cells up to 3 μm diam., inner layer composed of small, thin-walled, pale brown cells, up to 5 μm diam. Hamathecium of dense, 1–2 μm broad, long, pseudoparaphyses, embedded in mucilage, anastomosing between and above asci. Asci 62–90 × 7–10 μm (\( \bar{x} \) = 84 × 8.5 μm, n = 10), 8-spored, bitunicate, cylindro-clavate to clavate, with a short pedicel. Ascospores 2-seriate above and 1-seriate or overlapping 1-seriate below, 20–32 μm × 3–5 μm (\( \bar{x} \) = 27 × 4.2 μm, n = 10), hyaline to pale brown, fusoid, obtuse at the apex and tapering towards the lower end cell, mostly straight, sometimes slightly curved, 3-septate, slightly constricted at the septa, smooth-walled. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on water agar and germ tubes produced from ascospores within 12 h. Colonies on PDA reaching 40 mm in 7 days at 25 °C, margin feathery. Mycelium aerial, white, becoming green to yellow towards the edge from above. No asexual morph was produced on PDA after 60 days incubation.
Material examined: ITALY, Province of Forlì-Cesen [FC], Pian di Spino-Civitella di Romagna, on dead spines of Rosa canina L. (Rosaceae), 3 February 2015, E. Camporesi, IT2361 (MFLU 15-3394, holotype; KUN-HKAS 92414, isotype), ex-type living cultures, MFLUCC 15-0682, KUMCC 15-0080.
GenBank numbers: ITS = KU302779, LSU = KU302778, SSU = MN653132, TEF1-α = MN654114.
Notes: Based on a megablast search using the ITS sequence, the closest matches in NCBI’s GenBank nucleotide database were Neosetophoma shoemakeri Senwanna et al. (GenBank NR_161044; Identities 524/529 (99%), no gap), N. clematidis Wijayaw., Camporesi & K.D. Hyde (GenBank LC171715; Identities 523/530 (99%), no gaps). The highest similarities using the LSU sequence were Coniothyrium cerealis E. Müll. (GenBank MH873258; Identities 878/879 (99%), no gaps), Neosetophoma shoemakeri (GenBank MG844348; Identities 878/879 (99%), no gaps). The highest similarities using the SSU sequence were Poaceicolaagrostina Mapook, Camporesi & K.D. Hyde (GenBank NG_062198; Identities 1004/1004 (100%), no gaps). The highest similarities using the TEF1-α sequence were Neosetophoma shoemakeri (GenBank MG844352; Identities 855/873 (98%), no gaps).
The sexual morph taxon, Neosetophoma camporesii, is introduced based on phylogenetic analyses of combined ITS, LSU, SSU and TEF1-α sequence data, and distinct morphological characters (Figs. 64, 70). Phylogenetically, our isolate formed a distinct clade within Neosetophoma in Phaeosphaeriaceae distant from other four sexual morph species (MFLUCC 14-0528, MFLU16-1606, GZ13 and GZAAS 18-0100). Morphologically, N. camporesii is similar to Paraleptosphaeria nitschkei (Rehm ex G. Winter) Gruyter, Aveskamp & Verkley, but differs in its 3-septate, fusoid ascospores (obtuse at the apex and tapering at the end). Neosetophoma guiyangensis J.F. Zhang et al. and N. xingrensis J.F. Zhang et al. formed sister relationships in phylogenetic analysis with high bootstrap support (Fig. 70). Both species share the similar characters of fusiform, hyaline to pale brown guttulate ascospores, but N. guiyangensis has normally 3–5-septate ascospores and a furcate pedicel, while N. xingrensis has 1–3-septate ascospores and the pedicel is sessile (Hyde et al. 2018b). However, the most conspicuous difference between these four taxa is that N. camporesii has a clear ascomatal ostiole with a reddish tinge at the pore.
Paraloratospora Bundhun, Tennakoon, Phookamsak & K.D. Hyde, gen. nov.
Index Fungorum number: IF557115; Facesoffungi number: FoF 07195
Etymology: The generic epithet “Paraloratospora” refers to pheaosphaeriaceous taxa having phylogenetically close to the genus Loratospora.
Associated with living branches or leaves. Sexual morphAscomata solitary, scattered to aggregated, immersed to erumpent through host surface, noticeable as small black spots, uni-loculate, globose to subglobose, dark brown, glabrous, centrally ostiolate, with minute papilla, filled with hyaline periphyses. Peridium of unequal thickness, thicker near the apex and thinner at the sides towards the base, comprising 4–7 layers of dark brown, thick-walled pseudoparenchymatous cells of textura angularis, paler towards the inner layers. Hamathecium composed of dense, filamentous, septate pseudoparaphyses, slightly constricted at the septa, anastomosing above the asci, embedded in gelatinous matrix. Asci 8-spored, bitunicate, fissitunicate, cylindrical to subcylindric, sessile to subsessile, apex rounded, thick-walled, with a small ocular chamber. Ascospores overlapping 1–2-seriate, hyaline to light brown, or light yellowish, fusiform to ellipsoidal, with rounded ends, septate, the second cell from the apex swollen, smooth-walled, with or without a mucilaginous sheath. Asexual morph Undetermined.
Type species: Paraloratospora camporesii Bundhun, Jeewon & K.D. Hyde
Notes: Paraloratospora is introduced to accommodate phaeosphaeria-like taxa that have a close phylogenetic relationship with Wingfieldomyces Y. Marín & Crous, Loratospora and Sulcispora Shoemaker & C.E. Babc. and are phylogenetic distinct from Phaeosphaeriasensu stricto. Based on multi-gene phylogenetic analyses (Fig. 70) our strain MFLU 18-0915 formed an independent lineage and clustered with Phaeosphaeriasensu lato, P. caricicola (CBS 603.86), P. parvula (CBS 605.86), P. glyceriae-plicatae (CBS 101261), P. juncophila (CBS 575.86), P. eustoma (CBS 307.71), P. norfolcia (CBS 593.86) and P. gahniae (CPC 32454) with significant support (79% ML, 1.00 BYPP; Fig. 70). These species do not represent by the type stains, except for P. gahniae (CPC 32454). Therefore, we introduce the new genus Paraloratospora to accommodate our new species, Paraloratospora camporesii and Phaeosphaeria gahniae (CPC 32454) is also transferred to this new genus as Paraloratospora gahniae comb. nov. based on morphological characteristics coupled with phylogenetic support.
Paraloratospora has similar morphological characteristics with Phaeosphaeria and can be distinguished from Wingfieldomyces in having hyaline to light brown, or light yellowish, fusiform to ellipsoidal, 3–4-septate, smooth-walled ascospores, with the second cell from the apex is swollen and surrounded by a mucilaginous sheath, whereas the monotypic genus Wingfieldomyces has elongate fusoid, finely verruculose, red-brown, 2-septate ascospores and central cell somewhat swollen (Marin-Felix et al. 2019a). Loratospora differs in having ovoid or ampulliform asci, with pale yellowish, elongate fusiform ascospores and Sulcispora differs in having subglobose to clavate asci with reddish to dark brown, fusiform to subclavate ascospores, with 12–16 longitudinal furrows on surface and lacking mucilaginous sheath (Senanayake et al. 2018a). Thus, we believe it would taxonomically astute to place our isolate in a new genus for the time being (Paraloratospora). We recommend to collect more taxon sampling for Loratospora, Sulcispora and Wingfieldomyces for further clarify their phylogenetic affinities.
Paraloratospora camporesii Bundhun, Jeewon & K.D. Hyde, sp. nov.
Index Fungorum number: IF556993; Facesoffungi number: FoF 06980; Fig. 65
Paraloratospora camporesii (MFLU 18-0915, holotype). a Ascomata on host substrate. b Close-up of ascomata on host substrate. c Section of ascoma. d Section of ostiole. e Section through peridium. f, g Pseudoparaphyses. h–j Asci (Notes: i in congo red, j in cotton blue). k–m Ascospores (m stained in Indian ink). Scale bars: c = 100 µm, d, f–j = 20 µm, e = 10 µm, k–m = 10 µm
Etymology: Named in honour of Mr. Erio Camporesi who made an immense contribution to fungal investigation and collection.
Holotype: MFLU 18-0915
Associated with living branch of Lonicera xylosteum. Sexual morphAscomata 160–177 µm high, 138–145 µm diam. (\( \bar{x} \) = 169.1 × 140.6 µm, n = 7), solitary, scattered to aggregated, immersed to erumpent through host surface, noticeable as small black spots, mostly uni-loculate, globose to subglobose, dark brown, glabrous, centrally ostiolate. Ostioles up to 30 µm and 42 µm wide, central, with minute papilla, dark brown, comprising hyaline periphyses. Peridium of unequal thickness, thicker near the apex (17–21 µm) and thinner at the sides and base (12–16 µm), comprising 4–7 layers, outer part composed of dark brown thick-walled cells of textura angularis, inner layer made up of pale brown to hyaline thin-walled cells of textura angularis. Hamathecium 50–90 µm high, 2.6–4.5 µm wide, unbranched, septate, pseudoparaphyses, constricted at the septum. Asci 52–68 × 7–11 µm (\( \bar{x} \) = 55.3 × 9.8 µm, n = 13), 8-spored, bitunicate, fissitunicate, cylindrical, sessile to subsessile, apex rounded, thick-walled, with a small ocular chamber. Ascospores 20–24 × 3–5 µm (\( \bar{x} \) = 21.7 × 4.3 µm, n = 25), overlapping 1–2-seriate, 3-transversely septate, hyaline, fusiform with rounded ends, second cell from the apex swollen, all septa with black dots at ends, straight, smooth-walled, surrounded by a sheath. Asexual morph Undetermined.
Material examined: ITALY, Province of Forlì-Cesena, Strada San Zeno-Galeata, on living branch of Lonicera xylosteum L. (Caprifoliaceae), 23 March 2018, E. Camporesi, IT 3788 (MFLU 18-0915, holotype).
GenBank numbers: ITS = MN756639, LSU = MN756637, SSU = MN756635, TEF1-α = MN756633.
Notes: In multi-gene phylogenetic analyses, Paraloratospora camporesii (strain MFLU 18-0915) has a close phylogenetic relationship with Phaeosphaeria caricicola (CBS 133078) with significant support in BI analysis (67% ML, 1.00 BYPP; Fig. 70) and also group with other Phaeosphaeria sensu lato. Paraloratospora camporesii is similar to Phaeosphaeria caricicola but differs in size range and colour of ascospores. Phaeosphaeria caricicola has smaller ascospores (18–21 × 4 µm) and the ascospores are olivaceous (Shoemaker and Babcock 1989), while, Paraloratospora camporesii has hyaline ascospores. A nucleotide comparison of ITS and TEF1-α shows that our new species differs from Phaeosphaeria caricicola in 39/515 bp (7.6%) and more than 40 over 336 bp (> 11.9%) respectively.
Paraloratospora gahniae (Crous) Thiyagaraja, Bundhun & K.D. Hyde, comb. nov.
Index Fungorum number: IF557006; Facesoffungi number: FoF 06976
≡ Phaeosphaeria gahniae Crous, Persoonia 39: 441 (2017).
Holotype: AUSTRALIA, Victoria, Royal Botanic Gardens Victoria, Melbourne Gardens, on leaves of Gahnia aspera (Cyperaceae), 2 December 2016, P.W. Crous, CBS H-23320, ex-type culture CPC 32454 = CBS 143450.
Description: See Crous et al. (2017).
Notes: Phaeosphaeria gahniae has globose ascomata, bitunicate subcylindrical asci bearing pedicels and medium brown, guttulate ascospores, with 3–4 septa and second cell from apex slightly swollen (Crous et al. 2017). The fungus was isolated from leaves of Gahnia aspera (Cyperaceae). In their phylogenetic analyses, P. gahniae clustered with P. caricicola, P. avenaria f. sp. tritici and P. eustoma (Crous et al. 2017). The type species of Phaeosphaeria, P. oryzae I. Miyake, was not included in the analysis. In our multi-locus phylogenetic analyses, P. gahniae is sister to Phaeosphaeria norfolcia and clusters within Paraloratospora with significant support (79% ML, 1.00 BYPP; Fig. 70). It is thus transferred to Paraloratospora.
Phaeosphaeria I. Miyake
Notes: More than 200 epithets are listed under Phaeosphaeria in Index Fungorum (2020). With the molecular phylogeny, many previously known species were transferred to other phaeosphaeriaceous genera and related families (Quaedvlieg et al. 2013; Ariyawansa et al. 2015a, b; Li et al. 2015; Tennakoon et al. 2016). The most recent treatments for the genus are Phookamsak et al. (2014, 2017, 2019), Hyde et al. (2017, 2019a) and Tibpromma et al. (2017).
Phaeosphaeria chinensis K.K. Zhang et al., in Zhang et al., Phytotaxa 419(1): 32 (2019)
Facesoffungi number: FoF 06230; Fig. 66
Holotype: CHINA, ShenZhen, Tong long shan country park, dead leaves of unknown plant, 20 December 2018 (N 22° 34′ 44.13″ E 114° 0′ 23.20″), Keke Zhang, kk61-2 (SZU-001), ex-type living culture MFLUCC 19-0217, KUMCC 19-0161.
Saprobic on dead stems of Miscanthus gigantius (Poaceae), as raised, black, shiny globular structures on the host surface. Sexual morphAscomata 90–130 μm high, 110–120 μm diam. (\( \bar{x} \) = 109.5 × 115 μm, n = 5), solitary, gregarious, immersed under epidermis, becoming erumpent through host surface, subglobose, or polygonal at sides, uni-loculate, glabrous, ostiolate, with minute papilla; papilla 30–50 μm long (n = 5), carbonaceous, easily broken. Peridium 19–20 μm wide, thin-walled, of equal thickness, composed of few layers of dark brown to brown, pseudoparenchymatous cells of textura angularis. Hamathecium of dense, filamentous, septate, cellular pseudoparaphyses, embedded in a hyaline gelatinous matrix. Asci 45–50 × 10–12 μm (\( \bar{x} \) = 48 × 10.5 μm, n =20), 8-spored, bitunicate, fissitunicate, cylindric-clavate to obclavate, slightly broad at the base with short rounded pedicel, apically rounded. Ascospores 18–21 × 4.7–5.3 μm (\( \bar{x} \) = 20 × 5 μm, n = 40), overlapping 1–2-seriate, hyaline, fusiform, with acute ends, 3-septate, cell near the septum slightly larger, smooth-walled, guttulate. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on PDA within 12 h reaching 20 mm diam. after 7 days at 25 °C, circular, floccose, flat with even margin, initially pale yellow becoming greenish grey; reverse yellowish grey.
Material examined: TAIWAN, Chia Yi, Kwang Hwa, Miscanthus gigantius (Poaceae), 18 March 2018, A. Karunarathna, AKTW 63 (MFLU 18-2675), living culture, FU31023.
Known host and distribution: Phaeosphaeria chinensis was firstly found in ShenZhen, China. The authors were unable to provide the host identity (Zhang et al. 2019a).
GenBank numbers: ITS = MN937237, LSU = MN937219.
Notes: Phaeosphaeria chinensis was introduced by Zhang et al. (2019a). Our strain FU31023 is morphologically similar in all characters with P. chinensis. The similarity is further proved in phylogenetic analysis with high support (99% ML, 1.00 BYPP; Fig. 70). The nucleotide pairwise dissimilarity in ITS region is less than 1.5%. Hence considering the morphological similarities and less pairwise dissimilarity in ITS region, we introduce our strain FU31023 as Phaeosphaeria chinensis.
Phaeosphaeriopsis M.P.S. Câmara, M.E. Palm & A.W. Ramaley
Notes: Phaeosphaeriopsis was introduced by Câmara et al. (2003) to accommodate paraphaeosphaeria-like taxa based on phylogenetic analysis of the 18S rDNA sequence dataset. The genus is typified by P. glaucopunctata (Grev.) M.P.S. Câmara et al. and is characterised by scattered to clustered, gregarious, immersed to erumpent, globose to subglobose, ostiolate, with papillate or apapillate ascomata, brown to dark brown, thin-walled peridium, fissitunicate, broadly cylindrical to cylindric-clavate asci, with broad cellular pseudoparaphyses and pigmented, oblong to cylindrical, septate, smooth- to rough-walled, with punctate or verrucose ascospores (Phookamsak et al. 2014; Thambugala et al. 2014). Asexual morph of Phaeosphaeriopsis has been reported as coelomycetous coniothyrium-like or Phaeostagonospora (Câmara et al. 2003; Phookamsak et al. 2014; Thambugala et al. 2014). The sexual-asexual morph connection of Phaeosphaeriopsis has been reported by Câmara et al. (2003), Phookamsak et al. (2014) and Thambugala et al. (2014). Since 2014, many Phaeosphaeriopsis species were introduced by Crous et al. (2016a, b), Tibpromma et al. (2017) and Marin-Felix et al. (2019a). Seventeen species are accepted in this genus (Index Fungorum 2020). We report the sexual morph of Phaeosphaeriopsis pseudoagavacearum Crous & Y. Marín from Yucca gloriosa in Italy for the first time.
Phaeosphaeriopsis pseudoagavacearum Crous & Y. Marín, in Marin-Felix et al., Stud. Mycol. 94: 63 (2019)
Facesoffungi number: FoF 07196, Fig. 67
Phaeosphaeriopsis pseudoagavacearum (MFLU 17-2800). a Appearance of ascomata on the host surface. b Section through ascoma. c Section through peridium. d Asci embedded in pseudoparaphyses. e, f Asci. g, i Ascospores stained with Indian ink. h Ascospore. Scale bars: a = 500 μm, b = 100 μm, c–f = 20 μm, g = 10 μm, h, i = 5 μm
Holotype: FRANCE, Domaine la Fraysse, Valgorge, on leaves of Agave sp. (Asparagaceae), 15 July 2010, P.W. Crous, CBS H-23873, ex-type living culture, CBS 145370 = CPC 18383.
Saprobic on dead leaves of Yucca gloriosa. Sexual morphAscomata 180–260 μm high, 190–330 µm diam., scattered to clustered, gregarious, immersed, slightly raised to erumpent, visible as black dots on the host surface, globose to subglobose, uni- to bi-loculate, glabrous, ostiole central, with minute papilla. Peridium 15–60 μm wide, thin-walled of unequal thickness, composed of several cell layers, of flattened to broad, pseudoparenchymatous cells, brown to dark brown of textura angularis; thick at the apex (40–60 μm wide), arranged in a textura prismatica. Hamathecium composed of numerous, 1–1.7 μm wide, filamentous, septate, anastomosed pseudoparaphyses, embedded in a hyaline gelatinous matrix. Asci (85–)90–110(–119) × (9–)10–12(–14) μm (\( \bar{x} \) = 99.6 × 11.4 μm, n = 30), 8-spored, bitunicate, fissitunicate, cylindrical to subcylindric-clavate, subsessile to short pedicellate, blunt to rounded apex, with well-developed ocular chamber. Ascospores (18–)20–23(–25) × (4.5–)5–6.5 μm (\( \bar{x} \) = 21.8 × 5.6 μm, n = 30), overlapping 1–2-seriate, brown to dark brown, oblong to subcylindrical, slightly narrower towards the lower cells, 5-septate, not constricted at the septa, enlarged at the 4th cell from above, verruculose, surrounded by a distinct mucilaginous sheath. Asexual morph Undetermined.
Material examined: ITALY, Province of Arezzo [AR], near Stia, on dead land leaves of Yucca gloriosa L. (Asparagaceae), 1 November 2016, E. Camporesi, IT3148 (MFLU 17-2800).
Known host and distribution: Agave sp. (France), Yucca gloriosa (Italy) (Marin-Felix et al. 2019a; this study).
GenBank numbers: ITS = MN750613, LSU = MN750592, SSU = MN750607, TEF1-α = MN756837 (MFLU 17-2800A); ITS = MN750614, LSU = MN750593, SSU = MN750608, TEF1-α = MN756838 (MFLU 17-2800B).
Notes: Based on the NCBI BLASTn search of ITS sequence data, our newly generated sequences (MFLU 17-2800A and MFLU 17-2800B) have a 99.62% similarity with Phaeosphaeriopsis pseudoagavacearum (strain CBS 145370, GenBank no. MK539962), 98.62% similarity with P. obtusispora (Speg.) M.P.S. Câmara (strain CBS 246.64; GenBank no. KY090644) and a 98.04% similarity with P. agavacearum Crous & Thangavel (strain CPC 29122; GenBank no. NR_155689 and KY173430). Our collection morphologically resembles P. obtusispora and P. agavacearum in having 5-septate ascospores, enlarged at the 4th cell and verruculose in P. agavacearum. However, our collection can be distinguished from these two species based on the size range of ascomata, asci and ascospores as well as the host occurrences (Table 1). Phylogenetic analyses of a combined LSU, SSU, ITS and TEF1-α sequence dataset showed that our strains clade with P. pseudoagavacearum (93% ML, 0.99 BYPP; Fig. 70) and also cluster with P. agavacearum, P. obtusispora and P. grevilleae (Fig. 70). We, therefore, identify our collection as P. pseudoagavacearum. However, the morphological characteristics of our collection could not be compared with the type of P. pseudoagavacearum as the type is represented by its asexual morph. Thus, we report the sexual morph of P. pseudoagavacearum collected from Yucca gloriosa in Italy for the first time.
Septoriella Oudem.
Notes: Septoriella is typified by S. phragmitis Oudem. (Crous et al. 2015a). Species have been reported worldwide, from a range of hosts families including Juncaceae, Poaceae and Rosaceae, while Septoriella hirta is noted as an important secondary pathogen on grasses (Sprague 1950; Marin-Felix et al. 2019a; Farr and Rossman 2020). Septoriella is characterised by pycnidial, unilocular conidiomata, that produce subcylindrical to fusiform, euseptate, pale brown conidia with a mucoid appendage at each end (Crous et al. 2015a; Li et al. 2015; Marin-Felix et al. 2019a). In their recent phylogenetic analysis based on ITS and LSU combined data, Marin-Felix et al. (2019a) synonymized the genera Allophaeosphaeria Ariyaw., Camporesi & K.D. Hyde, Poaceicola W.J. Li, and Vagicola Chethana & K.D. Hyde under Septoriella. Marin-Felix et al. (2019a) however, did not take into account the characters of the sexual morphs. We therefore do not follow this treatment of Septoriella pending a more detailed study of Phaeosphaeriaceae.
Septoriella camporesii Goonas. & Hyde, sp. nov.
Index Fungorum number: IF656918; Facesoffungi number: FoF 06804; Fig. 68
Septoriella camporesii (MFLU 15-1259, holotype). a Appearance of conidiomata on the host surface. b Close up of conidiomata. c, d Section through conidiomata. e Pycnidial wall. f Conidia arising from conidiogenous cells. g–n Conidia. o, p Colony characteristics from above (o) and below (p). Scale bars: c, f = 50 μm, d, e, g–o = 20 μm
Etymology: Named in honour of the collector Erio Camporesi.
Holotype: MFLU 15-1259
Saprobic on dead leaf of Anthoxanthum odoratum, appearing as black ellipsoidal, raised spots on the host surface. Sexual morph Undetermined. Asexual morph Coelomycetous. Conidiomata 95–120 µm wide, 80–95 µm high, dark brown to black, pycnidial, solitary to gregarious or scattered, semi-immersed to erumpent, globose, unilocular, glabrous. Pycnidial walls 10–20 µm wide, in the upper and lateral parts, composed of outer dark brown and inner light brown to hyaline cells of textura prismatica, at the base, pale brown to hyaline cells of textura prismatica gradually merging with host tissue. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 2 × 3 µm, hyaline, phialidic, doliiform, discrete, determinate, smooth, arising from the inner layers of conidioma. Conidia 15.3–19.2 × 2.7–3.5 µm (\( \bar{x} \) = 17 × 3.2 μm, n = 30), hyaline when immature, becoming light brown to pale greenish-brown, oblong to subcylindrical, rounded at apex, slightly truncate at base, 1–3-septate at maturity, septa less conspicuous in immature spores, smooth-walled, greenish specks-like globules present in the cell matrix.
Culture Characteristics: Colonies showing moderate growth, circular, reaching 40 mm diam. in 4 weeks at 18–20 °C on PDA, medium dense and woolly, dull surface, convex, margins entire, from above white at the centre, becoming grey at the margins, reverse gray.
Material examined: ITALY, Province of Forlì-Cesena [FC], near Camposonaldo-Santa Sofia, on dead leaf of Anthoxanthum odoratum L. (Poaceae), 17 September 2012, E. Camporesi, IT 724 (MFLU 15-1259, holotype; KUN-HKAS 96289, isotype), ex-type living culture, KUMCC 16-0113.
GenBank numbers: ITS = MN648204, LSU = MN648201, SSU = MN648202.
Notes: Septoriella camporesii shares similar characteristics with other Septoriella species in having fusiform to subcylindrical pale brown, euseptate conidia with rounded ends. However, the conidia in S. camporesii lack distinct mucoid appendages at both ends, which distinguishes it from other species. It is most similar to S. subcylindrospora (W.J. Li et al.) Y. Marín & Crous in having smooth-walled, subcylindrical conidia with a truncate base, but the conidiomata of S. camporesii are semi-immersed to erumpent while in S. subcylindrospora are immersed to semi-immersed. In our phylogeny, S. camporesii groups within the clade represented by the type Septoriella phragmitis (ex-epitype strain CPC 24118). The new species has a close relationship with Loratospora luzulae Jayasiri, Camporesi & K.D. Hyde (MFLUCC 14-0826) and Septoriella germanica Crous, R.K. Schumach. & Y. Marín (CBS 145372), but with low bootstrap support (62% ML, 0.90 BYPP; Fig. 70). Loratospora luzulae needs to be revisited in future study.
Wojnowiciella Crous, Hern.-Restr. & M.J. Wingf., Persoonia 34: 201 (2015)
Notes: Wojnowiciella was introduced by Crous et al. (2015c) to accommodate W. eucalypti Crous, Hern.-Restr. & M.J. Wingf. as the type species. The members of Wojnowiciella are characterised in having non-papillate conidiomata lacking setae and having dark brown conidia (Crous et al. 2015c). Wojnowiciella species are probably cosmopolitan in distribution, since they have been recorded from both temperate and tropical countries (i.e. Australia, China, Colombia, Italy, South Africa) (Liu et al. 2015; Crous et al. 2015c, 2016b; Hernandez-Restrepo et al. 2016; Phookamsak et al. 2019). There are nine epithets for Wojnowiciella in Index Fungorum (2020). We follow the latest treatment and updated account of Wojnowiciella in Phookamsak et al. (2019). Wojnowiciella dactylidis is reported as a new host record from Galium sp. (Rubiaceae).
Wojnowiciella dactylidis (Wijayaw., Camporesi & K.D. Hyde) Hern.-Restr. & Crous, Sydowia 68: 221 (2016)
Facesoffungi number: FoF 00417; Fig. 69
Wojnowiciella dactylidis (MFLU 16-1366). a Appearance of conidiomata on host. b Close-up of conidioma. c, d Section of conidiomata. e Pycnidial wall. f Developing conidia attached to conidiogenous cell. g–k Conidia. l Germinating conidium. m Colony from above. n Colony from below. Scale bars: c, d = 75 µm, e = 10 µm, f = 20 µm, g–l = 20 µm
≡Wojnowicia dactylidis Wijayaw., Camporesi & K.D. Hyde, in Liu et al., Fungal Diversity: https://doi.org/10.1007/s13225-015-0324-y, [144] (2015)
Holotype: ITALY, Province of Forlì-Cesena, Teodorano, Meldola, on dead branch of Dactylis glomerata L. (Poaceae), 7 March 2013, E. Camporesi, NNW IT1101 (MFLU 14–0745), ex-type living culture, MFLUCC 13-0735, GUCC9.
Saprobic on dead stem of Galium sp. Sexual morph Undetermined. Asexual morphConidiomata 195–220 µm diam., 210–250 µm high, pycnidial, semi-immersed in the host tissue, scattered, solitary, non-papillate, brown. Pycnidial walls 22–28 µm thick, composed of several layers of light brown to dark brown cells, arranged in textura angularis to textura prismatica. Conidiophores absent. Conidiogenous cells formed on the inner layer of conidiomata, ampulliform, phialidic, smooth, pale brown. Conidia 30–42 × 5–7 µm (\( \bar{x} \) = 35.8 × 6.2 µm, n = 20), cylindrical, straight to slightly curved, gradually tapering towards the rounded apex, with rounded apex, initially hyaline to pale brown, after maturity golden brown, 7–10-euseptate, thin-walled, with pad-like appendages at the apex.
Culture characteristics: Colonies on PDA reaching 8 mm diam. after 2 weeks at 25–30 °C, colonies medium dense, circular, convex, surface slightly rough with edge entire, effuse, velvety to hairy, margin well-defined, colony from above: white to grayish at the margin, white at the centre reverse, white to grayish at the margin, white to greyish at the centre, mycelium white to greyish with tufting, not producing pigments in PDA.
Material examined: ITALY, Province of Forlì-Cesena [FC], Berleta - Santa Sofia, on dead aerial stem of Galium sp. (Rubiaceae), 17 May 2015, E. Camporesi, IT 2487 (MFLU 16-1366, HKAS93706), living Culture KUMCC 15-0560.
Known hosts and distribution: Dactylis glomerata L. (Poaceae) (Liu et al. 2015).
GenBank numbers: ITS = MN750612, LSU = MN750591, SSU = MN750606.
Notes: As morphological characters examined largely overlap with Wojnowiciella dactylidis (Wijayaw., Camporesi & K.D. Hyde) Hern.-Restr. & Crous, we therefore report our collection as a new host record of W. dactylidis from dead aerial stem of Galium sp. (Rubiaceae). Both collections share similar morphological characters, viz. semi-immersed, scattered, solitary conidiomata and pale brown, cylindrical, 7–10-septate conidia (Liu et al. 2015). Wojnowiciella dactylidis was initially introduced by Liu et al. (2015) as Wojnowicia dactylidis Wijayaw., Camporesi & K.D. Hyde, based on both morphology and phylogeny. Some Wojnowicia species (W. dactylidis, W. lonicerae Wijayaw., Camporesi & K.D. Hyde and W. spartii W.J. Li, Camporesi & K.D. Hyde) were transferred to Wojnowiciella by Hernández-Restrepo et al. (2016). Combined multi-gene (LSU, SSU, ITS and TEF1-α) based phylogenies also show that our collection clusters with other Wojnowiciella species, in particular W. dactylidis (MFLUCC 13-0735) with high support (89% ML, 0.99 BYPP; Fig. 70).
Phylogenetic tree generated from maximum likelihood (ML) analysis based on combined LSU, SSU, ITS and TEF1-α sequence dataset for the species from Phaeosphaeriaceae. Leptosphaeria doliolum (CBS 505.75) and Paraleptosphaeria dryadis (CBS 643.86) were used as the outgroup taxa. The dataset comprised 2977 characters including gaps (SSU: 1–954; LSU: 955–1760; TEF1-α: 1761–2383 and ITS: 2384–2977). The RAxML analysis of the combined dataset yielded a best scoring tree with a final ML optimization likelihood value of − 30528.834355. The matrix had 1084 distinct alignment patterns, with 29.72% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.247078, C = 0.227200, G = 0.267903, T = 0.257819; substitution rates AC = 1.221599, AG = 3.454967, AT = 2.544350, CG = 0.869235, CT = 7.100206, GT = 1.000000; gamma distribution shape parameter α = 0.665522. Support values for maximum likelihood (ML) higher than 75% and Bayesian posterior probabilities (BYPP) greater than 0.95 are given at the nodes
Tetraplosphaeriaceae Kaz. Tanaka & K. Hiray.
Notes: The family Tetraplosphaeriaceae was introduced based on dematiaceous hyphomycetes, a tetraploa-like asexual morph having four elongated setose appendages from a four lobed, verrucose columnar of the conidial body and is typified by Tetraplosphaeria Kaz. Tanaka & K. Hiray. Species of Tetraplosphaeriaceae have massarina-like sexual morph with hyaline 1(–3)-septate ascospores and/or tetraploa-like asexual morph with several setose appendages (Tanaka et al. 2009; Hyde et al. 2013). Following the reappraisal by Hyde et al. (2013), Tetraplosphaeria was treated as the synonym of Tetraploa. It has been predominantly recorded on bamboo worldwide. Tetraplosphaeriaceae was established with the inclusion of five new genera, Polyplosphaeria Kaz. Tanaka & K. Hiray., Pseudotetraploa Kaz. Tanaka & K. Hiray., Quadricrura Kaz. Tanaka, K. Hiray. & Sat. Hatak. Tetraplosphaeria and Triplosphaeria Kaz. Tanaka & K. Hiray., with their tetraploa-like hyphomycetous asexual morphs (Tanaka et al. 2009; Tibpromma et al. 2018; Wijayawardene et al. 2018a).
This study also supports the monophyly of the family and that of each genus as established earlier and expands the overall intergeneric relationships of Tetrplosphaeriaceae into seven well-supported generic lineages. In this study we introduce five new species in Tetraplosphaeriaceae based on fresh specimens, morphology and phylogenetic analyses of a combined ITS, LSU and TUB2 sequence data. Ernakulamia tanakae, Pseudotetraploa rajmachiensis, Tetraploa dwibahubeeja, T. pseudoaristata and T. thrayabahubeeja are introduced from India. Furthermore, the sexual morph of Tetraploa sasicola is reported on Pennisetum purpureum from Taiwan.
Ernakulamia Subram.
Notes: Ernakulamia was established by Subramanian (1994) for a fungus he earlier recognized as Petrakia cochinensis Subram. (Subramanian 1957). Ellis (1976) recombined Petrakia cochinensis to Piricauda cochinensis that was further reinstated as Ernakulamia cochinensis (Subram.) Subram. Two species of Ernakulamia have been reported, E. cochinensis (on Cocos nucifera from India) and E. krabiensis Jayasiri, E.B.G. Jones & K. D. Hyde (on Acacia sp. from Thailand) (Jayasiri et al. 2019). Delgado et al. (2017) analysed DNA sequence data from the ITS and TUB2 gene and proposed that E. cochinensis (collected on Astrocaryum standleyanum (Arecacaceae) from Panama) should be assigned to Tetraploapshaeriaceae (Pleosporales). Ernakulamia tanakae is the third addition to Ernakulmia. Ernakulamia is included in Tetraploaspaheriaceae allied to Polyplosphaeria.
Ernakulamia tanakae Rajeshkumar & K.D. Hyde, sp. nov.
Index Fungorum number: IF556700; Facesoffungi number: FoF 06338; Fig. 71
Etymology: Named after Prof. Kazuki Tanaka, for his contribution to Tetraplospheriaceae.
Holotype: AMH 10131
Saprobic on dead spathes of Cocos nucifera, dark brown. Sexual morph Undetermined. Asexual morphMycelia superficial or semi-immersed. Conidiophores absent. Conidigenous cells monotretic, integrated, terminal or intercalary, short, globose to subglobose or clavate, 8.2–9.3 × 6.2–8.5 µm. Conidia variable in shape, obconical to irregular, dark brown, thick-walled, dictyosporous, verruculose at basal part around attachment, 30–53 × 22–50 µm, with (4–)6–13-septate appendages. Conidial appendages 30–87 × 3–4.5 µm, apical or subapical, conidial base 7–15 µm.
Culture characteristics: Colonies on MEA at 25 ± 2 °C, slow growing, 9–12 mm diam. after 7 days; 38–55 mm diam. after 30 days, raised at centre, yellowish brown (5E5) to greyish brown (5F3). Outer mycelia velutinous, pale brown to grey (5F1). Colony margin irregular. Colony reverse grey (5F1) brownish grey (5F2) with white margin. Soluble pigments and exudates are absent. Colour codes and names used in descriptions are from Kornerup and Wanscher (1978).
Material examined: INDIA, Kerala, Kasaragod (12° 30′ 29″ N, 74° 59′ 02″ E, ± 50 msl.), on decaying spathe of Cocos nucifera (Arecacceae), July 2016, K. C. Rajeshkumar, AMH 10131(holotype), ex-type living culture NFCCI 4615; additional strains NFCCI 4616, NFCCI 4617.
GenBank numbers: ITS = MN937229, LSU = MN937211, TUB2 = MN938312 (NFCCI 4615); ITS = MN937227, LSU = MN937209, TUB2 = MN938310 (NFCCI 4616); ITS = MN937228, LSU = MN937210, TUB2 = MN938311 (NFCCI 4617).
Notes: In the phylogenetic analysis (Fig. 77), Ernakulamia tanakae is in a distinct lineage with a high support (95% ML) and sister to Polyplosphaeria xishuangbannaensis Tibpromma & K.D. Hyde (KUMCC 17-0187), Ernakulamia cochinensis (CCF 5738) and E. krabiensis (C372A). Present study distinguishes the muriform septations mentioned in the type description of E. cochinensis appears to be dubious and the descriptions in Ellis (1976) and Seifert et al. (2011) as dictyosporous are more appropriate. SEM studies revealed the surface texture was smooth with a thin peel-like outer wall of conidia, and micrographs show the presence of numerous internal hyphae. Ernakulamia tanakae closely resembles E. cochinensis and was also isolated from spathes of Cocos nucifera collected from Malabar region of Kerala, India. However, the conidia of E. tanakae are shorter than those of E. cochinensis (30–53 µm versus 30–70 µm). Ernakulamia krabiensis has shorter appendages than E. tanakae.
Pseudotetraploa Kaz. Tanaka & K. Hiray.
Notes: Pseudotetraploa, typified by P. curviappendiculata Kaz. Tanaka & K. Hirayama (Tanaka et al. 2009) contains three valid species: P. curviappendiculata, P. javanica (Rifai, Zainuddin & Cholil) Kaz. Tanaka & K. Hiray. and P. longissima (Sat. Hatak., Kaz. Tanaka & Y. Harada) Kaz. Tanaka & K. Hiray. (Index Fungorum 2020). These three species were previously described as Tetraploa (Hatakeyama et al. 2005), which were recombined to Pseudotetraploa, due to their unique characteristics such as distoseptate, two to three columnar, obpyriform conidial body and more curved and shorter setose appendages.
Pseudotetraploa rajmachiensis Rajeshkumar, K.D. Hyde & S. Lad, sp. nov.
Index Fungorum number: IF556701; Facesoffungi number: FoF 06340; Fig. 72
Etymology: Named after the place of collection in Rajmachi, Maharashtra, India
Holotype: AMH 10132
Saprobic superficial on dead branches. Sexual morph Undetermined. Asexual morphConidiophores absent. Conidiogenous cells micronematous, mononematous, monoblastic, integrated, usually undistinguishable from superficial hyphae. Conidia 2 to 3 columnar, pseudoseptate, 27.6–43 × 16–23.5 µm., pale brown to dark brown, ovoid to obclavate or obpyriform, almost smooth and minutely verruculose at base, 8.5–10.5 µm. Setose appendages at the apical part, appendages two in number, 16.5–79 µm long, pale brown at base 4.5–6 µm with a hyaline apex 2.5–5 µm., smooth, unbranched, straight or slightly curved, 4–10-septate, longer appendage is straight and shorter appendage is curved.
Culture characteristics: Colonies on MEA at 25 ± 2 °C, slow growing, 30–35 mm diam. after 30 days, floccose, centre yellowish brown (5E5) to light brown (5D4) forming two concentric zonations, margin regular, brownish grey (5D3). Colony reverse yellowish brown (5F4) to greyish brown (5F3) with grey to white margin (5B1–5A1). Soluble pigments and exudates are absent. Colour codes and names used in descriptions are from Kornerup and Wanscher (1978).
Material examined: INDIA, Maharashtra, Maval, Rajmachi (18° 49′ 29″ N, 73° 23′ 54″ E, ± 820 msl.), on decaying bamboo culms, Dendrocalamus stocksii (Poaceae), August 2016, K. C. Rajeshkumar, AMH 10132 (holotype), ex-type culture NFCCI 4618; additional strains NFCCI 4619, NFCCI 4620.
GenBank numbers: ITS = MN937222, LSU = MN937204, TUB2 = MN938305 (NFCCI 4618); ITS = MN937221, LSU = MN937203, TUB2 = MN938304 (NFCCI 4619); ITS = MN937223, LSU = MN937205, TUB2 = MN938306 (NFCCI 4620).
Notes: Pseudotetraploa rajmachiensis is a new species and first record of this genus from India. The species clusters with P. javanica (HC 4934) and P. curviappendiculata (HHUF 28582 and CBS 125426) with a statistical support of (100% ML; Fig. 77) in Pseudotetraploa. The new species differs from P. curviappendiculata in having smaller conidia and longer dimorphic (long and short) appendages. A synopsis of species of Pseudotetraploa is provided in Table 2.
Tetraploa Berk. & Broome
Notes: Tetraploa was erected by Berkeley and Broome (1850) to accommodate T. aristata. The genus is characterised by monoblastic, reduced conidiogenous cells, brown, euseptate, short cylindrical with 4 columnar conidial body, coarsely verrucose base having 4 prominent setose appendages at the apex. Tanaka et al. (2009) redefined Tetraploa as Tetraploasphaeria based on its massarina-like sexual morph and Tetraploasensu stricto asexual morphs. Hyde et al. (2013) resurrected and validated the genus name as Tetraploa (Wijayawardene et al. 2018a). Twenty Tetraploa species are recorded in the Index Fungorum (2020).
Tetraploa dwibahubeeja Rajeshkumar, K.D. Hyde & S. Lad, sp. nov.
Index Fungorum number: IF556702; Facesoffungi number: FoF 06341; Fig. 73
Etymology: Named after the emended genus epithet Dwibahubeeja; in Sanskrit means two-armed seed/conidium.
Holotype: AMH 10133
Saprobic on decaying spathes of Cocos nucifera inflorescence, yellowish brown to dark brownish. Sexual morph Undetermined. Asexual morphMycelia branched, semi-immersed or immersed, dark brown, euseptate. Conidiophores reduced to conidiogenous cells. Conidiogenous cells micronematous, mononematous, integrated, determinate, monoblastic, cylindrical, 3–5 µm. Conidia solitary, dry, acrogenous, smooth-walled, septate, rhexolytic, V-shaped, mostly bifurcate, rarely with a rudimentary tertiary arm, 80–406 × 3–6.25 µm, conidial base 10–12 µm wide, arms elongated, tip obtuse, cylindrical narrowing toward tip, up to 38 septa, apex obtuse.
Culture characteristics: Colonies on MEA at 25 ± 2 °C, 17–19 mm diam. after 7 days; 40–60 mm diam. after 30 days, centrally umbonate, brownish grey (6E2) to grey (6E1). Middle part mycelia velutinous, light brown (6D4) to greying brown (6D3), marginal mycelia raised brownish grey (6E2) to grey (6E1). Colony reverse dark brown at centre (6F8) brownish orange (6C8) towards margin. Soluble pigments and exudates are absent.
Material examined: INDIA, Maharashtra, Thane (19° 13′ 13″ N, 72° 57′ 17″ E, ± 23 msl.), on decaying spathes of Cocos nucifera (Arecacceae), July 2016, K.C. Rajeshkumar & Sneha Lad, AMH 10133 (holotype), ex-type living culture NFCCI 4621; additional strains NFCCI 4622, NFCCI 4623.
GenBank numbers: ITS = MN937226, LSU = MN937208, TUB2 = MN938309 (NFCCI 4621); ITS = MN937224, LSU = MN937206, TUB2 = MN938307 (NFCCI 4622); ITS = MN937225, LSU = MN937207, TUB2 = MN938308 (NFCCI 4623).
Notes: Dwibahubeeja, typified by D. indica N. Srivatava, A. K. Srivastava & Kamal, is a hypopodiate hyphomycete with bifurcate, two armed conidia in which basal cells of both arms are consistently fused (Srivastava et al. 1995). This genus was first identified on Calamus tenuis (Arecaceae), Barhani, Uttar Pradesh, India. In the present study, a morphologically similar species of Dwibahubeeja having longer conidial length and more septa was subjected to single conidial culturing and analysed phylogenetically. Phylogentic analyses based on concatenated ITS, LSU and TUB2 sequence data depict Tetraploa dwibahubeeja as sister taxon to T. sasicola (Kaz. Tanaka & K. Hiray.) Kaz. Tanaka & K. Hiray (AKTW 21, HHUF27566) with high support in Tetraploa (93% ML, 1.00 BYPP; Fig. 77). Hence, we include a new species in Tetraploa, with the bifurcate, two armed basally fused feature. The new species T. dwibahubeeja has longer conidia with more septa, up to 38 septa in the conidial arm when compared to the type species of Dwibahubeeja indica. Micrographs and SEM studies revealed that the basal cells in mature conidia are verruculose and having a rounded base.
Tetraploa pseudoaristata Rajeshkumar, K.D. Hyde & G. Anand, sp. nov.
Index Fungorum number: IF556703; Facesoffungi number: FoF 06339; Fig. 74
Etymology: The specific epithet “pseudoaristata” refers to ‘similar to Tetraploa aristata’ having quadriaristate morphology of conidia
Holotype: AMH 10134
Saprobic on decaying spathe of Cocos nucifera. Sexual morph Undetermined. Asexual morphMycelia dark brown to black, effuse, branched, septate, partly superficial. Setae or hypopodia absent. Conidiophores absent. Conidiogenous cells miconematous, dark brown, monoblastic, integrated, short, usually undistinguishable from superficial hyphae. Conidia short cylindrical, conidial body four columnar, coarsely verruculose, 22–31 × 15–20 µm, with four apical setose appendages, 23–107.5 × 2.4–5.2 µm, partly splitted conidia, where the two columns of the upper part of the main conidial body is totally splitted along with the two apical appendage but connected by the base.
Culture characteristics: Colonies on MEA at 25 ± 2 °C, 15–18 mm diam. after 7 days, velutinous to floccose, greenish grey (27F2) in the centre and to dull green towards margin (27E3), marnial mycelia white (27A1), thin, regular. Colony reverse grey (4F1) to brownish grey (4F2). Soluble pigments and exudates are absent.
Material examined: INDIA, Maharashtra, Thane (19° 13′ 13″ N, 72° 57′ 17″ E, ± 23 msl.), on decaying spathe of Cocos nucifera (Arecacceae), July 2016, K.C. Rajeshkumar & Sneha Lad, AMH 10134 (holotype), ex-type living culture NFCCI 4624; additional strains NFCCI 4625, NFCCI 4626.
GenBank numbers: ITS = MN937232, LSU = MN937214, TUB2 = MN938315 (NFCCI 4624); ITS = MN937230, LSU = MN937212, TUB2 = MN938313 (NFCCI 4625); ITS = MN937231, LSU = MN937213, TUB2 = MN938314 (NFCCI 4626).
Notes: Based on the concatenated phylogenetic analyses Tetraploa pseudoaristata aligned close to T. aristata (CBS 996.70) forming a well-supported adjacent clade with significant statistical support (100% ML, 1.00 BYPP; Fig. 77). The conidial morphology of new species is similar to that of T. aristata in having quadriaristate conidial body with four, apical, setose appendages. However, the conidium body in the new species (22–31 × 15–20 µm) is smaller than T. aristata (30–40 × 12–30 µm; MycoBank 148113). Tetraploa pseudoaristata is considered to be a cryptic species within Tetraploa having morphological similarity to T. aristata.
In the NCBI BLASTn search of ITS sequence, Tetraploa pseudoaristata (NFCCI 4624, NFCCI 4625, NFCCI 4626) has the highest similarity to Tetraploa yakushimensis (Kaz. Tanaka, K. Hiray. & Hosoya) Kaz. Tanaka & K. Hiray. (GenBank NR_119405.1; Identities = 472/506 (93%), with 12 gaps (2%)), T. yakushimensis (GenBank MH863690.1; Identities = 443/477 (93%), with 12 gaps (2%)), T. sasicola (GenBank NR_119404.1; Identities = 456/504 (90%), with 17 gaps (3%)) and Pseudotetraploa longissima (Sat. Hatak., Kaz. Tanaka & Y. Harada) Kaz. Tanaka & K. Hirayama (GenBank NR_119399.1; Identities = 442/502 (88%), with 20 gaps (3%)). The closest hits using the TUB2 sequence are Tetraplosphaeria yakushimensis (GenBank AB524870.1; Identities = 351/396 (89%), with 3 gaps (0%)), Tetraplosphaeria sasicola (GenBank AB524869.1; Identities = 337/394 (86%), with 7 gaps (1%)), and Pseudotetraploa curviappendiculata (GenBank AB524854.1; Identities = 295/359 (82%), with 10 gaps (2%)).
Tetraploa thrayabahubeeja Rajeshkumar, K.D. Hyde, G. Anand, sp. nov.
Index Fungorum number: IF556704; Facesoffungi number: FoF 06342; Fig. 75
Etymology: Named after the distinguishing 3 branched conidial body; thrayabahubeeja in Sanskrit means 3-armed seed/conidium
Holotype: AMH 10135
Saprobic on decaying spathe of Cocos nucifera. Sexual morph Undetermined. Asexual morphMycelia superficial or immersed. Conidiophores reduced to conidiogenous cells. Conidiogenous cells mononematous, integrated, short, monoblastic. Conidial body have 3 columns or rarely one with one apical appendage, euseptate, 20–32 × 15–20 µm, pale brown, smooth. Setose appendages at the apical part, appendages usually three in number rarely one or two, 23.5–46.5 µm long, pale brown at base 3.5–5.2 µm with a hyaline apex 2–3 µm, smooth, unbranched, straight, 2–4-septate.
Culture characteristics: Colonies on MEA at 25 ± 2 °C, 12–15 mm diam. after 7 days; 28–30 mm diam. after 15 days, velutinous, greyish brown (5E3) to yellowish brown (5E4). Colonies margin regular, grey (5D1). Colony reverse dark grey (5F1) with white (5A1) thin margin. Soluble pigments and exudates are absent.
Material examined: INDIA, Maharashtra, Thane (19° 13′ 13″ N, 72° 57′ 17″ E, ± 23 msl.), on decaying spathe of Cocos nucifera (Arecacceae), July 2016, K.C. Rajeshkumar & Sneha Lad, AMH 10135 (holotype), ex-type living culture, NFCCI 4627; additional strains NFCCI 4628, NFCCI 4629.
GenBank numbers: ITS = MN937235, LSU = MN937217, TUB2 = MN938318 (NFCCI 4627); ITS = MN937233, LSU = MN937215, TUB2 = MN938316 (NFCCI 4628); ITS = MN937234, LSU = MN937216, TUB2 = MN938317 (NFCCI 4629).
Notes: The phylogenetic analyses (Fig. 77) delineated Tetraploa thrayabahubeeja as an independent lineage close to other Tetraploa species within the Tetraploa clade with high support (98% ML, 1.00 BYPP). The morphology of the conidia in T. thrayabahubeeja is unique, having mostly 3 conidial body columns and three apical appendages. The conidial ornamentation is smooth to minutely roughened or verruculose. In most of the species of Tetraploa, the conidium body is coarsely verrucose and the ornamentation is reported as a prominent distinguishing character of the genus.
Tetraploa sasicola (Kaz. Tanaka & K. Hiray.) Kaz. Tanaka & K. Hiray., Fungal Diversity 63: 253 (2013)
Facesoffungi number: FoF07232; Fig. 76
Tetraploa sasicola (MFLU 19-2698) a, b Appearance of ascomata on the host (Pennisetum purpureum) (arrowhead in a). c Section through ascoma. d Section through ostiole. e Section through peridium. f Pseudoparaphyses. g–i Different developing stages of the asci. j–n Ascospores. o Germinating ascospore. Scale bars: c–e = 50 µm, f–k = 10 µm
≡ Tetraplosphaeria sasicola Kaz. Tanaka & K. Hiray., in Tanaka, Hirayama, Yonezawa, Hatakeyama, Harada, Sano, Shirouzu & Hosoya, Stud. Mycol. 64: 180 (2009)
Holotype: Japan, Hokkaido, Yoichi, Sawamachi (140° 46′ E, 43° 11′ N), on culms of Sasa senanensis, 7 July 2001, K. Tanaka, HHUF 27566, ex-type living culture KTC 563 (JCM 13167 = MAFF 239677).
Saprobic on dead leaves of Pennisetum purpureum (Poaceae), as raised, black, shiny globular structures on the host surface. Sexual morphAscomata 185–189 μm high, 186–188 μm diam. (\( \bar{x} \) = 187 × 187 μm, n = 5), solitary, gregarious, immersed under epidermis, becoming erumpent through host surface, subglobose, uni-loculate, glabrous, ostiolate, with minute papilla. Peridium 14–15 μm wide, thin-walled, of equal thickness, composed of few layers of dark brown to brown, pseudoparenchymatous cells of textura angularis. Hamathecium of dense, filamentous, septate, cellular pseudoparaphyses, embedded in a hyaline gelatinous matrix. Asci 97–109 × 22–23 μm (\( \bar{x} \) = 103 × 22.5 μm, n =20), 8-spored, bitunicate, clavate to obclavate, slightly broad at the upper half with short rounded pedicel, apically rounded. Ascospores 24–29 × 4–14 μm (\( \bar{x} \) = 26.5 × 9 μm, n = 40), overlapping 1-seriate, hyaline to pale yellowish brown, fusiform, with acute ends, 1-septate, near the septum slightly larger, smooth-walled, guttulate. Asexual morph Undetermined.
Culture characteristics: Spores germinated on PDA within 24 h. Spores germinated at both ends. Colonies on PDA, velvety, raised, gray, entire margin. Reddish to gray in reverse with pigmentation on PDA.
Material examined: TAIWAN, Chia Yi, Kwang Hwa, Pennisetum purpureum (Poaceae), 29 March 2018, A. Karunarathna, AKTW 21 (MFLU 19-2698), living culture, NCYUCC 19-0371.
Host and distribution: Sasa senanensis from Japan (Tanaka et al. 2009).
GenBank numbers: ITS = MN937236, LSU = MN937218.
Notes: Our strain is similar to Tetraploa sasicola with respect to all morphological characters. The strain is also phylogenetically associated with T. sasicola with very high support (100% ML, 1.00 BYPP; Fig. 77). Tetraploa sasicola was previously known as Tetraplosphaeria sasicola which was recorded on Sasa senanensis from Japan. Our strain is recorded on Pennisetum purpureum from Taiwan.
Phylogram generated from maximum likelihood analysis based on a combined LSU, ITS and TUB2 sequence dataset of taxa in Tetraplosphaeriaceae. Seventy strains are included in the combined sequence analyses. Cryptoclypeus ryukyuensis (HHUF 30509) and Lophiotrema nucula (CBS 627.86) were used as the outgroup taxa. Bootstrap support values for ML equal to or greater than 60% and Bayesian posterior probabilities equal to or greater than 0.90 BYPP are indicated at the nodes. Newly generated sequences are in red
Torulaceae Corda
Notes: We follow the latest treatment and updated accounts of Torulaceae in Su et al. (2016, 2018), Hyde et al. (2017, 2019a) and Li et al. (2017b).
Torula Pers.
Notes: Torula was introduced by Persoon (1795) and is typified by T. herbarum (Pers.) Link. The concept of Torula is restricted to the asexual morph which is characterised by dark brown to black colonies, effuse and velvety on host that the conidiophores are mostly reduced to conidiogenous cells, or with one brown supporting cell. Conidiogenous cells are terminal or lateral, monoblastic or polyblastic, with a thickened and heavily melanized wall on the base, thin-walled and frequently collapsing, becoming coronate on the apex, cupulate, brown, smooth to verruculose. Conidia are acrogenous, in branched chains, pale brown to dark brown, smooth to verrucose and each cell is globose to subglobose (Su et al. 2016; Li et al. 2017b). Currently 539 epithets are listed for Torula in Index Fungorum (2020). However, only 15 species have had their DNA sequence data analysed to reveal their phylogenetic placements in Torulaceae (Crous et al. 2015a; Su et al. 2016, 2018; Li et al. 2017b; Hyde et al. 2017, 2019a; Tibpromma et al. 2017). We introduce a novel species Torula camporesii from decaying herbaceous plant in Yunnan, China and a new host record on Malus sp. of Torula gaodangensis J. Yang & K.D. Hyde also reported from Yunnan, China for the first time.
Torula camporesii Phookamsak, E.F. Yang & K.D. Hyde, sp. nov.
Index Fungorum number: IF557230; Facesoffungi number: FoF 07198; Fig. 78
Etymology: Named in honor of Erio Camporesi who made an immense contribution to fungal investigation and collection.
Holotype: MFLU 20-0070
Saprobic on herbaceous litter. Sexual morph Undetermined. Asexual morphColonies visible as black dense, velvety, powdery on host. Mycelium 1.5–3.5 μm in wide, partly immersed to superficial on the substrate, composed of septate, branched, light brown hyphae. Conidiophores 9–18.5 × 3–5 μm (\( \bar{x} \) = 14 × 4 μm, n = 50), macronematous to semi- macronematous, mononematous, erect, straight, or slightly flexuous, without apical branches, light brown to brown, ellipsoid to subcylindrical, 0–1-septate, smooth-walled, with 1–3, globlose to subglobse cells, arising from prostrate hyphae. Conidiogenous cells 5.5–8.5 × 5–8 μm (\( \bar{x} \) = 7 × 6.5 μm, n = 30), mono- to polyblastic, integrated, terminal or intercalary, cupulate to globose or subglobose, brown to dark brown, smooth to verrucose thick-walled. Conidia (34–)40–70(–77) × 6–8(–10) μm (\( \bar{x} \) = 53.9 × 6.9 μm, n = 50), catenated, composed of moniliform cells, acrogenous, phragmosporous to scolecosporous, brown to dark brown, verruculose, wider cells in the middle, often smaller at apex, 4–20-septate, constricted at the septa, chiefly subcylindrical, produced branched chain, globlose to subglobose of each cell, basal cell slightly truncate, with a darkened scar at the base. Conidial secession schizolytic.
Culture characteristics: Colones on PDA, reaching around 40 mm diam., after 2 weeks at room temperature (20–25 °C), colonies medium dense, irregular, slightly effuse, rough, with standing tufts at the centre, umbonate edge, colonies from above, white to cream at the margin, pinkish white to white-brown at the centre, gradually turn to brown when mature, from below white to pink-white at the margin, irregular radiating with pink-brown at the middle, reddish brown to pinkish brown at the centre; not producing pigment in PDA.
Material examined: CHINA, Yunnan Province, Xishaungbanna, Mengla Country, Bubeng Field Station, Xishuangbanna Station for Tropical Rain Forest Ecosystem Studies (101° 35′ 4″ E, 21° 36′ 4″ N, ± 683 msl.), on decaying herbaceous plant, 7 May 2018, R. Phookamsak, XB001 (MFLU 20-0070, holotype), ex-type living culture, KUMCC 19-0112.
GenBank numbers: ITS = MN507400, LSU = MN507402, SSU = MN507401, RPB2 = MN507404, TEF1-α = MN507403.
Notes: Multi-gene phylogenetic analyses of a combined LSU, SSU, TEF1-α, RPB2 and ITS sequence dataset indicated that Torula camporesii forms a sister lineage with T. goaensis Pratibha & Prabhugaonkar in Torula with high support (100% ML, 1.00 BYPP; Fig. 80). Torula camporesii resembles T. goaensis in having phragmosporous to scolecosporous, brown, verruculose, 4–20-septate conidia. However, the species can be distinguished from T. goaensis in having shorter conidial chains (T. camporesii, (34–)40–70(–77) × 6–8(–10) versus 42–105 × 6–8.5 μm, T. goaensis) (Pratibha and Prabhugaonkar 2017). Torula camporesii has conidia that are brown to dark brown, long subcylindrical to ellipsoidal in chains, wider cells in the middle, often smaller towards the end cell, basal cell slightly truncate, with a darkened scar at the base. Torula goaensis has light brown to brown conidia, obtuse at the basal cell, and without a darkened scar at the base (Pratibha and Prabhugaonkar 2017). In NCBI BLASTn search based on ITS sequences, the closest match of T. camporesii is T. goaensis (MTCC 12620; GenBank no. NR_159045) with 97.14% similarity. A comparison of ITS nucleotide bases indicate that T. camporesii differs from T. goaensis in 15/522 bp (2.87%). Therefore, T. camporesii is introduced as a novel species based on the justification guildelines of Jeewon and Hyde (2016).
Torula gaodangensis J. Yang & K.D. Hyde, Fungal Diversity 87: 113 (2017)
Facesoffungi number: FoF 03288; Fig. 79
Holotype: CHINA, Guizhou Province, Anshun city, Gaodang village, 26° 4′ 16″ N, 105° 41′ 53″ E, on decaying wood submerged in Suoluo River, 19 October 2016, J. Yang, GD 24-3 (MFLU 17-0858); isotype: GZAAS 17-0011, ex-type living cultures MFLUCC 17-0234, GZCC 17-0051.
Saprobic on Malus sp. Sexual morph Undetermined. Asexual morphColonies immersed to superficial on host substrate, dark brown to black, sparse, difficult to see on host. Mycelium 0.8–1.5 μm wide, comprised branched, smooth, hyaline to pale brown hyphae. Conidiophores 1–4 × 1–2.5 μm (\( \bar{x} \) = 2 × 1.6 μm, n = 30) macronematous, mononematous, erect, straight or flexuous, pale brown to brown, ellipsoidal to ampulliform, or oblong, aseptate, unbranched, smooth and thin-walled, with a darkened scar at the apex connecting with conidiogenous cells. Conidiogenous cells 1–3 × 2–3 μm (\( \bar{x} \) = 2.4 × 2.8 μm, n = 30), monoblastic-acropetal, integrated, terminal, inflated, subglobose to oblate-sphaeroid, with flattened base, collapsing at apex, dark brown, verruculose. Conidia (4.5–)6–11(–12) × 2.5–3 μm (\( \bar{x} \) = 9.5 × 2.8 μm, n = 30), acrogenous, brown to dark brown, subcylindrical or vermiform, slightly wider at the middle cell, narrower towards the end cell, straight or curved, 2–8-septate, constricted at the septa, apically rounded, base truncate, with a darkened scar and central pore at the base, verruculose.
Culture characteristics: Colonies on PDA reaching 40 mm diam. after one week at room temperature (20–25 °C) in normal light, dense, circular, floccose to fluffy, surface smooth, edge entire, raised to low convex, effuse hyphae, slightly radiated, colony from above grey to brown, from below, brown at the margin, dark brown to black in the centre; not producing pigments in PDA.
Material examined: CHINA, Yunnan Province, Kunming City, Kunming Institute of Botany (102° 44′ 19″ E, 25° 8′ 27″ N; 1951 m ± 3.00 m), on dead branch of Malus sp. (Rosaceae), 18 August 2018, R. Phookamsak, KIB050 (MFLU 20-0071), living culture, KUMCC 19-0113.
Known host and distribution: On submerged decaying wood (Guizhou, China) (Hyde et al. 2017).
GenBank numbers: ITS = MN994339, LSU = MN994340, SSU = MN994341, TEF1-α = MT005777, RPB2 = MT005772.
Notes: In NCBI BLASTn search based on ITS sequences, the closest match with our strain is Torula gaodangensis J. Yang & K.D. Hyde (MFLUCC 17-0234; GenBank no. NR_164486) with 99.43% similarity. Our collection resembles T. gaodangensis in having monoblastic-acropetal, dark brown, subglobose to oblate-sphaeroid conidiogenous cells, with flattened base and acrogenous, brown to dark brown, subcylindrical or vermiform, 2–8-septate, base truncate, with a darkened scar and central pore at the base, verruculose conidia. Phylogenetic analyses of a combined LSU, SSU, TEF1-α, RPB2 and ITS sequence dataset also showed that our strain clades with the type strain of T. gaodangensis with significant support (70% ML, 1.00 BYPP; Fig. 80). We, hence, identify our strain as T. gaodangensis based on morphological and phylogenetic evidence. Hyde et al. (2017) introduced the species as a saprobe on submerged decaying wood in Suoluo River, from Guizhou Province, China. In this study, the species was found on dead branch of Malus sp. in terrestrial environment from Yunnan, China. Torula gaodangensis can survive in both aquatic and terrestrial habitats.
Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, TEF1-α, RPB2 and ITS sequence dataset representing genera of Torulaceae and other related families in Pleosporales. The updated sequence dataset was derived from Hyde et al. (2017, 2019a) and Li et al. (2017b). Sixty-eight strains are included in the combined analyses which comprise 4040 characters including gaps [LSU: 1–839 bp, SSU: 840–1776 bp, TEF1-α: 1777–2566 bp, RPB2: 2567–3418 bp, IST: 3419–4040). Occultibambusa bambusae (MFLUCC 13-0855) and Neooccultibambusa chiangraiensis (MFLUCC 12-0559) were selected as the outgroup taxa. Phylogenetic trees generated from maximum likelihood and Bayesian inference analyses were similar in overall topologies. The best scoring of the ML tree is selected to represent the phylogenetic relationships of taxa in Torulaceae and other related families, with the final ML optimization likelihood: − 31513.121691. The matrix had 1560 distinct alignment patterns, with 30.89% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.246674, C = 0.258646, G = 0.270512, T = 0.224169; substitution rates AC = 1.634530, AG = 3.727699, AT = 1.590702, CG = 1.072483, CT = 8.325214, GT = 1.000000; Tree-Length = 5.872247; gamma distribution shape parameter α = 0.552135. Bayesian posterior probabilities (BYPP) from MCMC were evaluated with a final average standard deviation of split frequencies less than 0.01. Bootstrap support values for maximum likelihood (ML) greater than 70% and Bayesian posterior probabilities (BYPP) greater than 0.95 are defined above the nodes as ML/BYPP. The type strains are indicated in bold and newly generated sequence are shown in blue
Trematosphaeriaceae K.D. Hyde et al.
Notes: Trematosphaeriaceae was introduced by Suetrong et al. (2011) to accommodate three genera Falciformispora K.D. Hyde, Halomassarina Suetrong et al. and Trematosphaeria Fuckel with Trematosphaeria pertusa Fuckel as the type species. The family is characterised by medium-sized globose to subglobose ascomata with a papilate ostiole, a relative wide coriaceous peridium, cylindrical cellular, septate pseudoparaphyses, cylindrical to clavate asci and ellipsoidal, fusiform to clavate, septate, hyaline to pigmented ascospores, with or without sheath, smooth- to rough-walled. Currently, six genera i.e., Bryosphaeria Döbbeler, Falciformispora, Hadrospora Boise, Halomassarina, Medicopsis Gruyter et al. and Trematosphaeria and 102 species are included in Trematosphaeriaceae (Wijayawardene et al. 2018a; Species Fungorum 2020). We follow the latest treatment and updated accounts of Trematosphaeriaceae in Hyde et al. (2019a). Based on phylogenetic analysis of ITS sequence data, a novel species Falciformispora uttaraditensis is introduced based on evidence of morphology and phylogenetic placement (Fig. 82).
Falciformispora K.D. Hyde
Notes: Falciformispora currently comprises four species including two human pathogens and two aquatic taxa which are confirmed in Trematosphaeriaceae based on molecular data (Hyde 1992; Suetrong et al. 2011; Ahmed et al. 2014; Hyde et al. 2019a).
Falciformispora uttaraditensis Boonmee & Huanraluek, sp. nov.
Index Fungorum number: IF557030; Facesoffungi number: FoF 06508; Fig. 81
Falciformispora uttaraditensis (MFLU 19-0485, holotype). a Appearance of ascomata on wood substrate. b Vertical section through ascoma. c Close up of peridium. f Pseudoparaphyses. e–g Asci. h, i Ascospores. j Ascospore with sheath in nigrosine reagent. k Germinated spore. l, m Culture on PDA after 4 weeks from surface and reverse. Scale bars: a = 500 µm, b = 100 µm, c = 20 µm, d = 20 µm, e–g = 50 µm, h–j = 20 µm, k = 50 µm
Etymology: Named after Uttaradit Province, Thailand, where this fungus was collected.
Holotype: MFLU 19-2361
Saprobic on the surface of submerged twigs in tidal creeks. Sexual morphAscomata 148–268 μm high × 110–224 μm diam. (\( \bar{x} \) = 208 × 179 μm, n = 5), immersed to erumpent, solitary, scattered on substrate, subglobose to globose, coriaceous, dark brown. Peridium 16–44 μm wide, 5 layers of dark brown cells of textura angularis, almost black at outside. Hamathecium with 2 μm wide, anastomosing, septate, branched pseudoparaphyses, hyaline. Asci 54–108 × 13–27 μm (\( \bar{x} \) = 73 × 18 μm, n = 4), 8-spored, bitunicate, cylindrical-clavate to broadly obpyriform, straight or slightly curved, sessile. Ascospores 28–59 × 6–14 μm (\( \bar{x} \) = 41 × 20 μm, n = 20), 2–3-seriate, fusiform to clavate, slightly curved, tapering towards narrow ends, 6-septate, obviously multi-guttulate when immature, hyaline, surrounded by a distinct mucilaginous sheath. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on MEA within 24 h at room temperature and germ tubes produced from the ends of the ascospore. Colonies on PDA reaching 2 cm diam. for one month at 25 °C. Initially aerial mycelium white, slightly raised, becoming grey to dark grey when old, convex with papillate on surface, crenated, green to dark brown from below, white to light yellow margin.
Material examined: THAILAND, Uttaradit Province, Phichai District, Thasak, Ban Cham Tok, on submerged twigs in tidal creeks, 29 December 2018, S. Boonmee, CTU6 (MFLU 19-2361, holotype), ex-type living culture, MFLUCC 19-0485.
GenBank numbers: ITS = MN809330.
Notes: Falciformispora uttaraditensis resembles F. aquatica D.F. Bao, K.D. Hyde & H.Y. Su in having immersed, coriaceous ascomata in wood tissue, dark pigmented, ostiolate, cylindro-clavate to broadly obpyriform asci, and fusiform to clavate, multi-septate, hyaline ascospores, surrounded by a distinct mucilaginous sheath (Hyde et al. 2019a). However, F. uttaraditensis differs from F. aquatica in having smaller ascomata, asci and ascospores. Furthermore, phylogenetic analyses indicate that F. uttaraditensis forms a basal lineage in the clade with high statistic support (92%ML, 0.99 BYPP; Fig. 82). To justify our new species, we follow the guidelines of Jeewon and Hyde (2016). Comparison of 546 nucleotides of the ITS sequences between F. uttaraditensis (MFLUCC 19-0485) and F. aquatica (MFLUCC 18-0212) reveals 20 nucleotide differences (3.8%), which is sufficient to delineate our new species.
Phylogram generated from maximum likelihood analysis based on ITS sequence data representing Trematosphaeriaceae and related families. Related sequences are taken from Hyde et al. (2019a). Twenty-five strains are included in the combined analyses which comprise 2505 characters 546 characters for ITS after alignement. Parathyridaria percutanea (CBS 868.95) in Thyridariaceae (Pleosporales) is used as the outgroup taxon. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood values of − 7765.257484 is presented. The matrix had 677 distinct alignment patterns, with 21.50% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.231191, C = 0.241799, G = 0.288729, T = 0.238282; substitution rates AC = 1.181282, AG = 2.193997, AT = 1.514916, CG = 0.832657, CT = 5.074084, GT = 1.000000; gamma distribution shape parameter α = 0.250521. Bootstrap values for maximum likelihood (ML) equal to or greater than 50% and clade credibility values greater than 0.95 (the rounding of values to 2 decimal proportions) from Bayesian-inference analysis labeled at the nodes. The newly generated sequence is indicated in bold and blue
Dothideomycetes, orders incertae sedis
Notes: Wijayawardene et al. (2018a) accepted 33 orders in this cosmopolitan class and treated another 27 orders as orders incertae sedis viz. Abrothallales, Acrospermales, Asterinales, Botryosphaeriales, Catinellales, Collemopsidiales, Dyfrolomycetales, Eremithallales, Jahnulales, Kirschsteiniotheliales, Lichenoconiales, Lichenotheliales, Microthyriales, Minutisphaerales, Monoblastiales, Muyocopronales, Natipusillales, Patellariales, Phaeotrichales, Stigmatodiscales, Strigulales, Superstratomycetales, Trypetheliales, Tubeufiales, Valsariales, Venturiales and Zeloasperisporiales.
Botryosphaeriales C.L. Schoch et al.
Notes: Botryosphaeriales was introduced for a single family Botryosphaeriaceae (Schoch et al. 2006). Currently there are six families: Aplosporellaceae, Botryosphaeriaceae, Melanopsaceae, Phyllostictaceae, Planistromellaceae and Saccharataceae, accepted in the Botryosphaeriales (Phillips et al. 2019).
Botryosphaeriaceae Theiss. & Syd.
Notes: The species belonging to Botryosphaeriaceae are endophytes, pathogens, and saprobes (Manawasinghe et al. 2016). They are reported on a wide range of hosts and broad geographical distribution (Chethana et al. 2016). There are over 100 species and 21 genera associated with this family (Phillips et al. 2013, 2019). We introduce two new host records belonging to Diplodia in family Botryosphaeriaceae.
Diplodia Fr.
Notes: In the combined gene phylogenetic tree of Diplodia, there are two distinct conidial morphologies which are also supported by phylogenetic analysis (Phillips et al. 2013). Recent taxonomic treatment for this genus is Jayasiri et al. (2019). Two new host records, Diplodia mutila and D. seriata are reported from Populus tremula and Onobrychis sp. in Italy.
Diplodia mutila (Fr.) Mont., Annls Sci. Nat., Bot., sér. 2 1: 302 (1834)
Facesoffungi number: FoF 05978; Fig. 83
≡ Sphaeria mutila Fr., Syst. mycol. (Lundae) 2(2): 424 (1823)
Epitype: PORTUGAL, Beira Litoral, Aveiro, Populus alba, 2012, A. Alves, LISE 96136, MBT176182, ex-epitype culture, CBS 136014.
Saprobic on dead aerial branch of Populus tremula. Sexual morph Undetermined. Asexual morphCoelomycetous. Conidiomata 600–680 μm diam., pycnidial, black, initially immersed, raised, becoming erumpent and opening through the host epidermis, sublgobose, uni- to bi-loculate, ostiolate, papillate. Pycnidial walls composed of 3–4 layers, dark brown to black, thick-walled cells of textura angularis, outer layer comprising 1–2 layers, dark brown to black cells, inner layer comprising 1–2 layers of hyaline cells. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 7–10 μm long × 2–5 μm wide, enteroblastic, phialidic, discrete, cylindrical to ampulliform, hyaline, aseptate, smooth-walled, proliferating at the same level giving rise to periclinal thickenings. Conidia 5–10 × 1–4 μm wide (\( \bar{x} \) = 8 × 3 µm, n = 20), initially hyaline, becoming dark brown with the age, ellipsoidal to oblong, aseptate thick- and smooth-walled, small granules.
Culture characteristics: Colonies on PDA covering a 90 mm diam. petri dish before 7 days at 28 °C, circular initially white mycelium, moderately aerial becoming pale grayish to dark and reverse gray to dark grayish green.
Material examined: ITALY, Province of Forlì-Cesena [FC], near Strada San Zeno-Galeata, on dead aerial branch of Populus tremula (Salicaceae), 26 March 2018, E. Camporesi, IT 3784 (MFLU 18-0912), living culture, JZB 3140011.
Known host and distribution: On dead aerial branch of Populus tremula (Salicaceae), Previously Diplodia mutila has been reported on Fraxinus excelsior, Olea europaea and Vitis vinifera from Italy (Farr and Rossman 2020).
GenBank numbers: ITS = MN944415.
Notes: The collection obtained from a dead aerial branch of Populus tremula was identified as Diplodia mutila with support from both morphology and phylogeny (Alves et al. 2014). Our strains clustered with the reference strain of Diplodia mutila (CBS 230.30), in the combined ITS and TEF1-α sequence phylogeny with significant support in MP analysis (85% ML; Fig. 84). Furthermore, our isolate showed 0.94% and 0.90% base pair difference with Diplodia mutila (CBS 230.30) in ITS and TEF1-α gene regions. This is the first record of Diplodia mutila from Populus tremula from Italy.
Phylogram generated from maximum parsimony analysis based on combined ITS and TEF1-α, sequence data representing Diplodia. Related sequences are taken from Dissanayake et al. (2017a) and Jayasiri et al. (2019). Forty-eight strains are included in the combined analyses which comprise 824 characters (529 characters for ITS and 292 characters for TEF1-α). Neofusicoccum luteum (CBS110299 and CBS110497) in Botryosphaeriaceae (Botryosphaeriales) are used as the outgroup taxa. Tree topology of the maximum parsimony analysis is similar to the maximum likelihood. The dataset consisted of 824 characters with 580 constant characters and 211 parsimony-informative and 33 parsimony-uninformative characters. The maximum number of trees generated was 1000, and the most parsimonious trees had a tree length of 458 (CI = 0.725, RI = 0.866, RC = 0.628, HI = 0.275). The best RAxML tree with a final likelihood values of − 3516.161706 is presented. The matrix had 278 distinct alignment patterns, with 6.13% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.206373, C = 0.300294, G = 0.260861, T = 0.232472; substitution rates AC = 1.148229, AG = 3.248235, AT = 1.117837, CG = 1.595230, CT = 4.204321, GT = 1.000000; gamma distribution shape parameter α = 0.482421. Maximum parsimony (MP) and maximum likelihood (ML) equal to or greater than 50% are given as MP/ML at the nodes. Ex-type strains are in bold and black, the new isolate is indicated in bold and blue
Diplodia seriata De Not., Micr. Ital. Dec. 4: 6. 1942
Facesoffungi number: FoF 03596; Fig. 85
Holotype: ITALY, on dead stems of Jasminium sp., 18 August 1837, De Notaris (HERB RO).
Epitype: PORTUGAL, Montemor-o-Novo, on dead stems of Vitis vinifera, 31 July 1997, A.J.L. Phillips, CBS-H 19809, ex-epitype culture CBS 112555.
Saprobic on dead aerial stem of Onobrychis sp. Sexual morph Undetermined. Asexual morphConidiomata pycnidial, stromatic, separate or aggregated and confluent, visible as black dots on host surface, immersed in the host, slightly raised, partially emergent at maturity, dark brown to black, ostiolate, apapillate. Pycnidial walls thick-walled, outer layers composed of dark brown cells, of textura angularis, inner layers composed of thin-walled, hyaline cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells hyaline, thin-walled, smooth, cylindrical, swollen at the base, discrete, producing a single conidium at the tip, indeterminate. Conidia 18–26 × 8–12 μm (\( \bar{x} \) = 21 × 10 μm, n = 30) initially hyaline, becoming dark brown, moderately thick-walled, aseptate, ovoid, widest in the middle, apex obtuse, base truncate or rounded.
Culture characteristics: Colonies reaching 90 mm diam. on PDA after 5 days in the dark at 25 °C. Initially, mycelia were white thick, superficial, branched, septate, hyaline, smooth, with age developing dense aerial mycelium. Initially both upper and reverse were white with age become black.
Material examined: ITALY, Province of Forlì-Cesena [FC], Civitella di Romagna, on the dead aerial stem of Onobrychis L., 21 April 2018, E. Camporesi, IT 3835 (MFLU 18-1120), living culture, JZB3140013.
Known host and distribution: Occuring on a wide range of hosts including Abies, Alnus sp., Citrus sp., Malus sp., Prunus sp., and Vitis sp. This species has been reported in different geographical localities. There are 271 records belonging to Diplodia seriata (Farr and Rossman 2020).
GenBank numbers: ITS = MN533807, TEF1-α = MN883826.
Notes: The Diplodia strain obtained in present study morphologically fits well within Diplodia. It developed a clade with Diplodia seriata representative strains (CBS 112555 and CBS 119049). The taxa identified in the present study are similar to the original description of Diplodia seriata (Phillips et al. 2013). However, in both MP and ML trees the branch length of current isolates was longer than in the other two Diplodia strains. Therefore we compared pairwise nucleotide difference between the type strain CBS 112555 and strain isolated in this study. 100% nucleotide similarities were observed for ITS across 512 nucleotides and 91% nucleotide similarities were observed in TEF1-α across 266 nucleotides. Diplodia species have wide range of hosts including economically important crops (Phillips et al. 2013; Dissanayake et al. 2017a). However this is the first time a Diplodia species has been reported on Onobrychis (Farr and Rossman 2020).
Muyocopronales Mapook, Boonmee & K.D. Hyde
Notes: The order was introduced by Mapook et al. (2016b), and comprises two families, Muyocopronaceae and Palawaniaceae, based on phylogenetic with molecular clock analyses and morphological comparison. See Mapook et al. (2016b) for the details.
Muyocopronaceae K.D. Hyde
Notes: Muyocopronaceae was introduced by Luttrell (1951) and reintroduced by Hyde et al. (2013) with a single genus Muyocopron. Muyocopronaceae was place in the order Muyocopronales to accommodate three new Muyocopron species in Thailand (Mapook et al. 2016b) and Arxiella Papendorf, Leptodiscella Papendorf, Mycoleptodiscus Ostaz., Muyocopron Speg., Neocochlearomyces Pinruan, Neomycoleptodiscus Hern.-Restr., J.D.P. Bezerra & Crous and Paramycoleptodiscus Crous & M.J. Wingf. were accepted in the family (Crous et al. 2016b, 2018a; Hernández-Restrepo et al. 2019). In this study, Setoapiospora is introduced as a new genus with its type species, Setoapiospora thailandica from Thailand, based on morphology and molecular data, together with description and illustrations (Fig. 86). A phylogenetic tree based on combined LSU, ITS, and SSU sequence data is presented in Fig. 87.
Setoapiospora thailandica (MFLU 20-0049, holotype). a, b Appearance of ascomata on substrate. c Squash mounts showing ascomata. d Section through of ascoma. e Setae. f Peridium. g Pseudoparaphyses. h, i asci. j–m Ascospores. Scale bars: a = 500 µm, b = 200 µm, d = 100 µm, c, h, i = 50 µm, e, f, j–m = 10 µm, g = 5 µm
Phylogram generated from maximum likelihood analysis based on combined dataset of LSU, ITS and SSU sequence data. Seventy-three strains are included in the combined sequence analysis, which comprise 2966 characters with gaps. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 18665.197487 is presented. The matrix had 1232 distinct alignment patterns, with 41.24% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.239788, C = 0.244858, G = 0.294052, T = 0.221302; substitution rates: AC = 1.516739, AG = 2.835597, AT = 1.527965, CG = 1.155606, CT = 5.870000, GT = 1.000000; gamma distribution shape parameter α = 0.229251. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. Newly generated sequences are in dark blue bold and type species are in bold. Lophium mytilinum (AFTOL-ID 1609) and Mytilinidion rhenanum (CBS 135.45) are used as outgroup taxa
Setoapiospora Mapook & K.D. Hyde, gen. nov.
Index Fungorum number: IF556905; Facesoffungi number: FoF 06793
Etymology: The generic epithet refers to two-celled spore with one cell smaller than the other (apiosporous) and ascomata with brown setose.
Saprobic on dead branches of wood. Sexual morphAscomata superficial to semi-immersed, solitary or scattered, carbonaceous, appearing as dark brown to black spots, with a poorly developed basal layer and an irregular margin. Ostiole central, with external dark brown setae. Peridium comprising dark brown cells of textura epidermoidea. Hamathecium with cylindrical to filiform, septate, pseudoparaphyses. Asci 8-spored, bitunicate, cylindrical, pedicellate, straight or slightly curved, with an ocular chamber. Ascospores 1-seriate, ellipsoid to broadly fusiform, hyaline, 1-septate, constricted at the septum, with a small lower cell and a large upper cell, widest at the centre and tapering towards the ends, with granular appearance. Asexual morph Undetermined.
Type species: Setoapiospora thailandica Mapook & K.D. Hyde
Notes: A phylogenetic analyses based on combined dataset of LSU, ITS and SSU sequence data show that Setoapiospora is closely related to Arxiella (Fig. 87). However, we could not compare the morphological characteristics of these genera as Setoapiospora is found as sexual morph in nature and an asexual morph is failed to develop in culture, while Arxiella was found as asexual morph in nature. Therefore, Setoapiospora is described here as a new genus based on phylogeny, together with a comparison of the ITS (+5.8S) gene region of Setoapiospora thailandica and Arxiella terrestris reveals 199 base pair differences (30.9%) across 644 nucleotides.
Setoapiospora thailandica Mapook & K.D. Hyde, sp. nov.
Index Fungorum number: IF556906; Facesoffungi number: FoF06794; Fig. 86
Etymology: Name reflects the country from where this species was collected, Thailand.
Holotype: MFLU 20-0049
Saprobic on dead branches of wood. Sexual morphAscomata 270–290 µm high × 390–415 µm diam. (\( \bar{x} \) = 280.5 × 400 µm, n = 5), superficial to semi-immersed, solitary or scattered, carbonaceous, solitary or scattered, appearing as dark brown to black spots, with a poorly developed basal layer and an irregular margin. Ostiole central, with external dark brown setae. Peridium 25–50(–60) µm wide, comprising dark brown cells of textura epidermoidea. Hamathecium comprising 1–2 µm wide, cylindrical to filiform, septate, pseudoparaphyses. Asci 85–160 × 13–24 µm (\( \bar{x} \) = 123 × 17 µm, n = 20), 8-spored, bitunicate, cylindrical, pedicellate, straight or slightly curved, with an ocular chamber. Ascospores 20–27 × 10–13 µm (\( \bar{x} \) = 25 × 11 µm, n = 25), 1-seriate, ellipsoid to broadly fusiform, hyaline, 1-septate, constricted at the septum, with a small lower cell and a large upper cell, widest at the centre and tapering towards ends, granular. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on MEA within 24 h at room temperature and germ tubes produced from the apex. Colonies on MEA irregular, initially mycelium white, slightly flattened, filamentous, cultures grayish to light brown in old, flattened on surface, pale brown to brown in reverse from the centre of the colony, creamy white at margin
Material examined: THAILAND, Chiang Rai Province, Huai Kang Pla waterfall, on dead branches of wood, 19 June 2015, A. Mapook, HKP1 (MFLU 20-0049, holotype), ex-type culture MFLUCC 17-1426.
GenBank numbers: ITS = MN638862, LSU = MN638847, SSU = MN638851, TEF1-α = MN648731.
Notes: In a BLASTn search of NCBI GenBank, the closest matches of the ITS sequence of Setoapiospora thailandica (MFLUCC 17-1426, ex-holotype) with 81.36% similarity, was to Mycoleptodiscus terrestris (strain IMI 159038, MK487755). The closest matches with the LSU sequence with 93.74% similarity, was to Mycoleptodiscus terrestris (strain CBS 231.53, MK487730), while the closest matches with the SSU sequences with 98.52% similarity, was to Neocochlearomyces chromolaenae (strain BCC 68250, NG_065766).
Tubeufiales Boonmee & K.D. Hyde
Notes: The order Tubeufiales was introduced by Boonmee et al. (2014b) to accommodate 19 genera in family Tubeufiaceae based on phylogenetic analyses of combined LSU and ITS sequences. Dictyospora Brahman., Y.Z. Lu, Boonmee & K.D. Hyde (Brahmanage et al. 2017), Helicotubeufia Y.Z. Lu & J.K. Liu (Liu et al. 2018a), Manoharachariella Bagyan., N.K. Rao & Kunwar (Doilom et al. 2017), Muripulchra Z.L. Luo, Hong Y. Su & K.D. Hyde, Neohelicomyces Z.L. Luo, Bhat & K.D. Hyde, (Luo et al. 2017), Neohelicosporium Y.Z. Lu, J.C. Kang & K.D. Hyde (Lu et al. 2017b), Neotubeufia Chaiwan, Boonmee, Y.Z. Lu & K.D. Hyde (Chaiwan et al. 2017) were added to Tubeufiaceae. Lu et al. (2018) reassessed Tubeufiales using multi-gene (including ITS, LSU, RPB2 and TEF1-α) phylogeny coupled with their morphology and determined 16 new genera including Chaetosphaerulina I. Hino, Artocarpomyces Subram and Helicodochium J.S. Monteiro, R.F. Castañeda, A.C. Cruz & Gusmão which were earlier not classified as tubeufialean taxa. In addition, two families Bezerromycetaceae (Bezerromycetales) and Wiesneriomycetaceae (Wiesneriomycetales) established by Bezerra et al. (2016), were transferred to order Tubeufiales by Liu et al. (2017a) based on divergence times estimates. Currently, Tubeufiales comprises three families Bezerromycetaceae (two genera), Tubeufiaceae (40 genera) and Wiesneriomycetaceae (four genera) (Bezerra et al. 2016; Liu et al. 2017a; Lu et al. 2018).
Tubeufiaceae M.E. Barr
Notes: Tubeufiaceae was initially assigned in Pleosporales based on the type genus, Tubeufia (Barr 1979). The natural classification of this family was uncertain at the early stage. Hyde et al. (2013) reviewed the family Tubeufiaceae with a description and a key to sexual and asexual genera. Boonmee et al. (2014b) established the order Tubeufiales to accommodate those genera in Tubeufiaceae and revised the taxonomic history of this family. The ribosomal genes LSU, SSU, ITS and protein-coding genes TEF1-α and RPB2 are reliable phylogenetic markers which can be useful in establishing inter-generic and interspecific relationships in Tubeufiaceae (Boonmee et al. 2011; Lu et al. 2018). However, Acanthophiobolus Berl., Bifrontia Norman, Chaetosphaerulina I. Hino, Podonectria Petch are still known only from their morphological features (Boonmee et al. 2011). In this study, we followed Lu et al. (2018) and Jayasiri et al. (2019) for species delimitation. A new genus Camporesiomyces is introduced to accommodate a novel species Camporesiomyces mali and two new combinations, C. patagoniensis and C. vaccinii.
Camporesiomyces D.P. Wei, Wanas. & K.D. Hyde, gen. nov.
Index Fungorum number: IF557070; Facesoffungi number: FoF 07074
Etymology: In honour of Erio Camporesi, for his invaluable contribution to mycology.
Saprobic on dead branches of wood. Sexual morphAscomata superficial or semi-immersed, solitary, unilocular or multi-loculate, black, subglobose to conical, ostiolate, with sparse setae surrounding the ostiole. Peridium comprising cells of textura angularis. Hamathecium comprising hyaline, filiform, pseudoparaphyses. Asci 8-spored, bitunicate, fissitunicate, cylindrical. Ascospores narrowly fusiform, hyaline, multi-septate. Asexual morph Hyphomycetous.
Type species: Camporesiomyces mali D.P. Wei, Wanas. & K.D. Hyde
Notes: The phylogenetic analyses based on combined LSU, ITS, TEF1-α and RPB2 dataset indicate Camporesiomyces mali, Acanthostigma patagonicum R.M. Sánchez, A.N. Mill. & Bianchin (BBB MVB 573) and Helicoma vaccinii Carris (CBS 216.90) group in a distinct clade sister to Helicosporium in family Tubeufiaceae (Fig. 89). Morphologically the type species of Camporesiomyces is distinguished from other genera in Tubeufiaceae by multi-loculate ascomata and narrowly fusiform ascospores. Acanthostigma patagonicum and Helicoma vaccinii were initially known only from their sexual and asexual morphs (Carris 1989; Tsui and Berbee 2006; Sanchez et al. 2012). Even though Boonmee et al. (2014b) considered A. patagonica and H. vaccinii as Helicosporium species, Lu et al. (2018) argued they are different to those in Helicosporium. Thus, the natural classification of these two taxa was uncertain. Camporesiomyces is introduced to accommodate the type species Camporesiomyces mali, Acanthostigma patagonicum and Helicoma vaccinii. Although the holotypes of Acanthostigma patagonicum and Helicoma vaccinii have not been rechecked, the DNA data derived from the ex-type strains, shows they belong to this new genus.
Camporesiomyces mali D.P. Wei & K.D. Hyde, sp. nov.
Index Fungorum number: IF557016; Facesoffungi number: FoF 07075; Fig. 88
Camporesiomyces mali (KUN-HKAS 102565, holotype). a Substrate. b Ascomata on substrate. c, d Transverse section of ascomata. e Setae on ascoma. f Peridium. g, h Asci. i Seta. j–l Ascospores. m Germinated ascospore. n Colony on PDA from above and below. Scale bars: c = 150 µm, d, e = 100 µm, m = 50 µm, f–l = 20 µm
Etymology: The specific epithet “mali” refers to the host genus Malus.
Holotype: KUN-HKAS 102565
Saprobic on decaying branches of Malus halliana Koehne. Sexual morphAscomata 200–300 μm (\( \bar{x} \) = 231 μm, n = 5), subimmersed, scattered, black, nearly globose, multi-loculate, ostiolate, setaceous. Setae 15–20 × 1–2 μm (\( \bar{x} \) = 17 × 1.5 μm, n = 5), short, dark brown to black, aseptate, attenuated toward the apex. Peridium 15–50 μm (\( \bar{x} \) = 23 μm, n = 20), comprising dark brown to pale-brown, irregular in thickness, thin-walled cells of textura angularis. Hamathecium comprising inconspicuous, filiform, hyaline pseudoparaphyses. Asci 50–100 × 10–25 μm (\( \bar{x} \) = 70 × 14 μm, n = 10), 8-spored, bitunicate, broadly clavate, thickened at apex, without ocular chamber. Ascospores 50–75 × 3–5 μm (\( \bar{x} \) = 62 × 4 μm, n = 30), hyaline, cylindrical, slightly curved, 10–12-septate, ends remaining cone-shaped, smooth-walled, guttulate when young.
Culture characteristics: Culture was obtained from germinated ascospores. Colonies slowly growing on PDA, brown, slightly raised, dense, with irregular margin, reaching 2 mm in 9 days at 25 °C.
Material examined: CHINA, Yunnan Province, Kunming City, Kunming Institute of Botany, on decaying wood branches of Malus halliana (Rosaceae), 19 August 2018, D.P. Wei, KIB1903 (KUN-HKAS 102565, holotype), ex-type living culture, KUMCC 19-0216.
GenBank numbers: ITS = MN792813, LSU = MN792811, TEF1-α = MN794018.
Notes: Our isolate has close affinity with Acanthostigma patagonicum (BBB MVB 573) and Helicoma vaccinii (CBS 216.90) by forming a distinct, strongly supported clade as basal to the remaining taxa in Helicosporium (100% ML, Fig. 89). Our isolate resembles A. patagonicum in having dark brown setae, clavate asci and inconspicuous pseudoparaphyses. But our isolate can be distinguished from A. patagonicum in having narrowly fusiform, symmetric ascospores, in contrast to elongate fusiform ascospores, with asymmetrical ends in A. patagonicum (Sanchez et al. 2012). Helicosporium vaccinii is only known from its asexual morph (Carris 1989). A comparison of ITS sequences between our isolate and A. patagonicum show 24 bp difference and 3 gaps within 330 bp nucleotides. When ITS sequences are compared with H. vaccinii, there are 52 bp difference and 17 gaps within 504 bp nucleotides. Thereby, we determined our isolate to be a novel species in the new genus Camporesiomyces.
Phylogram generated from maximum likelihood analysis based on combined LSU, ITS, TEF1-α, and RPB2 sequence dataset representing taxa in family Tubeufiaceae (Tubeufiales). Related sequences are taken from Lu et al. (2018). Sixty-six strains are included in the combined analyses which comprise 3219 characters (828 characters for LSU, 509 characters for ITS, 837 characters for TEF1-α and 1045 characters for RPB2) after alignment. The best scoring RAxML tree with a final likelihood value of − 31516.131561 is presented. The matrix had 1311 distinct alignment patterns, with 25.67% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.243290, C = 0.257008, G = 0.265413, T = 0.234289; substitution rates: AC = 0.995531, AG = 4.500491, AT = 2.148310, CG = 0.787501, CT = 8.890762, GT = 1.000000; gamma distribution shape parameter α = 0.791557. Bootstrap support values for ML equal to or greater than 70% and BYPP equal to or greater than 0.95 are given above the nodes. Newly generated sequences are in blue bold and type species are in black bold. Xiliomyces brasiliensis (URM7414) and Bezerromyces pernambucoensis (URM7412) in family Bezerromycetaceae (Tubeufiales) were used as the outgroup taxa
Camporesiomyces patagoniensis (R.M. Sánchez, A.N. Mill. & Bianchin) D.P. Wei & K.D. Hyde, comb. nov.
Index Fungorum number: IF557017; Facesoffungi number: FoF 07076
≡ Acanthostigma patagonicum R.M. Sánchez, A.N. Mill. & Bianchin. [as ‘patagonica’], Mycologia 104(1): 225 (2012)
Holotype: ARGENTINA. NEUQUÉN Province: Pque. Nacional Lanín, on Provincial Route Number 48, before reaching Playas de Yuco, on bark of Nothofagus alpina, 17 May 2007, M.V. Bianchinotti, R.M. Sánchez, MVB573.
Descriptions and illustrations: See Sánchez et al. (2012).
Notes: Sanchez et al. (2012) introduced Acanthostigma patagonicum on bark of Nothofagus spp. from Andean Patagonian forests in Argentina. It has a close morphological association with A. multiseptatum Promputtha & A.N. Mill. and A. scopulum (Cooke & Ellis) Peck in the ascospore characteristics. However, both species can be distinguished from A. patagonicum by their narrower ascospores (Sanchez et al. 2012). Also, Acanthostigma patagonicum is not closely related to any other Acanthostigma species in the phylogenetic analyses (Sanchez et al. 2012; Brahmanage et al. 2017; Chaiwan et al. 2017; Lu et al. 2018). Boonmee et al. (2014b) referred this as Helicosporium patagonicum in their phylogeny tree but did not formally synonymize it. Lu et al. (2018) argued that the morphological features of Acanthostigma patagonicum is not related to Helicosporium and retained the original name of the species. In our phylogenetic analysis, Acanthostigma patagonicum groups with Camporesiomyces mali and Helicoma vaccinia in a monophyletic clade with strong support. Hence, we transfered Acanthostigma patagonicum to Camporesiomyces.
Camporesiomyces vaccinii (Carris) D.P. Wei & K.D. Hyde, comb. nov.
Index Fungorum number: IF557018; Facesoffungi number: FoF 07077
≡ Helicoma vaccinii Carris, Mycotaxon 36(1): 29 (1989)
Holotype: USA, Georgia, Satilla River, Ware Co., on stems of Vaccinium elliotii, 9 July 1988, N. Vorsa, LMC 0043 (herb. BPI), ex-type living culture, ATCC 66068
Descriptions and illustrations: See Carris (1989).
Notes: Helicoma vaccinii was introduced by Carris (1989) from the stem scar of Vaccinium elliotii in Georgia, USA. Matsushima (1993) and Zhao et al. (2007) reported more collections of this species from Peru and China. Tsui et al. (2006) provided DNA based sequence data (ITS and LSU) from the ex-type strain (CBS 216.90) of the species. The phylogenetic analyses showed that H. vaccinii (CBS 216.90) is not related to any Helicoma taxa and constitute a close affinity to Acanthostigma patagonicum and the members of Helicosporium. Thus, Boonmee et al. (2014b) transferred this species to Helicosporium as H. vaccinii based on phylogenetic evidence, but did not formally synonymize it. Brahmanage et al. (2017), Chaiwan et al. (2017) and Lu et al. (2017a, b) also treated this species as Helicosporium vaccinii. Lu et al. (2018) compared its morphology as described by Carris (1989), Matsushima (1993) and Zhao et al. (2007), with both Helicosporium and Helicoma species. Because of its similarity to Helicoma, Lu et al. (2018) retained it as a Helicoma species. In this study, we observed a similar phylogenetic relationship to Lu et al. (2018). One of our new strains (Camporesiomyces mali) also grouped with strong support. Rather than keeping this species as an ambiguous taxon, we transfer it to Camporesiomyces until more samples are collected, analyzed and evaluated.
Venturiales Y. Zhang ter, C.L. Schoch & K.D. Hyde
Notes: The order Venturiales was established by Zhang et al. (2011) based on morphological characteristics and phylogenetic analyses. It comprises two families: Sympoventuriaceae and Venturiaceae (Zhang et al. 2011; Hyde et al. 2013). Members of this order includes saprobes and pathogens in plants, animals and humans. They are found in terrestrial, hot springs and aquatic environments.
Sympoventuriaceae Y. Zhangter, C.L. Schoch & K.D. Hyde
Notes: The family Sympoventuriaceae, typified by Sympoventuria, was introduced by Zhang et al. (2011) to accommodate Sympoventuria capensis Crous & Seifert, Veronaeopsis simplex (Papendorf) Arzanlou & Crous and fusicladium-like species. This family comprises both saprobes and pathogens of plants, and animals and have been reported from different environments, such as soil, hot springs, industrial effluents, terrestrial and aquatic habitats (Seyedmousavi et al. 2013; Samerpitak et al. 2014). Sympoventuriaceae is characterised by immersed, subglobose ascomata, that are black, papillate, ostioles, septate pseudoparaphyses, that are constricted at the septa and anastomosing and extending above the asci, 8-spored, bitunicate, fissitunicate, subcylindrical, pedicellate asci and hyaline, fusoid-ellipsoidal ascospores, constricted at the median septum. Asexual morphs are sympodiella-like, fusicladium-like and veronaeopsis-like (Crous et al. 2007a; Zhang et al. 2011; Samerpitak et al. 2014). Currently this family includes Acroconidiellina M.B. Ellis, Clavatispora Boonmee & K.D. Hyde, Fusicladium Bonord, Ochroconis de Hoog & Arx, Scolecobasidium E.V. Abbott, Sympoventuria, Veronaeopsis, and Verruconis (Wijayawardene et al. 2018a). Most members of this family are known by their asexual morphs (hyphomycetes). The species belonging to Acroconidiellina, Scolecobasidium, Veronaeopsis and Verruconis have hyphomycetous asexual morphs (Zhang et al. 2011; Samerpitak et al. 2014), while Sympoventuria, Ochroconis and Clavatisopora are known to produce sexual morphs (Crous et al. 2007a; Boonmee et al. 2014a; Machouart et al. 2014; Samerpitak et al. 2014).
Verruconis Samerp. et al.
Notes: Verruconis, typified by V. gallapova (W.B. Cooke) Samerp. & de Hoog, was introduced by Samerpitak et al. (2014) to accommodate thermophilic species. Verruconis is characterised by colonies that are brown, slow growing, with or without conidiophores; conidiogenous cells that are erect, pale brown, sparsely septate, with scattered denticles and rhexolytic conidial secession; conidia that are two-celled, verrucose to smooth-walled, clavate to cylindrical, produced sympodially. Currently, this genus includes six species supported by both morphological and molecular data. We introduce a new species Verruconis mangrovei from a marine habitat based on both morphology and multi-gene phylogeny.
Verruconis mangrovei Devadatha V.V. Sarma & E.B.G. Jones, sp. nov.
Index Fungorum number: IF557052; Facesoffungi number: FoF 07058; Fig. 90
Verruconis mangrovei (AMH-10004, holotype). a Superficial ascomata on the surface of the host. b, c, e, f Ascomata with short and long setae. g Ascomata with few layers of peridium. h Cellular pseudoparaphyses with asci. i, j Immature and mature asci with an ocular chamber and short pedicels. k–m Ascospores. n Germinating ascospore with germ tubes developing from the end cells. Scale bars: b–n = 10 μm
Etymology: With reference to mangrove habitat from where it was collected.
Holotype AMH-10004
Saprobic on dead mangrove wood. Sexual morphAscomata 42.5–120 μm high, 32.5–110 μm wide (\( \bar{x} \) = 82 × 71.5 μm, n = 20), superficial to semi-immersed, small, clustered, sometimes solitary, scattered, globose to subglobose, rarely ovate, coriaceous, pale brown to dark brown, setae septate, 3–7 μm diam., hyphophodia pale brown. Ostiole 7.5–24 μm high, 26–45 μm diam. (\( \bar{x} \) = 18 × 32 μm, n = 7). Setae 17–27 μm long, 3–5.5 μm diam. (\( \bar{x} \) = 21 × 4.5 μm, n = 6), septate, cylindrical or rounded, forked at the ends, smooth, pale brown to dark brown, 2–5 setae developing at the apical part of the ascomata. Peridium 6–9.5 (\( \bar{x} \) = 6.8 μm, n = 10) equal in thickness, composed of 2–3 layers of thin-walled, pale brown to dark brown cells of textura angularis. Hamathecium comprising numerous rarely branched, 1–2.5 μm cellular, hyaline, septate, pseudoparaphyses, embedded in a gelatinous matrix. Asci 50–80 × 17–32 μm, (\( \bar{x} \) = 62 × 22 μm, n = 20), 8-spored, bitunicate, broadly clavate, thick-walled with a short pedicel, apically rounded, with an ocular chamber. Ascospores 17.5–25.5 × 6.5–13 μm (\( \bar{x} \) = 22 × 9 μm, n = 30), 2–3-seriate, overlapping, hyaline when young, turning pale brown at maturity, broader and rounded at the apex, tapering towards the end, dictyosporous, ellipsoidal to oblong with obtuse ends, 4–5 transverse septate, highly constricted at the second septum, 1-longitudinal septum in each row except at the end cells, lack guttules and gelatinous sheaths.
Culture characteristics: Ascospores germinating on 50% SWA within 24 h, with germ tubes developing from the end cells of the ascospores. Colonies on MEA slow growing reaching 3–8 mm in diam., within 7 days and attain 10–15 mm diam., after 15 days of incubation at 25 °C. Colonies becoming pale brownish olive to brownish olive, reverse brownish, restricted growth, velvety, lobate margin and circular to irregular.
Material examined: INDIA, Tamil Nadu, Tiruvarur, Muthupet mangroves (10.4°N 79.5°E), on decaying wood of Excoecaria agallocha (Amaranthaceae), 24 December 2016, B. Devadatha, AMH-10004 (holotype), ex-type living culture, NFCCI-4390; ibid., Aegiceras corniculatum (Primulaceae), 28 November 2015, B. Devadatha AMH-10005, living culture, NFCCI-4391.
GenBank numbers: ITS = MN782361, LSU = MN241144, SSU = MN241147, TUB2 = MN848140 (NFCCI-4390); ITS = MN782362, LSU = MN241145, SSU = MN241148, TUB2 = MN848141(NFCCI-4391).
Notes: Verruconis mangrovei resembles Leptosphaerulina mangrovei Inderb. & E.B.G. Jones in having small ascomata, setae with forked apices, short ostiole, asci and muriform, ellipsoidal ascospores with 4–5 transverse septa. However, Verruconis mangrovei has superficial to semi-immersed ascomata, hyaline, cellular pseudoparaphyses and ascospores that lack mucilaginous sheaths. Leptosphaerulina mangrovei has erumpent ascomata, lacks pseudoparaphyses, ascospores have a mucilaginous sheath and produces a sexual morph on CMA (Inderbitzin et al. 2000). Leptosphaerulina mangrovei lacks sequence data to compare with Verruconis mangrovei at the molecular level. Verruconis mangrovei shares a few morphological similarities with Clavatispora thailandica Boonmee & K.D. Hyde in having superficial, setose, ostiolate ascomata, and muriform ascospores. However, C. thailandica is distinguished from Verruconis mangrovei in having asymmetrical, 4–7(–8) transverse and 1–3 vertical septa ascospores (Boonmee et al. 2014a). Verruconis heavae differs from V. mangrovei in having ellipsoidal to oblong, pale brown to dark brown, 1–3-septate conidia, with rounded apices, subacute bases and is an asexual morph from terrestrial habitats, while Verruconis mangrovei is a sexual morph from a marine habitat. Phylogenetic analysis (Fig. 91) showed that Verruconis mangrovei shares a sister relationship with Verruconis heveae with low statistical support.
RAxML tree based on combined dataset of LSU, SSU, ITS and TUB2 gene regions of Verruconis mangrovei along with taxa from Sympoventuriacaeae. Bootstrap support values for maximum likelihood (ML) equal to or greater than 75% and clade credibility values greater than 0.95 from Bayesian-inference analysis are given above each branch. Ex-type strains are in bold and black, the new isolates are in blue. The tree is rooted to Scolecobasidium excentricum CBS 469.95
Dothideomycetes, familyincertae sedis
Notes: Dothideomycetes is the largest class in the division Ascomycota and currently 36 families are classified under Dothideomyetes, families incertae sedis (Hyde et al. 2013; Wijayawardene et al. 2018a). We introduced a novel family Eriomycetaceae to accommodate a novel monotypic genus Eriomyces, Funbolia Crous & Seifert, Heleiosa Kohlm., Volkm.-Kohlm. & O.E. Erikss., Phellinocrescentia Crous & Decock and Pseudopassalora Crous placed in Dothideomyetes, families incertae sedis.
Eriomycetaceae Huanraluek & K.D. Hyde, fam. nov.
Index Fungorum number: IF557099; Facesoffungi number: FoF 07189
Etymology: Named after its type genus, Eriomyces.
Associated with living tree or saprobic on dead leaves and twigs in terrestrial habitat, or hyperparasite on other fungi. Sexual morphAscomata immersed beneath a raised dark brown to black pseudoclypeus, solitary or aggregated, coriaceous, subglobose to ampulliform, unilocular, ostiolate, apapillate or papillate, ostiole filled with periphyses. Peridium composed of several, small cell layers, of hyaline to heavily pigmented pseudoparenchymatous cells of textura angularis to textura epidermoidea, light coloured from the base towards the apex, with heavily pigmented cells at the base of ostiolar canal. Hamathecium composed of branched, anastomosed, cellular pseudoparaphyses. Asci 4–8-spored, or poly-spored, bitunicate, cylindrical to cylindric-clavate, with short to long pedicel, apically broadly rounded, with indistinct ocular chamber. Ascospores overlapping 1–2-seriate, or crowded, hyaline, oblong to allantoid or ellipsoidal to fusiform, aseptate to 1-septate, straight to slightly curved, thin- and smooth-walled, with guttules, with or without appendages. Asexual morph Coelomycetous, phoma-like, or hyphomycetous. Coelomycetous: Conidiomata pycnidial, dark brown to black, globose to subglobose, solitary or aggregated, uni- to multilocular, ostiolate, covering by brown, warty hyphae. Pycnidial walls composed of 3–6 layers of brown cells, of textura angularis, hyaline towards inner layers. Conidiophores reduced to conidiogenous cells. Conidiogenous cells phialidic with periclinal thickening, tightly aggregated, hyaline, subcylindrical, aseptate, smooth. Conidia hyaline, ellipsoidal to tear-drop-shaped, aseptate, smooth-walled, with guttutes. Hyphomycetous: Mycelium hyaline to pale brown, smooth-walled, aseptate to septate, branched hyphae, giving rise to conidiophores. Conidiophores solitary, micronematous to macronematous, straight to flexuous, cylindrical, unbranched or branched, hyaline to brown, smooth- to rough-walled, with finely verruculose, 1–2-septate or multi-septate. Conidiogenous cells terminal and lateral or integrated, occassionally polyblastic with 1–3 phialidic loci, hyaline to medium brown, subcylindrical to clavate, smooth- to rough-walled, with finely verruculose, subdenticulate with minute collarette, thickened scars along the rim at times. Conidia brown to medium brown, fusoid-ellipsoidal to subcylindrical, with truncate base, with or without thickened, darkened hilum, aseptate to multi-septate, verruculose to warty.
Type genus: Eriomyces Huanraluek, Thambugala & K.D. Hyde
Notes: Eriomycetaceae is introduced to accommodate the sexual and asexual monotypic genera which were previously treated in Dothideomycetes, genera incertae sedis. Five genera including Eriomyces (generic type), Funbolia, Heleiosa, Phellinocrescentia and Pseudopassalora are accommodated in this family based on phylogenetic analysis. These genera are represented by their type strains and form a well-resolved clade in our phylogenetic analyses with high support (98% ML, 1.00 BYPP; Fig. 93). Eriomyces and a sister genus Heleiosa are represented by their sexual morphs. These two genera share a similar morphology in having subglobose to ampulliform, unilocular, coriaceous ascomata, immersed in a clypeus, with light coloured ascomata, but having heavily pigmented cells at the base of ostiolar canal (Kohlmeyer et al. 1996). Eriomyces differs from Heleiosa in their asci and ascospores. Eriomyces has polysporous asci, with oblong to allantoid, aseptate ascospores, without a mucilagenous cap at each end. Heleiosa has 4–8-spored asci, with ellipsoidal to fusiform, 1-septate ascospores, with cilia-like subapical appendages at both ends (Kohlmeyer et al. 1996). Funbolia and Pseudopassalora have hyphomycetous asexual morphs, with brown conidia (Crous et al. 2011a, c). Funbolia differs from Pseudopassalora in having terminal and lateral, pale to medium brown conidiogenous cells, somewhat swollen, clavate to irregular, with aggregated loci in a rachis and dimorphic, 1- to multi-septate conidia. Pseudopassalora has integrated conidiogenous cells, reduced to loci on hyphae, or terminal on short conidiophores. The conidiogenous cells are cylindrical, hyaline, smooth, containing a solitary terminal, truncate locus, or polyblastic with 1–3 phialidic loci and ascospores are fusoid-ellipsoidal, aseptate or 1-septate (Crous et al. 2011a, c). Phellinocrescentia differs from other genera in this family in having a coelomycetous asexual morph. Based on phylogenetic analyses (Fig. 93), we tentative place these genera in the new family Eriomycetaceae pending further studies.
Eriomyces Huanraluek, Thambugala & K.D. Hyde, gen. nov.
Index Fungorum number: IF556538; Facesoffungi number: FoF 06151
Etymology: In honour of Erio Camporesi, a great collector, recognizing his invaluable contribution to mycology.
Saprobic on dead twigs of Hevea brasiliensis Müll. Arg. Sexual morphAscomata immersed beneath a raised dark brown to black pseudoclypeus, papilla erumpent through host surface, solitary or aggregated, scattered, black, coriaceous, unilocular, subglobose to ampulliform, or conical, with a raised neck. Ostioles central, papillate, periphysate. Peridium comprising several layers of heavily pigmented to hyaline, thick-walled, cells of textura epidermoidea. Hamathecium composed of branched or unbranched, cellular pseudoparaphyses. Asci polysporus, bitunicate, cylindrical to cylindric-clavate, with a long to short, furcate pedicel, apically broadly rounded. Ascospores overlapping, crowded, oblong to allantoild, aseptate, straight to slightly curved, hyaline. Asexual morph Undetermined.
Type species: Eriomyces heveae Huanraluek, Thambugala & K.D. Hyde
Notes: Eriomyces is introduced to accommodate a novel species from Hevea brasiliensis (rubber) in Thailand. Molecular phylogenetic analysis of LSU sequence data (Fig. 93) shows that Eriomyces heveae (MFLUCC 17-2232) clusters in Dothideomycetes together with Heleiosa barbatula (JK 5548I), Phellinocrescentia guianensis (CBS 138913), Pseudopassalora gouriqua (CPC 1811 and CBS 101954) and Funbolia dimorpha Crous & Seifert (CPC 14170 and CBS 126491). Eriomyces heveae resides in this clade as a sister species to Heleiosa barbatula.
Eriomyces heveae Huanraluek, Thambugala & K.D. Hyde, sp. nov.
Index Fungorum number: IF556539; Facesoffungi number: FoF 06152; Fig. 92
Eriomyces heveae (MFLU 18-0758, holotype). a Appearance of ascomata on host surface. b, c Vertical sections through ascomata. d Vertical sections through the apex of ascoma. e Peridium. f Pseudoparaphyses. g–j Asci. k–o Ascospores p Germinating ascospore. Scale bars: b, c = 50 µm, d, e = 25 µm, f–j = 20 µm, p = 10 µm, k–o = 5 µm
Etymology: The species epithet “heveae” refers to the host genus Hevea on which holotype occurs.
Holotype: MFLU 18-0758
Saprobic on dead twigs of Hevea brasiliensis Müll. Arg. Sexual morphAscomata 240–380(–445) μm high × 215–325 μm diam., immersed beneath a raised dark brown to black pseudoclypeus, papilla erumpent through host surface, solitary or aggregated, scattered, black, coriaceous, unilocular, subglobose to flask-shaped, or conical, with a raised neck. Ostiole central, papillate, periphysate. Peridium 30–56 μm wide, comprising several layers of heavily pigmented to hyaline, thick-walled, cells of textura epidermoidea, fusing at the outside with the host cells. Hamathecium composed of 1–3 μm wide, numerous, rarely septate, branched or unbranched, cellular pseudoparaphyses. Asci 52–102 × 9–13 μm (\( \bar{x} \) = 80 × 10.2 μm, n = 20), poly-spored, bitunicate, cylindrical to cylindric-clavate, with a long to short, narrow, thin-walled, furcate pedicel, apically broadly rounded. Ascospores 4.5–6.5 × 1–2 μm (\( \bar{x} \) = 5.5 × 1.4 μm, n = 50), crowded, ellipsoidal, aseptate, straight to slightly curved, hyaline, smooth-walled, with bipolar guttules. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on PDA within 24 h and germ tubes produced from one end. Colonies growing on PDA reaching 25 mm diam. after 12 days at 25 °C, circular, falt, slightly dense, surface white to iron-grey, reverse brownish to smoke-grey, white at the margin, smooth surface with entire to slightly curled edge.
Material examined: THAILAND, Sukhothai Province, Si Satchanalai District, on dead twigs of Hevea brasiliensis (Euphorbiaceae), 2 January 2017, N. Huanraluek, Sh1 (MFLU 18-0758, holotype); ibid., KUN-HKAS 97497, isotype, ex-type living culture MFLUCC 17-2232.
GenBank numbers: ITS = MH118116, LSU = MH109524.
Notes: Based on a megablast search of NCBIs GenBank nucleotide database using LSU sequences of strain MFLUCC 17-2232, the closest hit was Pseudopassalora gouriqua (CPC 1811; 95.47% similarity), followed by Heleiosa barbatula (JK 5548I; 95.70% similarity). In the present phylogeny (Fig. 93), Eriomyces heveae shows a close affinity to Heleiosa barbatula. Nevertheless, Eriomyces heveae differs from Heleiosa barbatula in having poly-spored asci and hyaline, oblong to allantoid, aseptate ascospores (Kohlmeyer et al. 1996). In addition, H. barbatula was described from Juncus roemerianus Scheele (Juncaceae) in salt marshes of the USA east coast, while Eriomyces heveae was reported from Hevea brasiliensis in Thailand. Therefore, considering the distinct morphological traits and phylogeny E. heveae is introduced as a new species. Ascospores of Eriomyces morphologically resemble those of Cytospora Ehrenb. and Eutypella (Nitschke) Sacc. in Sordariomycetes.
Phylogram generated from maximum likelihood analysis based on LSU sequence data representing Eriomyces (Dothideomyetes, families incertae sedis) and the closely related orders in Dothideomyetes. Related sequences are taken from Suetrong et al. (2009). Forty-seven strains are included in the combined analyses which comprise 886 characters after alignment. Sordaria fimicola (CBS 508.50) (Sordariomycetes) is used as the outgroup taxon. The best RAxML tree with a final likelihood values of − 7924.174629 is presented. Estimated base frequencies were as follows: A = 0.245455, C = 0.227992, G = 0.318413, T = 0.208140; substitution rates AC = 0.803611, AG = 2.255570, AT = 0.841920, CG = 1.068389, CT = 8.568242, GT = 1.000000. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. Bootstrap values for maximum likelihood (ML) equal to or greater than 70% and clade credibility values greater than 0.90 (the rounding of values to 2 decimal proportions) from Bayesian-inference analysis labeled on the nodes. Ex-type strains are in bold and black, the new isolate is indicated in bold and blue
Lecanoromycetes O.E. Erikss. & Winka
Notes: Lecanoromycetes is the largest class of lichenized fungi which comprises 15,131 species and 701 genera including two major subclasses Lecanoromycetidae and Ostropomycetidae (Lücking et al. 2017; Kraichak et al. 2018). This is the most evolved fungal class within Ascomycota and distributed worldwide with major adaptive characters (Ekanayaka et al. 2017).
Subclass Lecanoromycetidae P.M. Kirk et al.
Caliciales Bessey
Notes: The order Caliciales was established by Bessey in 1907. The taxa are lichenized and rarely parasitic (Ekanayaka et al. 2017). This order comprises the families Caliciaceae and Physciaceae and so far, 1276 species have been accepted (Lücking et al. 2017).
Caliciaceae Chevall.
Notes: The lichenized family Caliciaceae was introduced by Chevallier (1826) to accommodate lichenized and lichenicolous species (Jaklitsch et al. 2016). Presently this family comprises 35 genera (Lücking et al. 2017; Wijayawardene et al. 2017a). The taxa are characterised by producing prototunicate asci and a mazaedium (Prieto and Wedin 2017). The delimitation of this family remains uncertain (Aguirre-Hudson et al. 2007).
Pyxine Fr.
Notes: Pyxine was introduced by Fries (1825) with the type species P. sorediata (Ach.) Mont. (Yang et al. 2019b). Presently this genus comprises 75 species that occur on bark, wood, mosses or rocks (Mongkolsuk et al. 2012). Species in this genus are mainly distributed from pantropical to subtropical and few species extend to temperate or oceanic regions (Moberg 1983; Kalb 1987; Wei and Hur 2007; Elix 2009; Mongkolsuk et al. 2012; Yang et al. 2019b). Previously this genus was placed in the family Physciaceae. However, later it was moved to the family Caliciaceae (Wedin and Grube 2002; Crespo et al. 2004; Gaya et al. 2012; Prieto and Wedin 2017; Yang et al. 2019b). Taxa in this genus are characterised by adnate foliose thallus, dark brown hypothecium and generally two-celled, brown ascospores (Awasthi 1982; Kalb 1987, 2004; Elix 2009; Yang et al. 2019b). Molecular phylogenetic studies are lacking for this genus (Schmull et al. 2011; Gaya et al. 2012; Prieto and Wedin 2017; Yang et al. 2019b). We report a new record of Pyxine berteriana on dead bark from Yunnan, China for the first time.
Pyxine berteriana (Fée) Imshaug, Trans. Am. Microsc. 76: 254 (1957)
Facesoffungi number: FoF 07199; Fig. 94
Thallus corticolous, upper surface grey to greyish green, photobiont detected. Sexual morphAscomata apothecial, approximately 300–800 μm diam., black, circular to ellipsoidal, adnate, slightly erumpent from the thallus. Epihymenium 10–35 μm, olivaceous brown, Hymenium 65–100 μm, hyaline. Pseudoparaphyses 1–1.5 μm wide, brown at the tips, densely arranged, tip swollen with pigments, 0.2–0.8 μm wide, Hypothecium distinct, 40–110 μm height, dark brown. Asci 45–65 × 12–20 μm (\( \bar{x} \) = 55 × 16 μm, n = 40), 8-spored, bitunicate, fissitunicate, brown, clavate to obovoid, concave, tip blunted, narrowing towards the apex, poorly developed pedicel. Ascospores 12–20 × 4–7 μm (\( \bar{x} \) = 16 × 5.5 μm, n = 40), crowded, brown, smooth-walled, fusoid to obovoid, ends remaining cone-shaped, 0–1-septate, with large guttules, not constricted at the septa. Asexual morph Undetermined.
Material examined: CHINA, Yunnan Province, Baoshan, N 25° 16′ 19.3″, E 099° 18′ 47.8″, on dead bark of unidentified plant, 19 September 2018, V. Thiyagaraja, D4B1 (MFLU 19-0720).
GenBank numbers: ITS = MN792989, mtSSU = MN792788.
Notes: We followed the latest treatment and updated accounts of genus Pyxine in Yang et al. (2019b). Our specimen was collected from dead bark, while two other specimens (P. berteriana KUN 15-47921 and KUN 14-43730) grow on rock. A nucleotide base comparison of ITS gene region reveals 13/554 (2.3%) differences between our strain and P. berteriana (KUN 14 43730, KUN 15 47921). However, no significant differences were observed in the morphological comparison among these species. Phylogenetic analyses of a combined ITS and mtSSU sequence dataset reveal our strain clusters with P. berteriana (KUN 15-47921 and KUN 14-43730) with high statistical support (100% ML, 1.00 BYPP; Fig. 95). Therefore, we introduce P. berteriana (MFLU 19-0720) as a new record on dead bark in China.
Phylogram generated from maximum likelihood analysis based on combined ITS and mtSSU sequence dataset for Pyxine. Related sequences were referred to Yang et al. (2019b). Fourty-two strains are included in the combined genes sequence analyses which comprise total 1306 characters (519 characters for ITS, 787 characters for mtSSU) after alignment. Dirinaria applanata (MAF 9854 and MP48) (Caliciaceae, Caliciales) is used as the out group taxon. Single gene analyses were carried out and the topology of each tree with clade stability. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best sorting RAxML tree with a final likelihood value of − 7484.097920 is presented. The matrix had 517 distinct alignment patterns, with 34.16% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.290525, C = 0.207362, G = 0.235519, T = 0.266594; substitution rates AC = 1.131603, AG = 2.147524, AT = 1.030739, CG = 0.767273, CT = 3.756758, GT = 1.000000; gamma distribution shape parameter α = 0.418791. Bootstrap values for maximum likelihood (ML) equal to or greater than 70% and clade credibility values greater than 0.95 (the rounding of values to 2 decimal proportions) from Bayesian-inference (BI) analysis labeled on the nodes. The newly generated sequences are indicated in bold and blue
Class Leotiomycetes O.E. Erikss. & Winka
Notes: The class Leotiomycetes was erected when the superclass Leotiomyceta was split into seven classes by Eriksson and Winka (1997) with inoperculate discomycetes. Fungi in this class are ecologically diverse, including saprobes, endophytes, plant and mammalian pathogens, aquatic and aero-aquatic hyphomycetes, mycorrhizae, fungal parasites, root symbionts, and wood rot fungi (most are saprobes of a wide variety of substrates) (Wang et al. 2006a, b; Lantz et al. 2011; Takamatsu et al. 2015; Jaklitsch et al. 2016; Ekanayaka et al. 2019; Johnston et al. 2019). They are characterised by open apothecial fruiting bodies and unitunicate asci generally opening by apical perforation or pore to release their ascospores and are often referred to as “inoperculate discomycetes” (Nannfeldt 1932; Dennis 1968; Korf 1973; Pfister and Kimbrough 2001). Recent studies by Ekanayaka et al. (2019) and Johnston et al. (2019) have provided a backbone tree based on multi-gene analysis and morphological characteristics of this class.
Helotiales Nannf.
Notes: Helotiales was established by Nannfeldt in 1932 and was replaced with Leotiales by Carpenter in 1988. However, currently, Helotialessensu stricto and Leotialessensu stricto are recognised as two separate orders. Leotiales comprises only the Leotiaceaesensu stricto (Korf and Lizon 2000, 2001).
Heterosphaeriaceae Rehm
Notes: Heterosphaeriaceae is typified by Heterosphaeria Grev. and is the only representative genus of the family (Rehm 1888). The family can be found as saprobes on dead or dried plant material (Leuchtmann 1987). Nannfeldt (1932) placed the family in Helotiales and is unique in having a very thick, interwoven excipulum, cartilaginous hyphae, and isodiametric, and dark celled ascospores (Nannfeldt 1932).
Heterosphaeria Grev.
Notes: Heterosphaeria is typified by H. patella (Tode) Grev., the common strain that was found associated with the plant family Apiaceae. The genus was described as Sphaeria penetrans-patella (Tode 1791), however, Heterosphaeria is distinguishable in having globose apothecia when immature, becoming patelliform, depressed at maturity, with a coriaceous excipulum (Leuchtmann 1987). The asexual morph characters were described as Heteropatella Fuckel (Tulasne and Tulasne 1865), however, Jaklitsch et al. (2016) suggested that Heteropatella is a synonym of Heterosphaeria.
Heterosphaeria patella (Tode) Grev., Scott. crypt. fl. (Edinburgh) 2: 103 (1823)
Facesoffungi number: FoF 07095; Fig. 96
Heterosphaeria patella (MFLU 18-1491). a Apothecia on wood. b Close up of apothecia. c Vertical cross section. d Part of a cross section. e Filiform paraphyses. f Cylindrical asci in Melzer’s reagent. g–i Cylindrical asci. j J + , apical ring in Melzer’s reagent. k, l Ellipsoidal ascospores. Scale bars: a = 2000 µm, b = 100 µm, c = 50 µm, d = 20 µm e, j = 10 µm, f–i = 5 µm, k, l = 3 µm
≡ Sphaeria penetrans a patella Tode, Fung. mecklenb. sel. (Lüneburg) 2: 45 (1790)
Neotype: GERMANY, Altburg, Regensberg on a Daucus carota, 7 August 1987, leg. A.L. Leuchtmann 9634 (ZT 9634).
Saprobic on dead stem. Sexual morphApothecia 275–923 × 238–656 µm (x̄ = 585 × 487.5 µm, n = 10), when dry arising solitary, scattered on wood, sessile, superficial, deep cupulate, black, rough surface. Receptacle concave. Disc flat, black. Margin slightly curved, black. Hymenium inner mixed with asci and paraphyses, hyaline. Subhymenium forming a hyaline, small-celled tissue. Ectal excipulum 34.5–49 µm (x̄ = 41.5 µm, n = 10), composed of hyaline cells of textura globulosa, 4–6 layers, thin-walled cells. Medullary excipulum 58–157 µm (x̄ = 90 µm, n = 10), composed with hyaline cells of textura intricata, thick-walled. Paraphyses 1–2 µm (x̄ = 1.5 µm, n = 20), long-thin form, filiform, 1–3-septate, thin-walled, hyaline, swollen at the apex. Asci 55–65 × 6.5–9 µm (x̄ = 59 × 7.5, n = 20), 8-spored, unitunicate, inoperculate, cylindric-clavate, trapezoidal apex, thin-walled, blunt end, J + , ring. Ascospores 10–12 × 3–5 µm (x̄ = 10.5 × 4 µm, n = 10), 1-seriate overlapping, arranged in a whole length of asci, fusiform, ellipsoidal, hyaline, thin-walled, large oil drop at both ends, aseptate or 1-septate, smooth-walled. Asexual morph Undetermined.
Material examined: ITALY, Forlì-Cesena (FC), Corniolo, Santa Sofia, on dying stems of Anthriscus cerefolium (Apiaceae), 20 July 2018, E. Camporesi, IT 3978. (MFLU 18-1491)
GenBank numbers: ITS = MN860230, LSU = MN860231.
Notes: The phylogenetic results from different analysis (ML, MP and BI) show the same topology of Heterosphaeria patella strain MFLU 18-1491 that grouped as sister to H. patella (G.M. 2014-08-04-1) with low support in ML and MP analyses (51% ML, 58% MP; Fig. 97) but strong support in BI analysis (0.99 BYPP; Fig. 97). Heterosphaeria patella strain MFLU 18-1491 was found on Anthriscus cerefolium. This is the first report on this host. The morphology of our strain is very similar to H. patella in having ectal excipulum of cells of textura globulosa, 1–3-septate on paraphyses swollen at the apex, cylindric-clavate with J + apical ring and ellipsoidal, aseptate or 1-septate ascospores with large guttules at both ends. There are some different morphological features between our collection and H. patella (Leuchtmann 1987). Our strain has a medullary excipulum of cells of textura intricata, while H. patella (Leuchtmann 1987) has textura porrecta. Heterosphaeria patella (MFLU 18-1491) is 100% identical with H. patella (G.M. 2014-08-04-1) both in LSU and ITS. Therefore, we report a new host record and update sequence data for this species.
Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence dataset for family Heterosphaeriaceae and the closely related families in Helotiales. Sixteen strains are included in the combined genes sequence analyses which comprise total 1718 characters (1186 characters for LSU, 529 characters for ITS) after alignment. Blumeriella hiemalis CBS 146.35 and Thedgonia ligustrina CBS 547.71 (Drepanopezizaceae, Helotiales) are used as the outgroup taxa. Single gene analyses are carried out and the topology of each tree had clade stability. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best sorting RAxML tree with a final likelihood value of − 3604.645390 is presented. The matrix had 172 distinct alignment patterns, with 18.21% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.239, C = 0.235, G = 0.296, T = 0.230; substitution rates AC = 0.705719, AG = 1.044963, AT = 0.537347, CG = 0.872609, CT = 3.251127, GT = 1.000000; gamma distribution shape parameter α = 0.33. Bootstrap values for maximum likelihood (ML) and maximum parsimony (MP) equal to or greater than 50% and clade credibility values greater than 0.95 (the rounding of values to 2 decimal proportions) from Bayesian-inference analysis labeled on the nodes as ML/BYPP/MP. The newly generated sequences are indicated in bold and blue
Ploettnerulaceae Kirschst.
Notes: Ploettnerulaceae is typified by Ploettnerula Kirschst. that are mostly saprobic or pathogenic on plants. Some taxa i.e. Cadophora Lagerb. & Melin, Oculimacula Crous & W. Gams cause stem rot and wood decay on soybean or kiwifruit plants (Von Matheis 1977; Meyer and Luttrell 1986, 1987; Travadon et al. 2015; Vera and Murray 2016). Taxa in Ploettnerulaceae are characterised by apothecial ascomata that can be immersed or erumpent (Kirschstein 1924), cupulate, discoid or urn-sharped, sessile or substipitate. The ectal excipulum has pigmented cells of textura globulosa to angularis and medullary excipulum in ectal excipulum, hyaline cells of textura prismatica. Asci are 8-spored with a J + , ring at the apex and 0–3-septate, ellipsoid to long filliform ascospores with guttules (Kirschstein 1924; Smerlis 1966; Von Matheis 1977; Meyer and Luttrell 1986; Vera and Murray 2016). Asexual morphs of the family can be hyphomycetous or coelomycetous. Conidiophores are phialidic and hyaline to brown. Conidia are ellipsoid to rod-shaped or filliform and 0–1-septate (Marvanová and Bärlocher 2001; Goodwin 2002; Gönczöl and Révay 2003; Gramaje et al. 2011; Goncalves et al. 2012; King et al. 2013; Travadon et al. 2015; Duarte et al. 2016; Walsh et al. 2018). Currently, this family comprised 15 genera (Ekanayaka et al. 2019).
Rhexocercosporidium U. Braun
Notes: The genus typified by Rhexocercosporidium carotae (Årsvoll) U. Braun consists of soil-borne pathogens causing black spots on carrot (Daucus carota L.) roots and leaves (Årsvoll 1965) and ‘rusty root’ on ginseng plants (Panax quinquefolius L.) (Hildebrand 1935). Rhexocercosporidium carotae was described as Acrothecium carotae (Årsvoll 1965) and then de Hoog and Van Oorschot (1985) transferred it to Pseudocercosporidium Deighton based on its rhexolytic spore secession. However, it does not match the generic description of Pseudocercosporidium (Deighton 1973). Braun (1994) suggested that P. carotae should be separated from Pseudocercosporidium and introduced Rhexocercosporidium to accommodate this species. Based on the characteristics of the asexual morph, Shoemaker et al. (2002) classified Rhexocercosporidium in Leotiomycetes. Rhexocercosporidium presently comprises R. carotae and R. panacis Reeleder, only known from their asexual morph. The genus is characterised by different conidiophores from its vegetative hyphae and oboviod to cylindrical conidia with rhexolytic spore secession (Braun 1994; Shoemaker et al. 2002).
Rhexocercosporidium microsporum (Ekanayaka & K.D. Hyde) Phutthacharoen & K.D. Hyde, comb. nov.
Index Fungorum number: IF556282; Facesoffungi number: FoF 07523
≡ Cadophora microspora Ekanayaka & K.D. Hyde, in Ekanayaka et al., Mycosphere 10(1): 10.5943/mycosphere/10/1/7, [310–489] (2019)
Holotype: UK, Calbourne Stream, Isle of Wight, on Apiaceae stem, 11 May 2015, E.B.G. Jones, GJ 154b (MFLU 18-2672).
Description and illustration: See Ekanayaka et al. (2019).
GenBank numbers: ITS = MK584939, LSU = MK591966.
Notes: Rhexocercosporidium microsporum was introduced by Ekanayaka et al. (2019) as Cadophora microspora Ekanayaka & K.D. Hyde. We noted the strain of the Cadophora microspora does not group with the generic type of Cadophora in phylogenetic analyses (Fig. 99) and it has a close phylogenetic affiliation with Rhexocercosporidium. Morphology of R. microsporum is similar to Rhexocercosporidium sp. (MFLU 17-1594; Phutthacharoen et al. in prep.) in their apothecial, excipulum, asci and ascospores characteristics (Ekanayaka et al. 2019). Therefore, based on morpho-phylo evidence, we believe it is taxonomically correct to transfer Cadophora microspora to Rhexocercosporidium.
Rhexocercosporidium senecionis Phutthacharoen & K.D. Hyde, sp. nov.
Index Fungorum number: IF556988; Facesoffungi number: FoF 06820; Fig. 98
Rhexocercosporidium senecionis (MFLU 17-1635, holotype). a Fresh apothecia on wood. b Close up of apothecia. c, d Part of a cross section of apothecium. e, f Cylindrical septate hair. g Filiform paraphyses. h Cylindrical asci in Melzer’s reagent. i–l Cylindrical asci. m J + ring in Melzer’s reagent. n Fusoid ascospores. Scale bars: a, b = 100 µm, c, d = 50 µm, e–g = 20 µm, h–l = 10 µm, m = 5 µm, n = 3 µm
Etymology: The specific epithet “senecionis” refers to the plant genus Senecio.
Holotype: MFLU 17-1635
Saprobic on dead stems. Sexual morphApothecia 150–300 × 100–200 µm (\( \bar{x} \) = 222.3 × 159 µm, n = 10), when dry arising singly or gregarious scattered on wood, sessile, rough surface, erumpent, black, with black setae surrounded. Receptacle concave. Disc flat, black. Margins light brown rounded, rough. Hairs up to 30–60 µm length (\( \bar{x} \) = 43.3 µm, n = 10), hyaline and dark brown, composed of thick-walled with septate. Ectal excipulum 30–50 µm (\( \bar{x} \) = 39.3 µm, n = 10) composed of thin-walled, 4–5 cell layers, lighter inwards, composed of cells of textura angularis. Medullary excipulum 15–25 µm (\( \bar{x} \) = 20.1 µm, n = 10), composed with hyaline cells of textura epidermoidea. Hymenium hyaline, inter mixed with asci and paraphyses. Paraphyses 2–3 µm wide (\( \bar{x} \) = 2.4 µm, n = 20), filiform, thin-walled, blunt end, aseptate. Asci 30–50 × 5–10 µm (\( \bar{x} \) = 39.5 × 7.5 µm, n = 20), 8-spored, unitunicate, inoperculate, cylindrical, thin-walled, short pedicellate, blunt apices, J + . Ascospores 5–10 × 2–3 µm (\( \bar{x} \) = 8.4 × 2.8 µm, n = 20), multi-seriate, fusiform to filiform, hyaline with 2–3 guttules, aseptate. Asexual morph Undetermined.
Material examined: ITALY, Forlì-Cesena (FC), Campigna, Santa Sofia, on dying stems of Senecio nemorensis (Asteraceae), 3 September 2017, E. Camporesi, IT 3478b (MFLU 17-1635, holotype).
GenBank numbers: ITS = MN688635, LSU = MN688633.
Notes: Phylogenetically our novel strain MFLU 17-1635 has a strong sister relationship to Pirottaea senecionis (CBS 307.58), but this latter strain is not related to any type material. Thus we introduce our collection as a new species. Further fresh collections of Pirottaea senecionis are needed to confirmed their actual phylogenetic placement with DNA based sequence data. Rhexocercosporidium senecionis is distinct from other Rhexocercosporidium taxa by its apothecia that are surrounded by black setae and a light brown margin. Rhexocercosporidium sp. (MFLU 15-2755) also has a close phylogenetic relationship to Rhexocercosporidium senecionis (Fig. 99). But, it has a different medullary excipulum. Rhexocercosporidium senecionis has textura epidermoidea in the medullary excipulum while Rhexocercosporidium sp. (MFLU 15-2755) has textura oblita. Rhexocercosporidium sp. (MFLU 16-0559) also clusters with other taxa in Rhexocercosporidium. It differs from R. senecionis by the features of medullary excipulum and the paraphyses that are swollen at the apex. Ascospores of MFLU 16-0559 are ellipsoidal and smooth whereas R. senecionis has fusiform to filiform ascospores with several guttules. Two other species of Rhexocercosporidium (R. carotae and R. panacis) are known only from their asexual morph and a comparison with R. senecionis is impossible.
Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence data representing family Ploettnerulaceae and the closely related families in Helotiales. Sixty-two strains are included in the combined analyses which comprise 1523 characters (858 characters for LSU, and 665 characters for ITS) after alignment. Polydesmia pruinosa (CBS 111545) and Polydesmia pruinosa (TNS-F12764) in Amorphothecaceae (Erysiphales) are used as the outgroup taxa. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RAxML tree with a final likelihood values of − 9306.895864 is presented. The matrix had 627 distinct alignment patterns, with 25.84% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.242172, C = 0.226812, G = 0.281300, T = 0.249716; substitution rates AC = 3.027442, AG = 3.912241, AT = 1.864690, CG = 0.983026, CT = 9.977836, GT = 1.000000; gamma distribution shape parameter α = 0.496928. Bootstrap values for maximum likelihood (ML) equal to or greater than 50% and clade credibility values greater than 0.90 (the rounding of values to 2 decimal proportions) from Bayesian-inference analysis labeled on the nodes. Ex-type strains are in bold and black, the new isolate is indicated in bold and blue
Vibrisseaceae Korf
Notes: Korf erected Vibrisseaceae in 1990 in order to accommodate the genera Vibrissea Fr., Chlorovibrissea L.M. Kohn and Leucovibrissea (A. Sánchez) Korf. Members of the family are characterised by aquatic or semi-aquatic habitat, sessile to long stipitate apothecia, disc colour ranging from whitish, yellowish, brownish, olivaceous to blackish green, an ectal excipulum composed of cells of textura globosa, textura angularis to textura prismatica and filiform ascospores (Korf 1990). Phialocephala and Acephala are the two asexual genera of Vibrisseaceae. A new genus, Srinivasanomyces is introduced based on its morphological distinctiveness supported by strong phylogenetic support.
Srinivasanomyces S. Rana & S.K. Singh, gen. nov.
MycoBank number: MB830717; Facesoffungi number: FoF 06119
Etymology: Generic epithet named after M.C. Srinivasan, who is a well-known mycologist of India, to commemorate his immense contribution in mycology.
Isolated from dead bark of Prunus cerasoides. Sexual morph Undetermined. Asexual morphColonies flat, circular, margin irregular smooth, slightly cottony, slow growing. Hyphae simple to branched, sometimes in bundles, smooth-walled, subhyaline, septate. Chlamydospores abundant, solitary or in chains, globose to subglobose, smooth-walled, pigmented, subhyaline to light olivaceous. Produces multiple asexual morphs. Conidiophores produced from lateral hyphae, indeterminate, intercalary, simple to densely branched, septate, smooth-walled, subhyaline to light olivaceous. Conidiogenous cells subhyaline to light olivaceous, lateral to terminal, phialidic, simple to dentate, solitary to densely produced, monoblastic to polyblastic. Phialides variable, solitary or in dense clusters forming globose conidial heads, subhyaline to light olivaceous, smooth-walled, collarette, cylindrical to ampulliform. Conidia produced in gleosporic mass, sometimes directly from superficial lateral hyphae, subhyaline to light olivaceous, smooth-walled, pyriform to obpyriform, globose to subglobose, fusoid, clavate, solitary to catenate, with 1–2 guttules.
Type species: Srinivasanomyces kangrensis S. Rana & S.K. Singh
Notes: The proposed genus Srinivasanomyces morphologically resembles some features in Phialocephala. However, it differs in having variably-shaped conidia that are pyriform to obpyriform, globose to subglobose, fusoid, or clavate. It produces dense globose clusters of conidial heads and the conidiophores are formed in an indeterminate, intercalary, simple to dense globose to subglobose clustered mass. Srinivasanomyces differs from other genera based on phylogenetic analysis. In a megablast analysis, the ITS sequence of Srinivasanomyces was nearly 88% similar with known taxa. Phylogenetically, it formed a separate distinct clade with strong bootstrap support.
Srinivasanomyces kangrensis S. Rana & S.K. Singh, sp. nov.
MycoBank number: MB830718; Facesoffungi number: FoF 06589; Fig. 100
Srinivasanomyces kangrensis (AMH 10175, holotype). a Colony morphology on PDA (front view). b Reverse view of colony. c Vegetative hyphae and chlamydospores. d Conidia produced from lateral hyphae. e, f Conidiophores produced from lateral hyphae with conidia. g Arrow showing conidia produced from phialide. h Tufts of slimy gleosporic mass of conidia with densely produced phialides. i Magnified view of gleosporic mass of conidia (arrow showing reduced phialides). j Conidia (arrow shows truncate and oblique base). k SEM of hyphae. l SEM of magnified view gleosporic mass of conidia (arrow showing densely produced phialides). m SEM of tufts of slimy gleosporic mass of conidia. Scale bars: c–j = 20 μm, k–m = 5 μm
Etymology: The specific epithet refers to the place of collection
Holotype: AMH-10175
Isolated from dead bark of Prunus cerasoides. Sexual morph Undetermined. Asexual morphHyphae thin, simple to branched, sometimes forming bundles, smooth-walled, subhyaline, septate, sometimes constricted near septa, 0.9–3.1 µm (\( \bar{x} \) = 1.8 µm, n = 30) wide. Chlamydospores abundant, solitary or in chains, globose to subglobose, smooth-walled, pigmented, subhyaline to light olivaceous, 3.1–7.8 × 2.4–5.7 µm (\( \bar{x} \) = 5.2 × 3.9 µm, n = 30). Produces multiple morphs (synanamorphs) of Diplococcium and Phialocephala. Conidiophores produced from lateral hyphae, indeterminate, intercalary, simple to densely branched, septate, smooth-walled, subhyaline to light olivaceous. Conidiogenouscells subhyaline to light olivaceous, lateral to terminal, phialidic, densely produced, monoblastic to polyblastic. Phialides variable, solitary or in dense clusters forming globose conidial heads 12.3–45.9 × 8.1–37.6 µm (\( \bar{x} \) = 18.7 × 16.1 µm, n = 30), subhyaline to light olivaceous, smooth-walled, collarette, cylindrical to ampulliform, 3.7–58.5 × 0.8–3 µm (\( \bar{x} \) = 13.7 × 1.4 µm, n = 30). Conidia produced in gleosporic mass, sometimes directly from superficial lateral hyphae, width 1.9–2.3 µm (\( \bar{x} \) = 2.1, n = 10), subhyaline to light olivaceous, smooth-walled, pyriform to obpyriform, globose to subglobose, fusoid, clavate, solitary to catenate, 1–2 guttules.
Culture characteristics: Colonies on PDA reaching to 15–20 mm diam. after 3 weeks at 25 °C; colonies from above greyish white (7B1) to fawn/light brown (7D4), flat, circular, margin irregular smooth, slightly cottony; colony from reverse, dark brown (8F4).
Material examined: INDIA, Himachal Pradesh, Kangra District, Simbal (31.9754 N” 76.6507 E”), from dead bark of Prunus cerasoides, 10 April 2018, S. Rana, AMH 10175 (holotype), ex-type living culture, NFCCI 4505.
GenBank numbers: ITS = MK478471, LSU = MK478470.
Notes: Srinivasanomyces kangrensis differs from other taxa based on the sequence analysis as well as morphology (Figs. 100, 101). In the megablast analysis, an ITS sequence of S. kangrensis, was 88.03% (368/418) similar with 6 gaps (1.4%) with Phialocephala cladophialophoroides Madrid et al. (HM87, type), 87.63% (418/477) similar with 7 gaps (1.4%) with P. dimorphospora W.B. Kendr. (CBS 300.62, type), 87.86% (420/478) similar with 7 gaps (1.4%) with Acephala applanata Grünig & T.N. Sieber (109321, type) and 86.84% (416/479) similar with 9 gaps (1.87%) with A. macrosclerotiorum Münzenb. & Bubner (CBS 123555, type). Srinivasanomyces kangrensis differs from Phialocephala oblonga (C.J.K. Wang & B. Sutton) Tanney, Seifert & B. Douglas (Tanney et al. 2016) in having dense globose clusters of conidial heads, while in P. oblonga, conidia are produced laterally from compact cylindrical synnemata. Phialocephala dimorphospora (Kendrick 1961) produces conidiophores from lateral superficial hyphae forming penicillate heads, which bears numerous phialides, while the conidiophores of Srinivasanomyces kangrensis are indeterminate, intercalary, in simple to dense globose to subglobose clusters. In Phialocephala catenospora Tanney & B. Douglas (Tanney et al. 2016) dark brown septate conidia are produced from short lateral stalks in long branched catenate chains and from solitary or groups of phialides. Phialocephala nodosa Tanney & B. Douglas (Tanney et al. 2016) produces irregular dark brown microsclerotia. The phialides of P. aylmerensis Tanney & B. Douglas (Tanney et al. 2016) are produced as solitary loose clusters, while conidiophores of P. victorin Vujan. & St-Arn. (Vujanović et al. 2000) arise singly or in groups, are mono- to tripenicillate, and form terminally or laterally on the hyphae. Proliferated phialides have an extremely expanded collarette.
Phylogram generated from Maximum likelihood analysis for Srinivasanomyces kangrensis NFCCI 4505 (AMH 10175) using combined ITS and LSU sequence data based on the Tamura-Nei model (Tamura and Nei 1993). Bootstrap support values are indicated at the nodes and values below 50% are not shown. Phylogenetics analyses were conducted in MEGA7 (Kumar et al. 2016). The novel taxon is shown in blue colour and type taxa used are depicted in bold
Class Pezizomycetes O.E. Erikss. & Winka
Notes: This class produces apothecial ascomata with a varity of forms (Ekanayaka et al. 2018), but the main feature of the class is operculate asci (Ekanayaka et al. 2018). The morphology of each family and detailed phylogeny were provided by Ekanayaka et al. (2018).
Pezizales J. Schröt.
Notes: As the single order in the class Pezizomycetes, Pezizales was separated to six clades by Ekanayaka et al. (2018) with each clade provided with a detailed molecular analysis (Ekanayaka et al. 2018).
Helvellaceae Fr.
Notes: This family was established by Fries (1822) and is typified by Helvella L. with Helvella mitra L. as type species. This family includes epigeous or hypogeous species. Ascomata are apothecial (Ekanayaka et al. 2018). Six genera are accepted within this family and four genera have available sequence data (Ekanayaka et al. 2018). Within Pezizomycetes, this family has a close relationship with Tuberaceae in phylogenetic tree (Ekanayaka et al. 2018)
Helvella L.
Notes: This is a well-known genus worldwide introduced by Linnaeus (1753). This genus is characterised by stipitate, cupulate, saddle-shaped, lobed to irregular apothecia, with a cream to black hymenium, glabrous, pubescent to villose receptacle surface, terete to sulcate, solid to hollow or lacunose stipe and uni-guttulate ascospores (Dissing 1966; Abbott and Currah 1997; Hyde et al. 2016; Zhao et al. 2016). One new Helvella species is established based on two combined genes in this study.
Helvella subtinta M. Zeng, Q. Zhao & K.D. Hyde, sp. nov.
Index Fungorum number: IF556789; Facesoffungi number: FoF 06591; Fig. 102
Etymology: The epithet refers to it is similar to Helvella tinta.
Holotype: KUN-HKAS 104666
Symbiotic in the coniferous forests. Sexual morphApothecia semi-hypogeous, scattered to gregarious. Pileus 0.5–2.5 cm high, 1–5 cm broad, cupulate, black, margin cracked slightly, hymenium glabrous, dark brown to black. Receptacle surface black, pubescent. Stipe 2–3 cm long, 0.5–1 cm broad, black, externally sulcate. Medullary excipulum 150–230 µm broad, of textura intricata, hyaline, composed of 3–4 µm broad hyphae. Ectal excipulum 50–120 µm broad, of textura angularis, composed of 18–23 × 11–15 internal cells, hyaline to brownish, 20–28 × 12–14 μm elongated terminal cells, brown, outermost cells catenuliform in long fascicled tufts. Stipitipellis 40–100 µm, of textura angularis, comprised of 12–17 × 9–12 μm, hyaline to brownish, internal cells, terminal cells 19–26 × 9–13 μm, brown, elongated. Asci 208–262 × 13–16 µm, 8-spored, pleurorhynchous, subcylindrical to clavate, J-, apical ring. Paraphyses 3–4 µm broad in the middle, filiform, exceeding the asci, apex enlarged, 4–6 µm broad, brown to dark brown, J-. Ascospores [20/1/1, in H2O] (14.6–)15.3–18.1(–19.6) × (10–)10.2–11.8(–12.5) (Q = 1.32–1.81, Q = 1.52 ± 0.14), 1-seriate, ellipsoid, smooth-walled with a large oil droplet. Asexual morph Undetermined.
Material examined: CHINA, Yunnan Province, Shangri-La, on sand soil in the coniferous forests, alt. 4131 m, 16 August 2018, M. Zeng, ZM 34 (KUN-HKAS 104666, holotype; MFLU 19-1557, isotype).
GenBank number: ITS = MN449991, LSU = MN449992.
Notes: This species is characterised by its semi-hypogeous, black cupulate apothecia, black hymenium, sulcate stipe, dark brown paraphyses and pleurorhynchous asci. The phylogenetic tree (Fig. 103) indicates a closed relationship with Helvella tinta Q. Zhao, B. Feng & K.D. Hyde. Helvella tinta has cupulate to irregularly cupulate pileus with white to smoky grey stipe and mottled hymenium with patches of paler pigmentation hymenium (Hyde et al. 2016). Across the LSU sequences, there are 52 notable nucleotides include 6 gaps differences among the 890 analysed nucleotides between H. tinta (HKAS 82560) and H. subtinta (HKAS 104666).
Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence dataset for Helvella species. Related sequences are taken from Hyde et al. (2016) and Zhao et al. (2016). Fifty-one strains are included in the combined analyses which comprise 1090 characters (551 characters for LSU, 539 for ITS) after alignment. Wynnella subalpina (HKAS45750) is used as the outgroup taxon. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RAxML tree with a final likelihood values of − 7795.733971 is presented. The matrix had 495 distinct alignment patterns, with 29.28% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.225857, C = 0.272487, G = 0.320790, T = 0.180866; substitution rates AC = 1.046286, AG = 1.656238, AT = 0.972263, CG = 0.793285, CT = 2.850648, GT = 1.000000; gamma distribution shape parameter α = 0.290387. Bootstrap values for maximum likelihood (ML) equal to or greater than 70% and clade credibility values greater than 0.90 (the rounding of values to 2 decimal proportions) from Bayesian-inference analysis labeled on the nodes. The new species are indicated in blue
Pyronemataceae Corda
Notes: This family is the largest of the families within Pezizomycetes and it has various ascomata (Hansen et al. 2013; Pfister 2015). In micromorphological, species have J- asci, smooth to ornamented ascospores with diversiform shapes (Pfister 2015; Ekanayaka et al. 2018). Unfortunately, within this family, approximately half genera are lack of molecular data (Ekanayaka et al. 2018).
Wilcoxina Chin S. Yang & Korf
Notes: This genus was introduced by Yang and Korf (1985). The genus is characterised by small sessile cupulate apothecia with brown hairs, white to orange brown hymenium, hyaline to yellowish ascospores (Yang and Korf 1985; Shi et al. 2016). Four species are accepted in the genus, Wilcoxina alaskana Kempton, Chin S. Yang & Korf, W. mikolae (Chin S. Yang & H.E. Wilcox) Chin S. Yang & Korf, W. rehmii Chin S. Yang & Korf and W. sequoiae (W. Phillips) T. Schumach.
Wilcoxina verruculosa M. Zeng, Q. Zhao & K.D. Hyde, sp. nov.
Index Fungorum number: IF556790; Facesoffungi number: FoF 06592; Fig. 104
Wilcoxina verruculosa (KUN-HKAS 104676, holotype). a–d Fruiting bodies visible in natural field. e Appearance of apothecia on host substrate. f Vetical section of apothecium. g Receptacle surface of pileus. h Asci and paraphyses. i–l Asci (Notes: k in congo red, l Asci in cotton blue). m–p Ascospores (Notes: o, p Ascospore in cotton blue). Scale bars: e = 500 μm, f = 300 μm, g–l = 50 μm, m–p = 50 μm
Etymology: The epithet refers to its ascospores with small warty.
Holotype: KUN-HKAS 104676
Saprobic on dead wood. Sexual morphApothecia scattered to gregarious, sessile to subsessile. Disc 1–2 μm high, 0.2–0.3 μm broad, cupulate, margin black brownish, with brown setaceous hairs. Hymenium glabrous, grey white. Receptacle surface brown, with brown setaceous hairs, 90–280 μm long, 7–13 μm broad in the middle, septate, tapering. Medullary excipulum 30–70 µm broad, of textura intricata, hyaline, composed of 2–4 µm broad hyphae. Ectal excipulum 66–162 µm broad, of textura angularis to textura globulosa, composed of 27–37 × 19–26 brown cells. Asci 222–251 × 12–15 µm, 8-spored, cylindrical to clavate, J-. Paraphyses 2–4 µm broad in the middle, filiform, septate, apex enlarged slightly, 3–6 µm broad, hyaline, J-. Ascospores [20/1/1, in H2O] (17.9–)18.5–20.3(–20.9) × (8–)8.4–9.9(–10.4) (Q = 1.8–2.4, Q = 2.12 ± 0.14), 1-seriate, ellipsoid, rough-walled with indistinguishable small warts, one big oil droplet in the centre. Asexual morph Undetermined.
Material examined: CHINA, Sichuan Province, S301, on dead wood, alt. 3559 m, 20 August 2018, M. Zeng, ZM 48 (KUN-HKAS 104676, holotype; MFLU 19-1558, isotype).
GenBank numbers: LSU = MN449993, TEF1-α = MN447536.
Notes: This species is characterised by its cupulate apothecia with setaceous hairs, greyish white hymenium, and rough ascospores with a single guttule. All other Wilcoxina species have smooth ascospores without oil (Yang and Korf 1985; Shi et al. 2016). Phylogenetic analyses of a combined LSU and TEF1-α sequence dataset revealed our new taxon clusters with Wilcoxina mikolae (Chin S. Yang & H.E. Wilcox) Chin S. Yang & Korf (strains WS 36, RMD 2144) and and W. verruculosa Chin S. Yang & Korf (strain HKAS 104676) with high support (98% ML, 1.00 BYPP; Fig. 105). Across the LSU sequences, there are 24 notable nucleotides include 1 gap difference among the 859 analysed nucleotides between W. mikolae (WS 36) and W. verruculosa (HKAS 104676).
Phylogram generated from maximum likelihood analysis based on a combined LSU and TEF1-α sequence dataset for species in Pyronemataceae. Related sequences are taken from Ekanayaka et al. (2018). Seventy-five strains are included in the combined analyses which comprise 2424 characters (1034 characters for LSU, 1389 characters for TEF1-α) after alignment. Otidea alutacea (KH.09.178) was used as the outgroup taxon. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RAxML tree with a final likelihood values of − 26615.652303 is presented. The matrix had 1310 distinct alignment patterns, with 41.94% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.247487, C = 0.234384, G = 0.282118, T = 0.236012; substitution rates AC = 1.099765, AG = 3.168904, AT = 2.408118, CG = 0.827304, CT = 8.780410, GT = 1.000000; gamma distribution shape parameter α = 0.255537. Bootstrap values for maximum likelihood (ML) equal to or greater than 70% and clade credibility values greater than 0.90 (the rounding of values to 2 decimal proportions) from Bayesian-inference analysis labeled on the nodes. The new species are indicated in blue
Class Sordariomycetes O.E. Erikss. & Winka
Notes: Sordariomycetes is the second largest classes of Ascomycota that comprises a diverse range of fungi characterised by perithecial ascomata and inoperculate unitunicate asci (Kirk et al. 2008; Maharachchikumbura et al. 2015). Species of Sordariomycetes are terrestrial, while some can be found in aquatic habitats (Samuels and Blackwell 2001; Jones et al. 2009). Sordariomycetes include plant pathogens, endophytes, saprobes, epiphytes, and fungicolous, lichenized or lichenicolous taxa (Hyde et al. 2014; Maharachchikumbura et al. 2016; Sun et al. 2019). The class Sordariomycetes comprises seven subclasses (Wijayawardene et al. 2018a). A monogrpah of the family was provided by Hyde et al. (2020b).
Subclass Diaporthomycetidae Senan. et al.
Diaporthales Nannf.
Notes: The order Diaporthales accommodates saprobic, endophytic or pathogenic species with abundant asexual morphs in nature (Castlebury et al. 2002; Rossman et al. 2007; Senanayake et al. 2017, 2018b). Based on recent fresh and herbarium morphology and multi-gene phylogeny, 30 families were accepted in Diaporthales viz. Apiosporopsidaceae Senan. et al., Apoharknessiaceae Senan. et al., Asterosporiaceae Senan. et al., Auratiopycnidiellaceae Senan. et al., Coryneaceae Corda, Cryphonectriaceae Gryzenh. & M.J. Wingf., Cytosporaceae Fr., Diaporthaceae Höhn., Diaporthosporellaceae C.M. Tian & Q. Yang, Diaporthostomataceae X.L. Fan & C.M. Tian, Dwiroopaceae K.V. Xavier et al., Erythrogloeaceae Senan. et al., Gnomoniaceae G. Winter, Harknessiaceae Crous, Juglanconidaceae Voglmayr & Jaklitsch, Lamproconiaceae C. Norphanphoun et al., Macrohilaceae Crous, Melanconidaceae G. Winter, Melanconiellaceae Senan. et al., Neomelanconiellaceae Crous, Phaeoappendicosporaceae Crous & M.J. Wingf., Prosopidicolaceae Senan. & K.D. Hyde, Pseudomelanconidaceae C.M. Tian & X.L. Fan, Pseudoplagiostomataceae Cheew. et al., Schizoparmaceae Rossman, Stilbosporaceae Link, Sydowiellaceae Lar.N. Vassiljeva, Synnemasporellaceae X.L. Fan & J.D.P. Bezerra, Tirisporellaceae Suetrong and Tubakiaceae U. Braun et al. (Senanayake et al. 2017, 2018b; Crous et al. 2018a, 2019b; Fan et al. 2018; Xavier et al. 2019).
Cryphonectriaceae Gryzenh. & M.J. Wingf.
Notes: Cryphonectriaceae (Diaporthales) was established to accommodate five genera Endothia Fr., Cryphonectria complex, Amphilogia Gryzenh., H.F. Glen & M.J. Wingf., Chrysoporthe Gryzenh. & M.J. Wingf. and Rostraureum Gryzenh. & M.J. Wingf. by Gryzenhout et al. (2006). Twenty-six genera were subsequently added to the family based on morphological and phylogenetic support by different authors (Gryzenhout et al. 2004, 2005b; Nakabonge et al. 2006; Chen et al. 2013a, 2016; Jiang et al. 2019b; Wijayawardene et al. 2018a; Ferreira et al. 2019). We introduce a novel genus Eriocamporesia as a monotypic genus to accommodate E. aurantia based on morphological characteristics and phylogenetic support.
Eriocamporesia R.H. Perera, Samarak. & K.D. Hyde, gen. nov.
Index Fungorum number: IF556791; Facesoffungi number: FoF 06959
Etymology: Named after the collector Erio Camporesi
Associated with twigs of dicotyledonous plant. Sexual morphAscostromata on host single, appearing as yellow pustules, pulvinate, semi-immersed in plant tissue, orange, KOH + purple; ascostromatic tissue pseudoparenchymatous, covering the top of the ascomatal bases. Ascomata perithecial, usually valsoid, embedded in stromata at irregular levels, fuscous black, necks emerge at stromatal surface as dark orange brown ostioles covered with orange stromatal tissue to form papillae extending above stromatal surface. Peridium of 5–7 layers of flattened, elongate, brown-walled cells of textura angularis. Paraphyses lacking. Asci 8-spored, unitunicate, oblong ellipsoidal to fusoid to ellipsoidal, apedicellate, floating freely in the perithecial cavity, thin-walled, apex simple. Ascospores 2-seriate, cylindrical to allantoid, occasionally ellipsoidal, ends round, curved or sometimes straight, aseptate, hyaline, smooth-walled, without appendages. Asexual morphConidiostromata appearing as yellow pustules on the host, occur as separate structures, pulvinate, semi-immersed, orange. Conidiomata multi-locular, with locules often convoluted, apapillate, with 1-ostiolar opening, orange, KOH + purple; stromatic tissue pseudoparenchymatous. Paraphyses lacking. Pycnidial walls of 2–4 layers of flattened, elongate, hyaline, cells of textura angularis. Conidiophores cylindrical, rarely septate or branching, hyaline. Conidiogenous cells phialidic, cylindrical with attenuated apices, hyaline. Conidia hyaline, cylindrical or allantoid, ends round, aseptate, hyaline, smooth-walled, without appendages.
Type species: Eriocamporesia aurantia R.H. Perera, Samarak. & K.D. Hyde
Notes: DNA sequence evidence shows Eriocamporesia is a member of Cryphonectriaceae and distinct from other genera in the family (Fig. 108). Eriocamporesia resembles other genera in Cryphonectriaceae by orange ascostromata and conidiostromata which turned purple in 3% KOH, and phialidic conidiogenous cells (Gryzenhout et al. 2006; Jiang et al. 2019b). Eriocamporesia shares similar morphology with Holocryphia in having orange stromata, valsoid, fuscous black ascomata and apedicellate asci (Chen et al. 2013a). However, it can be segregated from Holocryphia by the lack of paraphyses in the conidiomata and allantoid conidia, while Holocryphia has paraphyses and cylindrical conidia. Phylogenetic analyses based on a combined LSU, ITS and TUB2 sequence dataset shows that Eriocamporesia forms a distinct lineage basal to the genera Endothia and Aurantioporthe G.L. Beier & Blanchette in Cryphonectriaceae. Considering both molecular and morphological data we introduce Eriocamporesia as a new genus.
Eriocamporesia aurantia R.H. Perera, Samarak. & K.D. Hyde, sp. nov.
Index Fungorum number: IF556792; Facesoffungi number: FoF 06960; Figs. 106, 107
Sexual morph of Eriocamporesia aurantia (MFLU 19-0943, holotype). a Herbarium material. b Ascostromata on host. c Longitudinal section of ascostroma. d Section of peridium. e Peridium in face view. f–j Asci. k–o Ascospores. p Germinating ascospore. Scale bars: b, c = 200 μm, d–j = 20 μm, k–o = 10 μm, p = 20 μm
Asexual morph of Eriocamporesia aurantia (MFLU 19-0943, holotype). a Conidiostromata on host. b Longitudinal section of conidiostroma. c Section of conidioma. d Section of peridium. e, f Conidiophores with conidia (f in Congo red). g Conidia. Scale bars: a = 100 μm, b = 500 μm, c, d = 20 μm, e–g = 10 μm
Etymology: Name reflects orange stromata.
Holotype: MFLU 19-0943
Associated with twigs of dicotyledonous plant. Sexual morphAscostromata on host single, appearing as yellow pustules, 450–670 μm high, 870–1000 μm diam., pulvinate, semi-immersed in plant tissue, orange, KOH + purple; ascostromatic tissue pseudoparenchymatous, covering the top of the ascomatal bases. Ascomata 450–700 μm high, 120–330 μm diam., perithecial, usually valsoid, embedded in stromata at irregular levels, fuscous black, necks emerge at stromatal surface as dark orange brown ostioles covered with orange stromatal tissue to form papillae extending above stromatal surface. Peridium 12–24 μm wide, composed of 5–7 layers of flattened, elongate, brown-walled, cells of textura angularis. Paraphyses lacking. Asci 30–43 × 6.7–8.7 µm (\( \bar{x} \) = 37.6 × 7.8, n = 14), 8-spored, unitunicate, oblong ellipsoidal to fusoid to ellipsoidal, apedicellate, floating freely in the perithecial cavity, thin-walled, apex simple. Ascospores 9.5–14.5 × 2.5–3.3 μm (\( \bar{x} \) = 11.6 × 2.8, n = 30), 2-seriate, cylindrical to allantoid, occasionally ellipsoidal, ends round, curved or sometimes straight, aseptate, hyaline, smooth-walled, without appendages. Asexual morphConidiostromata 445–630 μm high, 1500–1800 μm diam., appearing as yellow pustules on the host, occur as separate structures, pulvinate, semi-immersed, orange. Conidiomata multi-locular, with locules often convoluted, apapillate, with 1-ostiolar opening, orange, KOH + purple; stromatic tissue pseudoparenchymatous. Paraphyses lacking. Pycnidial walls 4–7 μm wide, composed of 2–4 layers of flattened, elongate, hyaline, cells of textura angularis. Conidiophores 16.3–23.6 μm long, 1.6–2.3 wide, cylindrical, rarely septate or branching, hyaline. Conidiogenous cells 8.8–9.7 μm long, 1.1–1.6 wide, phialidic, cylindrical with attenuated apices, hyaline. Conidia 3.7–5.7 × 1–1.5 μm (\( \bar{x} \) = 4.9 × 1.2, n = 30), hyaline, cylindrical or allantoid, ends round, aseptate, hyaline, smooth-walled, without appendages.
Culture characteristics: Ascospores germinated on PDA within 12 h, colonies on PDA reaching 35–38 mm diam. after 2 weeks at 25 °C, mycelium partly superficial, partly immersed, slightly effuse, with irregular edge, initially white, becoming orangish with time.
Material examined: THAILAND, Chiang Mai Province, Mae Rim District, on twigs of dicotyledonous plant, R.H. Perera, 9 September 2017, Rim19 (MFLU 19-0943, holotype), ex-type living culture, MFLUCC 17-2625.
GenBank numbers: ITS = MN699135, LSU = MN699130.
Notes: Eriocamporesia aurantia clusters within Cryphonectriaceae as a separate lineage (Fig. 108). BLASTn search of the ITS rDNA region of our new isolate showed the highest similarity to Amphilogia gyrosa (Berk. & Broome) Gryzenh., H.F. Glen & M.J. Wingf. strain 91123101 (94% similarity, 465/494 bp), Endothia gyrosa (Berk. & Broome) Höhn. strain AFTOL-ID 1223 (94% similarity, 531/564 bp), E. radicalis (Schwein. ex Fr.) Tul. & C. Tul. strain CBS 116.13 (94% similarity, 549/585 bp), Chrysoporthe inopina Gryzenh. & M.J. Wingf. strain CMW 12731 (94% similarity, 549/585 bp) and strain CBS 116.13 (94% similarity, 543/580 bp). Eriocamporesia aurantia shares similar asexual morph characters to Amphilogia (Gryzenhout et al. 2005a; Senanayake et al. 2017). However, it can be distinguished from Amphilogia species in having valsoid perithecia, and aseptate, cylindrical to allantoid ascospores, while Amphilogia has diatrypoid perithecia, and 1–3-septate, fusoid to ellipsoid ascospores (Gryzenhout et al. 2005a; Senanayake et al. 2017). Considering both molecular and morphological data, we introduce E. aurantia as a novel species.
Phylogram generated from maximum likelihood analysis based on combined LSU, ITS and TUB2 sequence data representing Cryphonectriaceae. Related sequences are taken from Senanayake et al. (2017). Fifty-four strains are included in the combined analyses which comprise 1830 characters (866 characters for LSU, 639 characters for ITS, 323 characters for TUB2) after alignment. Coniella tibouchinae (CPC 18512) and C. quercicola (CBS 904.69) in Schizoparmaceae is used as the outgroup taxa. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RAxML tree with a final likelihood values of − 7862.571574 is presented. The matrix had 525 distinct alignment patterns, with 29.64% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.239368, C = 0.254757, G = 0.280128, T = 0.225747; substitution rates AC = 2.087607, AG = 3.025115, AT = 2.060161, CG = 1.371584, CT = 7.165898, GT = 1.000000; gamma distribution shape parameter α = 0.727546. Bootstrap values for maximum likelihood (ML) equal to or greater than 50% and clade credibility values greater than 0.90 (the rounding of values to 2 decimal proportions) from Bayesian-inference analysis labelled on the nodes. Ex-type strains are in bold and black, the new isolate is indicated in bold and blue
Cytosporaceae Fr.
Notes: We follow the latest treatment and updated accounts of Cytosporaceae in Norphanphoun et al. (2017, 2018) and Fan et al. (2018, 2019).
Cytospora Ehrenb.
Notes: Cytospora was introduced by Ehrenberg (1818) in the family Cytosporaceae order Diaporthales (Wijayawardene et al. 2018a). The sexual morph of Cytospora is characterised by erumpent, pseudostromatic, perithecial, uni- to multilocular ascostromata, with converging beak-like, ostiolar necks, peridium composed of pseudoparenchymatous cells, of textura angularis, septate paraphyses, 8-spored, unitunicate, clavate to obovoid asci, with J-, apical rings and hyaline, elongated-allantoid, smooth-walled ascospores (Adams et al. 2004; Lawrence et al. 2018; Norphanphoun et al. 2017, 2018). The asexual morph is characterized by the fruiting bodies comprised of stromata (conidiomata) with intricate chambers, filamentous branched conidiophores and hyaline allantoid conidia, exuding from the fruiting bodies with yellow, orange or red colour (Norphanphoun et al. 2017; Fan et al. 2019). Cytospora species are mostly plant pathogens (Norphanphoun et al. 2017; Fan et al. 2019). The genus comprises 120 accepted species (Wijayawardene et al. 2017a) and new species have been added (Fan et al. 2018, 2019; Lawrence et al. 2018). We introduce two new Cytospora species, C. fusispora and C. rosigena, based on phylogenetic and morphological evidence.
Cytospora fusispora M. Niranjan & V.V. Sarma, sp. nov.
Index Fungorum number: IF556638; Facesoffungi number: FoF 06282; Fig. 109
Etymology: The specific epithet “fusispora” refers to fusiform shape of the ascospores.
Holotype: AMH 10073
Saprobic on unidentified twig. Sexual morphAscostromata erumpent to superficial, associated with the host tissues, apically brown of textura angularis cells and basally medulla consisting loosely connected pale brown hyphae, wider at base and narrower towards apices. Ascomata 200–250 × 210–290 µm (\( \bar{x} \) = 232.2 × 248.8 µm, n = 25), perithecial, immersed to erumpent, solitary to gregarious, carbonaceous, subglobose, ostiolate, papillate. Necks 160–270 µm long, with textura angularis cells, soft at base and carbonaceous towards narrow apices. Peridium 13.2 µm, dark brown to brown with several layers of cells of textura angularis. Paraphyses filamentous, septate, wider at base, narrower towards apex, septate and unbranched. Asci 20–29 × 4.2–5.6 µm (\( \bar{x} \) = 23.4 × 5.1 µm, n = 25), 8-spored, unitunicate, cylindric-clavate, apically flat with J-, apical ring in Lougal’s solution, wider in the middle, short or apedicellate. Ascospores (3.9–)4–5.3 × (1.6–)1.7–2.2 µm (\( \bar{x} \) = 4.6 × 1.9 µm, n = 25), overlapping 1–2-seriate, hyaline, aseptate, ovoid to fusoid, acute ended, smooth-walled. Asexual morph Undetermined.
Culture characteristics: White colonies, radial, initially hyaline, 45 mm in one-week old culture in MEA at 28 °C.
Material examined: INDIA, Andaman and Nicobar Islands, South Andaman, Mount Harriet, Gun point, recorded on unidentified decaying twig, 7 December, 2017, M. Niranjan & V.V. Sarma (AMH 10073, holotype; Ajrekar Mycological Herbarium (AMH), Agharkar Research Institute (ARI), ex-type living culture NFCCI-4372, National fungal culture collection centre (NFCCI), ARI, Pune, India.
GenBank numbers: ITS = MN227694, LSU = MN250032.
Notes: Most Cytospora species were published based on the asexual morph and rarely based on the sexual morph (Norphanphoun et al. 2017). The sexual morph of Cytospora mostly have multi-loculate ascostroma, sessile asci and allantoid ascospores (Adams et al. 2006). The new species, C. fusispora fits well with Cytospora in terms of having only 2–4 locules, sessile to subsessile asci and fusoid ascospores. The molecular phylogenetic tree constructed based on ITS, LSU, RPB2 and ACT sequence matrix shows that it has a sister relationship with Cytospora paratranslucens Norph. et al. (Fig. 111). Hence, based on both morphological characters and molecular sequence analyses a new species Cytospora fusispora is introduced.
Cytospora rosigena Chaiwan, Wanas., Bulgakov & K.D. Hyde, sp. nov.
Index Fungorum number: IF557061; Facesoffungi number: FoF 07096; Fig. 110
Cytospora rosigena (MFLU 17-2131, holotype). a Host substrate. b Conidiostromata on host substrate. c Horizontal section of conidiostroma showing the arrangement of conidiostroma. d Vertical section of conidiostroma. e Pycnidial wall. f, g Conidiogenous cells (f = stained in congo red). h–l Conidia (l = stained in congo red). Scale bars: d = 300 µm, e = 20 µm, f, g = 15 µm, h–l = 5 µm
Etymology: The specific epithet “rosigena” refers to borne on Rosa.
Holotype: MFLU 17-2131
Saprobic or weak pathogenic on Rosa sp. Sexual morph Undetermined. Asexual morphConidiostromata 700–1000 × 470–580 µm diam., pycnidial, scattered, immersed to erumpent, multi-loculate. Locules composed of numerous interconnecting, chambers arranged radially or irregularly within a continuous mass of ectostromatic tissue, pycnidial, solitary or clustered, immersed in the host when young, semi-erumpent at maturity, dark brown to black, globose, ostiolate. Ostioles 50–100 µm. Pycnidial walls comprising a few to several layers of cells of textura angularis, with innermost layer thin, brown, outer layer dark brown to black. Conidiophores unbranched, reduced to conidiogenous cells. Conidiogenous cells blastic, enteroblastic, phialidic, formed from the innermost layer of pycnidial wall, hyaline, smooth-walled. Conidia 5.8–7.7 × 1.2–1.3 µm (\( \bar{x} \) = 6.7 × 1.25 µm, n = 30), unicellular, elongate-allantoid, slightly curved, hyaline, smooth-walled.
Culture characteristics: Conidia germinating on water agar and germ tubes produced from conidia within 12 h from lower end. Colonies growing on MEA, circular, with flat surface, edge entire, reaching 8 cm in 7 days at 25 °C, white to pale brown in PDA medium. Mycelium superficial and partially immersed, branched, septate, hyaline grey to black, smooth.
Material examined: RUSSIA, Rostov region, Shakhty City, artificial ravine forest belt, on dead and dying branches of Rosa sp., T.S. Bulgakov, 11 May 2017, T-1829 (MFLU 17-2131, holotype), ex-type living culture, MFLUCC 18-0921.
GenBank numbers: ITS = MN879872, LSU = MN879873.
Notes: Cytospora rosigena is most similar to C. ribis in its conidiomata having multi locules (Yang et al. 2015). The new taxon is closely related to the group of C. ribis and C. tanaitica (Fig. 111). These formed a close relationship with C. ribis with conidiomata but they have different host plants (Yang et al. 2015): C. ribis is a pathogen on twigs and branches of Ulmus pumila from Xining City, Qinghai Province in China, but C. rosigena grows on twigs and branches of Rosa sp. from Rostov region in Russia.
Phylogram generated from maximum likelihood analysis based on combined ITS, LSU, RPB2 and ACT sequence data representing Cytospora. Related sequences are taken from Fan et al. (2019). Seventy-nine strains are included in the combined analyses which comprise 4340 characters (842 characters for LSU, 1030 characters for SSU, 512 characters for ITS, 926 characters for TEF1-α, 1030 characters for RPB2) after alignment. Diaporthe vaccinia (CBS160.32) was used as the outgroup taxon. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RAxML tree with a final likelihood values of − 38637.764110 is presented. The matrix had 1968 distinct alignment patterns, with 33.91% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.245895, C = 0.251442, G = 0.271420, T = 0.231244; substitution rates AC = 1.592217, AG = 3.496391, AT = 1.418709, CG = 1.209916, CT = 8.405355, GT = 1.000000; gamma distribution shape parameter α = 0.569582. Bootstrap values for maximum likelihood (ML) equal to or greater than 70% and clade credibility values greater than 0.90 (the rounding of values to 2 decimal proportions) from Bayesian-inference analysis labeled on the nodes. Ex-type strains are in bold and black, the new isolate is indicated in bold and blue
Diaporthaceae Höhn. ex Wehm.
Notes: The fungal family Diaporthaceae contains a wide range of taxa which are endophytes, pathogenic and saprobic worldwide (Udayanga et al. 2011, 2012). Ten genera are accepted in Diaporthaceae (Senanayake et al. (2018b).
Diaporthe Nitschke
Notes: The species belonging to genus Diaporthe are well known as plant pathogenic taxa which are mainly cause “Phomopsis disease” (Udayanga et al. 2011). They also have been recorded as endophytes and saprobes on a wide range of hosts. There are over 200 species associated with the family and many more have been introduced (Manawasinghe et al. 2019; Marin-Felix et al. 2019b). Thus the genus needs revision to determine species boundaries. The most recent taxonomic treatment for this genus are Guarnaccia et al. (2018), Marin-Felix et al. (2019b) and Manawasinghe et al. (2019).
Diaporthe camporesii Manawasinghe & K.D. Hyde, sp. nov.
Index Fungorum number: IF557087; Facesoffungi number: FoF 06801; Fig. 112
Diaporthe camporesii (JZHB320143, holotype). a Material examined. b Appearance of conidiomata on host. c ection through conidioma stained in congored. d Pycnidial wall. e Conidiogenous cells with attached conidia (on host). f, g Conidia (on host). h Conidiogenous cells with attached conidia on PDA. i Upper view of colony on PDA. j Reverse view of colony on PDA. Scale bars: c = 100 μm,d = 20 μm, e–h = 10 μm
Etymology: Name reflects the collector of the specimens, Erio Camporesi.
Holotype: MFLU 18-0135
Saprobic on Urtica dioica. Sexual morph Undetermined. Asexual morphConidiomata on host pycnidial, ampulliform, scattered, immersed. Pycnidial walls multi-layered, comprising 2–3 layers, with heavily pigmented, thick-walled, comprising dark brown cells of textura angularis, with lighter cells towards the inside, with inner layer composed of 2–5 layers, hyaline, thin-walled cells of textura angularis. Pycnidia in culture black, immersed, solitary or merged, exudated. Conidiophores reduced to conidiogenous cells. Conidiogenous cells enteroblastic, integrated, clustered, hyaline, smooth-walled. Paraphyses not observed. Alpha conidia, on culture 3–6 × 1.4–3 μm (\( \bar{x} \) = 4.7 × 2.5 μm, n=30), hyaline, smooth-walled, uni- or bi-guttulate. Beta conidia not observed.
Culture characteristics: Colonies on PDA 50 mm diam., after 7 days at 25 °C, cream to white cottony mycelium, regular margin, immersed and semi-immersed, thin mycelium. Initially reverse white and with age it becomes yellowish.
Material examined: ITALY, Province of Forlì-Cesena [FC], Meldola, on the dead aerial stem of Urtica dioidca L. (Urticaceae), 23 December 2017, E. Camporesi, IT 3638 (JZHB320143, holotype), ex-type living culture JZB320143.
GenBank numbers: ITS = MN533805, TEF1-α = MN984254, TUB2 =MN561316.
Notes: Diaporthe camporesii is morphologically similar to other species of Diaporthe. In the phylogenetic tree constructed using combined ITS, TEF1-α and TUB2 genes (Fig. 116) D. camporesii formed a separate subbclade with D. ganjae (CBS 180.91), D. sambucusi (CFCC 51986) and D. compacta (LC3083). Morphologically D. camporesii is closely related to the D. ganjae with conidial dimensions. For D. camporesii and D. ganjae (CBS 180.91), 50% nucleotide difference was observed for ITS across 29 nucleotides, 5% nucleotide difference was observed in TUB2 across 419 nucleotides and 48% nucleotide difference was observed in TEF1-α across 235 nucleotides. Therefore we identify the present taxon as a novel species in genus Diaporthe. Diaporthe camporesii was isolated from twigs and branches of Urtica dioica, and three other species of Diaporthe are associated with Urtica spp. (Farr and Rossman 2020).
Diaporthe cynaroidis Marinc., M.J. Wingf. & Crous, CBS Diversity Ser. (Utrecht) 7: 39 (2008)
Facesoffungi number: FoF 07201; Fig. 113
Diaporthe cynaroidis (MFLU 14-0847). a Ascostromata (black arrow) and conidiomata on substrate (white arrow). b Cross section of ascoma. c Peridium. d–f Asci. g–i Ascospores. j Cross section of conidioma. k Conidiogenous cells attached to conidia. l–n Conidia. Scale bars: b = 200 μm, c = 15 μm, d–f = 20 μm, g–i, k = 10 μm, j = 100 μm, l–n = 5 μm
Holotype: SOUTH AFRICA, Western Cape Province, Betty’s Bay, on leaf litter of Protea cynaroides, 26 June 2000, S. Marincowitz, S.L. 522 (PREM 56862), ex-type living culture CBS 122676 = CPC13180.
Saprobic on dead stem of Eupatorium cannabinum L. Sexual morphClypeus appears as black, wide patches. Ascomata 280–300 μm high, 180–200 μm wide (\( \bar{x} \) = 290 × 190 μm, n = 10), solitary or rarely aggregated, erumpent, globose to subglobose, black, coriaceous, ostiolate, papillate. Papilla 135–138 μm high, 110–140 μm wide, (\( \bar{x} \) = 136 × 115 μm, n = 10), internally covered by hyaline periphyses. Peridium 8–13 μm (\( \bar{x} \) = 10 μm, n = 10), comprising thick-walled, brown, compressed cells of textura angularis. Paraphyses a cellular mass of tissues. Asci 45–60 × 11–14 μm (\( \bar{x} \) = 51 × 13.5 μm, n = 20), 8-spored, unitunicate, clavate to fusiform, sessile, apex rounded, with a characteristic, bilobed, J-, apical ring. Ascospores 14–18.5 × 4–5 μm (\( \bar{x} \) = 16 × 4.2 μm, n = 20), 2-seriate, fusiform to elongate ellipsoid, 1-median septate, with each cell containing two guttules, hyaline, smooth-walled. Asexual morphConidiomata 125–140 μm high, 265–300 μm diam., at base (\( \bar{x} \) = 135 × 280 μm, n = 10), pycnidial, pyriform, initially immersed, erumpent at maturity, globose to pyriform, black, coriaceous, elongated neck, often with yellowish white, conidial cirrus extruding from ostiole. Pycnidial walls 34–36 μm diam. (\( \bar{x} \) = 35 μm, n = 10), parenchymatous, consisting of 4–7 layers of pale brown, thick-walled cells of textura angularis. Conidiophores 4–6 × 4.5–8 μm (\( \bar{x} \) = 4.6 × 6.6 μm, n = 20), ampulliform, straight to sinuous, unbranched, hyaline, smooth. Conidiogenous cells 8–14 × 1.5–3 μm (\( \bar{x} \) = 11.2 × 2.2 μm, n = 20), phialidic, terminal, cylindrical, slightly tapering towards the apex. Aparaphysate. Alpha conidia 5.8–7.5 × 2.5–3.5 μm (\( \bar{x} \) = 6.4 × 2.8 μm, n = 10), aseptate, hyaline, smooth, ovate to ellipsoidal, base subtruncate, often bi-guttulate. Beta conidia not seen.
Culture characteristics: Colonies growing on PDA attaining 2 cm diam. within a week when incubated at 18 °C, circular, entire, flat, white, tightly attached to the media, aerial mycelia less or sparse, forming few, erumpent, globose, black, viscous droplets after a week.
Material examined: ITALY, Province of Forlì-Cesena, Rio dell’Eremo, Cesena, dead aerial stem of Eupatorium cannabinum L. (Asteraceae), 16 December 2014, E. Camporesi, IT 1909 (MFLU 14-0847), living culture MFLUCC 14-0847.
Known host and distribution: Protea cynaroides, Vitis sp. (Italy, South Africa) (Farr and Rossman 2020).
GenBank numbers: ITS = MN764319, LSU = MN764357.
Notes: Marincowitz et al. (2008) introduced Diaporthe cynaroidis from leaf litter of Protea cynaroides (Proteaceae). This species was isolated from South Africa and only asexual morph was reported from its type collection. We illustrate the both sexual and asexual morph of D. cynaroidis from our collection obtained from Italy and this is the first illustration of sexual morph. It is also the first record of D. cynaroidis in Italy.
Diaporthe foeniculina (Sacc.) Udayanga & Castl., in Udayanga, Castlebury, Rossman & Hyde, Persoonia 32: 95 (2014)
Facesoffungi number: FoF 02183; Fig. 114
≡ Phoma foeniculina Sacc., Michelia 2(no. 6): 95 (1880)
Epitype: PORTUGAL, Madeira, Serra da Agua, at base of 2-yr-old stem of Foeniculum vulgare, August 2001, A.J.L. Phillips, LISE 94791, ex-epitype culture CBS 111553 = DP0391; MBT175961.
Saprobic on dead aerial stem of Asparagus spp. Sexual morph Undetermined. Asexual morph Coelomycetous. Conidiomata 100–200 μm high, 80–200 μm diam., pycnidial, solitary, scattered or gregarious, globose to subglobose, semi-immersed to immersed, sometimes erumpent, unilocular, thin-walled, visible as small round to oval dark brown to black dots on the host surface. Pycnidial walls composed 3–4 layers, similarly dense at the apex and base, outer 2–3 layers dark brown to black, inner 1–2 layers hyaline, with thin-walled cells of textura angularis. Conidiogenous cells 4–7 μm long × 2–4 μm wide, hyaline, phialidic, globose. Conidia 5–10 × 1– 4 μm wide (\( \bar{x} \) = 8 × 3 µm, n = 20), ellipsoidal to cylindrical, rounded at both ends, hyaline, straight or slightly bent at the middle, 1-septate to aseptate, thin- and smooth-walled, guttulate.
Culture characteristics: Colonies on PDA reaching 80 mm diam. after 14 days at 25 °C, circular colony, white, without any diffusible pigments.
Material examined: ITALY, Province of Forlì-Cesena [FC], Trivella-Predappio, on dead aerial stem of Asparagus spp., 21 November 2017, E. Camporesi, IT 3339b (MFLU 17-0997), living culture, JZB 320145, JZB 320146.
Known hosts and distribution: Occuring on various hosts worldwide (e.g. Citrus, Foeniculum vulgare, Glycine max, Malus domestica, Prunus and Vitis; Farr and Rossman 2020).
GenBank numbers: ITS = MN944411.
Notes: The isolate obtained from dead aerial stem of Asparagus spp. is identified as Diaporthe foeniculina with support from both morphology and phylogeny (Figs. 114, 116). Diaporthe foeniculina has been reported in many economically important host plants in worldwide (Farr and Rossman 2020). This is the first record of D. foeniculina from Asparagus from Italy.
Diaporthe nigra Brahmanage & K.D. Hyde, sp. nov.
Index Fungorum number: IF556927; Facesoffungi number: FoF 07202; Fig. 115
Etymology: Name refers to the host epithet, [Ballota] nigra.
Holotype: JZBH320170
Saprobic on dead aerial stem of Ballota nigra L., visible as raised, black spots or, black necks immerging through the host surface. Sexual morph Undetermined. Asexual morphConidiomata pycnidial or multi-loculate, scattered, semi-immersed, globose or irregular, black. Pycnidia walls consisting brown cells of textura angularis in surface view. Conidiophores reduce to conidiogeneous cells or rarely short, 8–25 × 1–3 μm (\( \bar{x} \) = 20 × 1.5 μm), compressed cylindrical, aseptate, branched, densely aggregated, cylindrical, straight to sinuous rarely reduced to conidiogenous cells, hyaline, smooth-walled. Alpha conidiogenous cells 190–295 × 120–175 μm (\( \bar{x} \) = 270 × 160 μm), phialidic, subcylindrical to ampulliform, hyaline, with visible periclinal thickening, and a flared collarette. Alpha conidia 17–28 × 7–7.5 μm (\( \bar{x} \) = 25 × 7.2 μm), enteroblastic, ovate to ellipsoidal, base obtuse to subtruncate, aseptate, straight, bi-guttulate, hyaline, smooth-walled. Beta and Gamma conidia not observed.
Culture characteristics: Colonies on PDA, reaching 30 mm diam. after 2 weeks at 25 °C, flat, circular, margin entire, white with radially arranged minute mycelium clots later becoming pale yellow, reverse whitish. Odour not pronounced. Sporulated on PDA after 2 months incubation period in dark, at 25 °C.
Material examined: ITALY, Province of Forlì-Cesena [FC], Predappio, Tontola, on dead aerial stem of Ballota nigra L. (Lamiaceae), 13 March 2018, E. Camporesi, IT 3773 (JZBH320170, holotype), ex-type living culture, JZB320170.
GenBank numbers: ITS = MN653009, TEF1-α = MN892277, TUB2 = MN887113.
Notes: Asexual morphs of Diaporthe have mostly similar morphologies (Dissanayake et al. 2017b; Perera et al. 2018a). Based on multi-gene phylogenetic analysis of combined ITS, TUB2, CAL and TEF1-α sequence data of Diaporthe species (85% ML, 0.90 BYPP; Fig. 116), our strain (JZB320170) clusters with D. baccae (CBS 136972), but differs in alpha conidial size (17–28 × 7–7.5 μm) compared to the ex-type strain D. baccae (CBS 136972) ((6–)7–9 × 2–3 μm).
Phylogram generated from maximum likelihood analysis based on combined ITS, TUB2, CAL and TEF1-α sequence data of Diaporthe species. Bootstrap support values for ML greater than 50% and Bayesian posterior probabilities greater than 0.90 are given near nodes. The tree is rooted with Diaporthella corylina CBS 121124. Ex-type strains are in bold and black. The newly generated sequences are indicated in blue
Myrmecridiaceae Crous
Notes: Crous et al. (2015c) established the family Myrmecridiaceae to accommodate Myrmecridium Arzanlou, W. Gams & Crous (Arzanlou et al. 2007), which is typified by M. schulzeri (Sacc.) Arzanlou, W. Gams & Crous. (previously as Acrotheca acuta Grove). It is characterised by obovoid or fusiform conidia with a narrowly truncate base, hyaline mycelium, and relatively unpigmented, pimple-like denticles (Arzanlou et al. 2007; Crous et al. 2011b; Tibpromma et al. 2017). A second genus Neomyrmecridium in Myrmecridiaceae was introduced by Crous et al. (2018a). Some Myrmecridiaceae species are saprobic on plant tissues (Crous et al. 2012, 2015c, 2018a), while some are isolated from soil (Arzanlou et al. 2007; Jie et al. 2013; Peintner et al. 2016) and submerged wood in freshwater (Réblová et al. 2016a; Tibpromma et al. 2017).
Neomyrmecridium Crous
Notes: Crous et al. (2018a) introduced Neomyrmecridium to accommodate N. asiaticum Crous, N. septatum Crous (type species) and N. sorbicola (Crous & R.K. Schumach.) Crous. Species in the genus are characterised by solitary, unbranched conidiophores, polyblastic conidiogenous cells with several denticles at the apex, and fusoid-ellipsoid, septate conidia with upper two-thirds encased in mucoid sheath. We introduce the fourth species of Neomyrmecridium based on both morphology and phylogeny.
Neomyrmecridium guizhouense N.G. Liu, K.D. Hyde & J.K. Liu, sp. nov.
Index Fungorum number: IF557234; Facesoffungi number: FoF 06704; Fig. 117
Etymology: Name refers to Guizhou Province, China, the place where this fungus was collected.
Holotype: GZAAS 20-0001
Saprobic on decaying wood. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies on natural substrate effuse, greyish white, velvety. Mycelium immersed, composed of brown, branched, septate hyphae. Conidiophores 75–140 × 2–4.5 μm (\( \bar{x} \) = 98.4 × 3.3 μm, n = 15), macronematous, mononematous, solitary, erect, straight or slightly flexuous, subcylindrical, unbranched, septate, medium brown at base, paler towards apex, smooth, thick-walled. Conidiogenous cells 2.2–4.3 μm (\( \bar{x} \) = 3.2 μm) wide, polyblastic, terminal, integrated, subcylindrical, subhyline, finely verrucose, with several denticles at apex, not thickened nor darkened. Conidia 8.9–12.7 × 2.8–4.8 μm (\( \bar{x} \) = 10.4 × 4.1 μm, n = 30), solitary, subhyaline to pale brown, (2–)3-septate, rarely constricted at septum, smooth-walled, guttulate, fusoid-ellipsoid, apex obtuse or tapering, with a subtruncate hilum at base.
Culture characteristics: Conidia germinating on water agar within 24 h. Germ tubes produced from one or both ends. Mycelia superficial, circular, with entire edge, mycelia dense at centre, sparse towards circumference, yellowish white to yellow from above, yellow at centre, paler towards circumference from below.
Material examined: CHINA, Guizhou Province, Dushan, on decaying wood in a bank of a small freshwater, 6 July 2018, N.G. Liu, DS002 (GZAAS 20-0001, holotype), ex-type living culture, GZCC 20-0008.
GenBank numbers: ITS = MT002305, LSU = MT002307, SSU = MT002308, RPB2 = MT023016, TEF1-α = MT023013.
Notes: Neomyrmecridium guizhouense resembles N. asiaticum in having pale brown, (2–)3-septate conidia. However, N. guizhouense has smaller conidia (8.9–12.7 × 2.8–4.8 versus 15–16 × 4.5 μm) and longer conidiophores (75–140 versus 50–100 μm) than those of N. asiaticum. The conidia of N. guizhouense are also smaller than those of N. septatum (8.9–12.7 × 2.8–4.8 versus 14–16 × 4 μm). Phylogenetic analyses of a combined LSU, SSU, ITS and RPB2 sequence dataset showed that N. guizhouense forms a sister lineage with N. septatum with high support (100% ML, 1.00 BYPP; Fig. 120). We did not observe the generic character that upper two-thirds of conidia are encased in mucoid sheath, but phylogenetic analyses confirmed that our taxon belongs to Neomyrmecridium (Fig. 120). Therefore, N. guizhouense is introduced as a new species.
Phomatosporales Senan. et al.
Notes: The order Phomatosporales was introduced by Senanayake et al. (2016) to accommodate Phomatosporaceae based on morphology and multi-gene phylogenetic analyses. The order is characterised by globose or subglobose ascomata with a small blackened clypeus thin-walled, long, cylindrical asci with minute apical rings and small, globose, unicellular, hyaline cylindrical or ellipsoidal to fusiform ascospores and sporothrix-like asexual morphs. This order includes saprobes from terrestrial and aquatic habitats (Senanayake et al. 2016).
Phomatosporaceae Senan., Maharachch. & K.D. Hyde
Notes: Von Arx (1951) invalidly introduced the family Phomatosporaceae, while Senanayake et al. (2016) validated it to accommodate three genera Phomatospora Sacc., Lanspora K.D. Hyde & E.B.G. Jones and Tenuimurus Senan., Camporesi & K.D. Hyde based on morphology and molecular data. This family includes species that are saprobes on submerged wood or decaying twigs in terrestrial or aquatic environments. Phomatosporaceae is mainly characterised by perithecial, solitary to gregarious, immersed or erumpent globose or subglobose ostiolate, papillate ascomata that are immersed under a small blackened clypeus. The peridium is composed of brown pseudoparenchymatous cells, paraphyses are hypha-like or absent, asci are unitunicate, cylindrical, thin-walled, short-pedicellate or sessile with J-, apical ring, ascospores are ellipsoidal to fusiform or cylindrical, septate to aseptate with guttules, striations or appendages and asexual morphs are sporothrix-like (Rappaz 1992; Fournier and Lechat 2010; Senanayake et al. 2016).
Lanspora K.D. Hyde & E.B.G. Jones
Lanspora was introduced by Hyde and Jones (1986) to accommodate Lanspora coronata K.D. Hyde & E.B.G. Jones. This monotypic genus was placed in Halosphaeriaceae (Microascales) based on its fibrous, smooth, appendaged ascospores, which closely resemble Eiona Kohlm. Subsequently, Réblová et al. (2016a) transferred the genus to Sordariomycetidae incertae sedis. Both Phomatospora and Tenuimurus can be distinguished from Lanspora in having ascospores that are ellipsoidal to fusiform, bi-guttulate, with striations and appendages (Senanayake et al. 2016). A new species Lanspora cylindrospora is introduced here based on significant differences in morphology and multi-gene phylogeny.
Lanspora cylindrospora Devadatha, V.V. Sarma & E.B.G. Jones, sp. nov.
Index Fungorum number: IF556222, Facesoffungi number: FoF 07059; Fig. 118
Lanspora cylindrospora (AMH-10017, holotype). a Ascomata immersed in the decaying wood of Suaeda monoica.b, c Longitudinal sections of ascoma. d Section of peridium comprising inner hyaline to pale brown cells of textura angularis.e Ascomata covered by a clypeus. f, g, i Immature and mature asci. h, j–o Ascospores. q Germinating ascospore. Scale bars: b = 100 μm, c = 50 μm, d–q = 10 μm
Etymology: In reference to cylindrical shape of the ascospores
Holotype: AMH-9948
Saprobic on decaying woody stems of Suaeda monoica.Sexual morphAscomata 95–185 µm high, 100–190 µm diam. (\( \bar{x} \) = 132 × 133 µm, n = 10), immersed beneath a clypeus, globose to subglobose, solitary to gregarious, coriaceous, brown to black. Ostioles 35–60 µm long, 30–70 µm diam. (\( \bar{x} \) = 46 × 44 µm, n = 10), brown, short. Periphyses filamentous, short 0.5–2 µm (\( \bar{x} \) = 1.5 µm, n = 10). Peridium 12–15 µm (\( \bar{x} \) = 13 µm, n = 10), equal in thickness, wide both at the base and sides, comprising two layers, inner stratum with 3–4 layers of hyaline to light brown of textura angularis and outer stratum with 2–3 layers of light brown to brown pseudoparenchymatous cells, forming textura angularis, fusing with the host tissues. Paraphyses not observed. Asci 30–65 × 3–8 µm (\( \bar{x} \) = 51 × 5 µm, n = 30), 8-spored, unitunicate, cylindrical to obclavate, short pedicellate, with a J-, subapical ring. Ascospores 6–10 × 1.5–2.7 µm (\( \bar{x} \) = 8 × 2 µm, n = 50), 1–2-seriate, overlapping, unicellular, cylindrical, straight, hyaline, smooth- to rough-walled, lacking guttules, appendages or sheaths. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on SWA within 24 h, germ tubes arising from apical end of the ascospore. Colonies on MEA, slow-growing, reaching 30–40 mm diam. after 25 days of incubation at room temperature, hyaline to bright yellow at centre and creamy at margin, reverse cream, moderate, slimy, hairy, undulate, surface raised, irregular.
Material examined: INDIA, Tamil Nadu, Tiruvarur, Muthupet mangroves, on intertidal wood of Suaeda monoica Lam. (Amaranthaceae), (10.4°N 79.5°E), 24 December 2016, B. Devadatha, AMH-10017 (holotype), ex-type living culture, NFCCI-4391; ibid., 25 February 2018, B. Devadatha, PUHD104, living culture NFCCI-4427.
GenBank numbers: ITS = MN168889, LSU = MN168891, SSU = MN169053, RPB2 = MN795090, TEF1-α = MN795088 (NFCCI-4391); ITS = MN168890, LSU = MN168892, TEF1-α = MN795089 (NFCCI-4427).
Notes: Our preliminary BLAST search, based on LSU sequence data, revealed that Lanspora cylindrospora is closely related to Lanspora and Phomatospora. Further, combined LSU, SSU, ITS, TEF1-α and RPB2 phylogenetic analyses showed that it clusters with L. coronata K.D. Hyde & E.B.G. Jones (Fig. 119). Lanspora cylindrospora shares similar morphological affinities with L. coronata such as immersed, globose to subglobose ascomata, absence of paraphyses and presence of hyaline ascospores (Hyde and Jones 1986). However, it is distinct in having smaller ascomata and ascospores, cylindrical to obclavate asci with subapical rings, straight to cylindrical, smooth-walled ascospores lacking guttules, and appendages or sheaths (Hyde and Jones 1986). Lanspora coronata also has larger ascomata and ascospores, with cylindrical or oblong ventricose asci, ascospores that are ellipsoidal, with longitudinal wall striations and crown-like appendages at both ends (Hyde and Jones 1986). The marine species Phomatospora nypae K.D. Hyde is clearly distinguishable from L. cylindrospora in having ellipsoidal, hyaline, 1-seriate ascospores that have faint striations and are covered with a mucilaginous sheath and appendages at both ends (Hyde 1993). Tenuimurus clematidis Senan., Camporesi & K.D. Hyde can be distinguished from L. cylindrospora in having ascospores that are ellipsoidal to fusiform, bi-guttulate and occurs in terrestrial habitats (Senanayake et al. 2016). Furthermore, LSU, TEF1-α and RPB2 base pair differences of L. cylindrospora and L. coronata are 1.87% (out of 745 bp), 20.5% (out of 949 bp), 8.1% (out of 783 bp), respectively, which indicates they are different species, according to the base pair differences prescribed by Jeewon and Hyde (2016). Hence the new species, Lanspora cylindrospora is introduced.
Phylogram generated from maximum likelihood analysis based on combined dataset of LSU, SSU, ITS, TEF1-α and RPB2 sequence data. Bootstrap support values for maximum likelihood (ML, MP equal to or greater than 75%) and clade credibility values greater than 0.94 BYPP from Bayesian-inference analysis are given above each branch. Ex-type strains are in bold and black, the new isolates are in blue. The tree is rooted to Leotia lubrica AFTOL-ID1
Diaporthomycetidae, genus incertae sedis
Notes: The subclass Diaporthomycetidae was introduced by Maharachchikumbura et al. (2015). Based on phylogenetic analyses, Maharachchikumbura et al. (2015, 2016) included Platytrachelon Réblová in the Diaporthomycetidae, genera incertae sedis. Wijayawardene et al. (2018a) placed Aquapteridospora Jiao Yang, K.D. Hyde & Maharachch., Aquaticola W.H. Ho et al., Clohiesia K.D. Hyde, Dictyosporella Abdel-Aziz, Fusoidispora Vijaykr., Jeewon & K.D. Hyde, Pseudoannulatascus Z.L. Luo, Maharachch. & K.D. Hyde and Pseudostanjehughesia J. Yang & K.D. Hyde under Diaporthomycetidae, genera incertae sedis. We introduce a new genus Pseudoconlarium under Diaporthomycetidae, genus incertae sedis based on morphology and phylogeny.
Pseudoconlarium N.G. Liu, K.D. Hyde & J.K. Liu, gen. nov.
Index Fungorum number: IF557094; Facesoffungi number: FoF 06705
Etymology: Referring to the morphologically similar genus Conlarium.
Saprobic on decaying wood. Sexual morph Undetermined. Asexual morphColonies on natural substrate effuse, scattered, punctiform. Mycelium partly immersed, partly superficial. Conidiophores micronematous or semi-macronematous, mononematous, septate, branched, flexuous, subhyaline to pale brown. Conidiogenous cells monoblastic, integrated, terminal, subhyaline to pale brown, determinate, doliiform. Conidia solitary, brown, irregularly globose to subglobose, or trapeziform, muriform, constricted at the septa, smooth-walled.
Type species: Pseudoconlarium punctiforme N.G. Liu, K.D. Hyde & J.K. Liu
Notes: Pseudoconlarium resembles Conlarium in having punctiform colonies, micronematous or semi-macronematous conidiophores, and subglobose and muriform conidia. However, in our phylogenetic analyses based on combined LSU, SSU, ITS, TEF1-α sequence data (Fig. 120), Pseudoconlarium did not fit in with any orders or families in the subclass Diaporthomycetidae and its phylogenetic placement is unstable. Thus, we introduce a new genus and treat it as Diaporthomycetidae genus incertae sedis in this study.
Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, ITS and TEF1-α sequence dataset representing Diaporthomycetidae (Sordariomycetes). Ninety-one strains are included in the combined analyses which comprise 4690 characters (1203 characters for LSU, 1646 characters for SSU, 712 characters for ITS, 1129 characters for TEF1-α) after alignment. Diatrype disciformis (AFTOL-ID 927) in Diatrypaceae (Xylariales) is used as the outgroup taxon. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RAxML tree with a final likelihood values of − 54946.205991 is presented. The matrix had 2477 distinct alignment patterns, with 49.48% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.248881, C = 0.237137, G = 0.282063, T = 0.231918; substitution rates AC = 1.401634, AG = 2.637969, AT = 1.281525, CG = 1.264278, CT = 5.804548, GT = 1.000000; gamma distribution shape parameter α = 0.240704. Bootstrap values for maximum likelihood (ML) equal to or greater than 75% and clade credibility values greater than 0.95 from Bayesian-inference analysis labeled on the nodes. The new isolates are indicated in bold and blue
Pseudoconlarium punctiforme N.G. Liu, K.D. Hyde & J.K. Liu, sp. nov.
Index Fungorum number: IF557095; Facesoffungi number: FoF 06706; Fig. 121
Etymology: Name refers to the punctiform colonies on natural substrate.
Holotype: MFLU 19-2855
Saprobic on decaying wood. Sexual morph Undetermined. Asexual morphColonies on natural substrate effuse, scattered, punctiform. Mycelium partly immersed, partly superficial, composed of brown, septate, branched hyphae. Conidiophores micronematous to semi-macronematous, mononematous, septate, branched, flexuous, subhyaline to pale brown. Conidiogenous cells monoblastic, integrated, terminal, subhyaline to pale brown, determinate, doliform. Conidia 17–33 × 13–24 μm (\( \bar{x} \) = 23.9 × 18.6 μm, n = 30), solitary, median brown, irregularly globose or subglobose, trapeziform, muriform, constricted at the septa, smooth-walled.
Culture characteristics: Conidia germinating on water agar within 48 h. Germ tubes produced from the base of conidia. Mycelia superficial, circular, with entire edge, sparse, white to greyish white from above, grey from below.
Material examined: CHINA, Guizhou Province, Dushan, on decaying wood in a bank of a small freshwater, 6 July 2018, N.G. Liu, DS020 (MFLU 19-2855, holotype), ex-type living culture, GZCC 20-0009.
Genbank numbers: ITS = MT002306, LSU = MN897833, SSU = MN901116, TEF1-α = MT023010.
Notes: Based on a blast search in NCBI, the closest hit using the LSU sequence are Conlarium dupliciascosporum F. Liu & L. Cai (GenBank no. JN936993; Identities = 774/816 (95%), Gaps 2/816). Conlarium duplumascosporum was introduced by Liu et al. (2012) and its asexual morph was produced in culture. Morphologically, C. duplumascosporum and Pseudoconlarium punctiforme share similar conidial size (15.5–35 × 11–26.5 versus 17–33 × 13–24 μm), but P. punctiforme has more conidial septa than those of Conlarium duplumascosporum.
Subclass Hypocreomycetidae O.E. Erikss. & Winka
Glomerellales Chadef. ex Réblová et al.
Notes: The ordinal name “Eu-Glomérellales” was introduced by Chadefaud (1960), but was not validly published. Réblová et al. (2011) proposed the order Glomerellales to accommodate the family Australiascaceae Réblová & W. Gams, Glomerellaceae Locq. ex Seifert & W. Gams and Reticulascaceae Réblová & W. Gams based on morphology as well as ITS, LSU, SSU and RPB2 sequence data analyses. Maharachchikumbura et al. (2016) established the taxonomic position of the family Plectosphaerellaceae W. Gams, Summerb. & Zare in the order Glomerellales. Tibpromma et al. (2018) introduced a monotypic family, Malaysiascaceae Tibpromma & K.D. Hyde to this order based on morphology and phylogeny.
Glomerellaceae Locq. ex Seifert & W. Gams
Notes: Glomerellaceae is a monotypic family comprising pathogens, endophytes and saprobes (Maharachchikumbura et al. 2016), and was accepted in the order Glomerellales by Réblová et al. (2011). The family is characterised by a Colletotrichum asexual morph and a Glomerella sexual morph (Maharachchikumbura et al. 2016).
Colletotrichum Corda
Colletotrichum is the type genus of Glomerellaceae, and it was established by Corda (1831) based on C. lineola Corda (Cannon et al. 2012; Jayawardena et al. 2016). The genus contains 895 epithets (Index Fungorum 2020), with 14 species complexes (Damm et al. 2019). However, less than 250 species have molecular data and are accepted in this genus.
Colletotrichum hederiicola Jayaward. & K.D. Hyde, sp. nov.
Index Fungorum number: IF556899; Facesoffungi number: FoF 06768; Fig. 122
Etymology: Named after host, where the fungus was first discovered.
Holotype: MFLU 15-0689
Saprobic on dead branch of Hedera helix. Sexual morph Undetermined. Asexual morphConidiomata black, acervulus, circular to oval, submerged, solitary or aggregated, conidiophores formed on a cushion of roundish and medium brown cells. Setae not observed. Conidiophores hyaline to pale brown, septate, branched. Conidiogenous cells phialidic, hyaline to pale brown, cylindrical to ovoid or ampulliform, 5.3–11.5 × 2.3–5 μm (\( \bar{x} \) = 7 × 4 μm, n = 15). Conidia hyaline, aseptate, smooth-walled, cylindrical, obtusely rounded apex and a truncated base, guttulate, 7–18 × 2–5 μm (\( \bar{x} \) = 10 × 4 μm, n = 40; L/W ratio = 3.6). Appressoria single, medium brown, globose and rarely lobate, 5–9 × 4–7 μm (\( \bar{x} \) = 6 × 5 μm, n = 5; L/W ratio = 1.3).
Material examined: ITALY, Province of Forlì-Cesena [FC], Fiumana di Predappio, on dead branch of Hedera helix (Araliaceae), 28 October 2014, E. Camporesi, IT 2201 (MFLU 15-0689, holotype).
GenBank numbers: ITS = MN631384, CHS-1 = MN635794, ACT = MN635795.
Notes: The gloeosporioides species complex is mainly known as plant pathogens (Weir et al. 2012; Jayawardena et al. 2016) and some species can be endophytes as well as saprobes (Jayawardena et al. 2016). Species in this genus have a wide host distribution. We were unable to obtain a culture for this strain and the herbarium material was used for direct DNA extraction. Our strain falls within the Colletotrichum gloeosporioides species complex and forms a separate branch with 78% ML and 1.00 BYPP and it has a close affinity with C. henanense F. Liu & L. Cai (Fig. 123). However, our strain differs from C. henanense in having larger conidia with obtusely rounded apex and a truncate base as well as globose appressoria. Colletotrichum hederiicola differs from C. gloeosporioides var. hederae in having conidia with obtusely rounded apex and a truncate base. Colletotrichum gloeosporioides var. hederae has setae while our strain lacks setae. Colletotrichum hederiicola differs from closely related species in CHS-1 and ACT sequence data.
Phylogenetic tree generated by maximum parsimony analysis of combined ITS, GAPDH, CHS-1, ACT and TUB2 sequence data of gloeosporioides species complex. Related sequences were obtained from GenBank. Forty-four strains are included in the analyses, which comprise 1831 characters including gaps. The tree is rooted with C. boninense (CBS 123755). The maximum parsimonious dataset consisted of 1266 constant, 261 parsimony-informative and 304 parsimony-uninformative characters. The parsimony analysis of the data matrix resulted in the maximum often equally most parsimonious trees with a length of 998 steps (CI = 0.696, RI = 0.757, RC = 0.527, HI = 0.304) in the first tree. Bayesian posterior probabilities ≥ 0.90 BYPP and MP bootstrap values ≥ 50% are shown at the nodes. The scale bar indicates 10 changes per site. The ex-type strains are in bold
Hypocreales Lindau
Notes: Species belonging to Hypocreales are highly diverse in tropical and subtropical regions (Põldmaa 2011; Maharachchikumbura et al. 2015). This order comprises 15 families (Sun et al. 2017; Zhuang and Zeng 2017; Wijayawardene et al. 2018a).
Bionectriaceae Samuels & Rossman
Notes: Rossman et al. (1999) introduced Bionectriaceae based on Bionectria Speg. (current name: Clonostachys Corda) as the type genus. Currently, there are 40 accepted genera in the family (Wijayawardene et al. 2018a; Voglmayr and Jaklitsch 2019). Species of Bionectriaceae are associated with fresh or dead plant material, lichens, dung and, are parasitic on other fungi (Maharachchikumbura et al. 2016). Bionectriaceae species are characterised by variously coloured, perithecial or rarely cleistothecial ascomata, aseptate to multi-septate or rarely muriform ascospores and acremonium-like, gliocladium-like, gyrostroma-like or penicillium-like asexual morphs (Rossman et al. 1999; Maharachchikumbura et al. 2016; Voglmayr and Jaklitsch 2019).
Acremonium Link
Notes: Acremonium accommodates saprobic species including important plant pathogens and agents of opportunistic infections in humans (Gams 1971; Summerbell 2003). Acremonium species are difficult to distinguish on morphological characters because they are poorly differentiated in asexual morphs. Molecular phylogenetic analyses have demonstrated that this genus is polyphyletic (Summerbell et al. 2011). A novel species Acremonium chiangraiense is introduced in Acremoniumsensu lato.
Acremonium chiangraiense J.F. Li, R.H. Perera & Phookamsak, sp. nov.
Index Fungorum number: IF556891; Facesoffungi number: FoF 07055; Fig. 124
Acremonium chiangraiense (MFLU 14-0202, holotype). a, b Colonies on palm leaves. c–i, l, m Conidia in chains. j, k. Conidiophores with conidiogenous cells. n–v Conidia. w Germinated conidium. x Culture characteristics on PDA. Scale bars: a = 500 µm, b = 100 µm, d, f, w = 20 µm, e–l = 10 µm, c, m, n–v = 5 µm, x = 0.5 cm
Etymology: Name reflects the location from which it was collected, Chiang Rai, Thailand.
Holotype: MFLU 14-0202
Saprobic on moisted leaves of palm. Sexual morph Undetermined. Asexual morph Hyphomycetous. Colonies effuse, white to light grey, velvety. Mycelium mostly immersed, partly superficial, consisting of septate, branched, thin-walled, white racquet hyphae. Conidiophores 71.2–83 × 8.6–11.2 µm (\( \bar{x} \) = 75.1 × 10 µm, n = 5) micronematous, ampulliform, subhyaline, smooth, thin-walled. Conidiogenous cells 2.5–2.6 × 2.1–2.5 µm (\( \bar{x} \) = 2.4 × 2.3 µm, n = 5), phialidic, globose, subhyaline, smooth. Conidia 11.9–15.3 × 2.9–4.5 µm (\( \bar{x} \) = 14 × 3.9 µm, n = 100), catenate, in branched chains, abundant, ellipsoid-fusiform, narrow and truncate at both ends, hyaline to green-yellow, smooth, thick-walled, aseptate.
Culture characteristics: Conidia germinating on PDA within 14 h and germ tubes produced from all cells. Colonies growing on PDA, hairy, pale white, reaching 5 mm in 15 days at 30 °C, mycelium partly superficial, partly immersed, slightly effuse, radially striate, with irregular edge, pale white; not producing pigmentation on PDA.
Material examined: THAILAND, Chiang Rai Provice, Khun Korn Waterfall, on dead moisted leave of palm, 9 January 2014, J.F. Li, H-10b (MFLU 14-0202, holotype; KUN-HKAS, isotype), ex-type living culture MFLUCC 14-0397.
GenBank numbers: ITS = MN648324, LSU = MN648329.
Notes: Acremonium chiangraiense fits well in Acremonium in having ampulliform conidiophores with phialidic conidiogenous cells and bearing conidia in chains. In phylogenetic tree based on LSU gene, A. chiangraiense forms a separated clade sister to A. luzulae (Fuckel) W. Gams in Acremoniumsensu lato (Bionectriaceae) with signifucant support (94% ML, 0.95 BYPP; Fig. 125). In a comparison of ITS nucleotides, A. chiangraiense has 2.8% (16/568 bp; no gaps) difference from A. luzulae. Acremonium chiangraiense differs from A. luzulae in having smooth conidiophores and larger conidia (11.9–15.3 × 2.9–4.5 µm); A. luzulae has verrucose conidiophores and smaller conidia (3.9–9.3 × 1.5–2.9 µm; Dickinson 1968).
Phylogram generated from the best scoring of the RAxML tree based on LSU sequence dataset of taxa in Acremoniumsensu stricto and related genera in Bionectriaceae. Dactylonectria macrodidyma (CBS 112615), Ilyonectria capensis (CBS 132815), Cylindrocladiella parva (CBS 114524), Nectria cinnabarina (CBS 125165), Hypocrea rufa (DAOM JBT1003), Trichoderma viride (CBS 119326), Hypocrea nigricans (NBRC 30611), Neomyrothecium humicola (CBS 310.96), Paramyrothecium roridum (CBS 357.89) and Stachybotrys chartarum (CBS 182.80) were selected as the outgroup taxa. The best RAxML tree with a final likelihood value of − 5389.526823 is presented. RAxML analysis yielded 261 distinct alignment patterns and 5.49% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.243131, C = 0.224240, G = 0.309793, T = 0.222836, with substitution rates AC = 1.062627, AG = 2.954134, AT = 1.259961, CG = 0.466986, CT = 7.967948, GT = 1.000000. The gamma distribution shape parameter alpha = 0.146261. Tree-Length = 1.834559. Bayesian posterior probabilities (BYPP) from MCMC were evaluated with final average standard deviation of split frequencies = 0.008123. Bootstrap support values for maximum likelihood (ML, left) equal to or greater than 60% is given above or below the nodes. BYPP (right) equal to or greater than 0.95 are given above or below the nodes. Ex-type strains are in black bold and newly generated sequences is indicated in blue bold
Clonostachys Corda
Notes: Corda (1839) introduced Clonostachys based on C. araucaria Corda. Clonostachys species are characterised by penicillate, sporodochial or dimorphic conidiophores and phialidic conidiogenous cells, producing hyaline conidia (Schroers 2001). Rossman et al. (2013) linked the sexual morphic genus Bionectria with Clonostachys and, Bionectria was synonymised under Clonostachys.
Clonostachys eriocamporesiana R.H. Perera & K.D. Hyde, sp. nov.
Index Fungorum number: IF556896; Facesoffungi number: FoF 06963; Fig. 126
Clonostachys eriocamporesiana (MFLU 18-2713, holotype). a Herbarium material. b, c Ascomata on host. d Section of ascoma. e Section of ostiole. f Section of peridium. g Catenophysis. h–k Asci. l–p Ascospores. q Germinating ascospore. r, s Colony on PDA. Scale bars: b, c = 200 µm, d = 100 µm, e, f = 50 µm, g–k = 20 µm, l–q = 10 µm
Etymology: Named after the collector Erio Camporesi.
Holotype: MFLU 18-2713
Hyperparasitic on ascomata of botryosphaeriaceous fungus, occurring on dead stems of Chromolaena odorata.Sexual morphStromata well-developed, erumpent. Ascomata 160–260 × 160–285 μm (\( \bar{x} \) = 200 × 218 μm), perithecial, solitary or densely crowded in groups, globose to subglobose, not collapsing or sometimes slightly laterally pinched when dry, light orange, KOH-, slightly papillate, surface smooth to slightly roughened, but not distinctly warted. Ostiole periphysate. Peridium 32–40 μm wide, two regions: outer region 21–30 μm wide, comprising unevenly thickened hyaline to brownish yellow cells of textura globosa, becoming larger towards the periphery, most cells containing a vacuole; inner region 10–16 μm wide, consists with, flat, hyaline cells of textura angularis. Catenophyses up to 9.5 wide, septate, hyaline. Asci 58–66 × 7.3–10.3 μm (\( \bar{x} \) = 61.4 × 8.8 μm, n = 20), 8-spored, unitunicate, narrowly clavate, apex truncate with somewhat rounded edges, short stalked, apical ring J-. Ascospores 10.5–15.2 × 3.6–4.4 μm (\( \bar{x} \) = 12.5 × 4 μm, n = 30), 2-seriate to 1-seriate, median or submedian, 1-septate, equilateral or inequilateral, not constricted at the septum, ellipsoidal, 2–4-guttulate, hyaline, spinulose when mature. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on PDA within 12 h. Colonies growing on PDA, reaching up to 26 cm in 7 days at 28 °C, surface effused, smooth, margin entire to undulate, aerial mycelium abundant, white, reverse white, yellowish at the centre.
Material examined: THAILAND, Phayao Province, Phachang Noi Pong District, hyperparasite on ascomata of botryosphaeriaceous fungus on dead stems of Chromolaena odorata (L.) R.M. King & H. Rob. (Asteraceae), 9 November 2017, R.H. Perera, Bion 21 (MFLU 18-2713, holotype), ex-type living culture MFLUCC 17-2620.
GenBank numbers: ITS = MN699132, TEF1-α = MN699964, TUB2 = MN699965.
Notes: The new isolate clusters with ex-type of Clonostachys wenpingii (J. Luo & W.Y. Zhuang) Z.Q. Zeng & W.Y. Zhuang (HMAS 172156) and C. byssicola Schroers (CBS 364.78) with low statistical support. However, C. eriocamporesiana differs from C. byssicola in having smaller asci (58–66 versus 68–80 μm) and ascospores (10.5–15.2 × 3.6–4.4 versus 16.8–18.2 × 4.8–5.4 μm) (Schroers 2001). Clonostachys eriocamporesiana differs from C. wenpingii in having light orange ascomata versus yellow ascomata and larger asci (58–66 × 7.3–10.3 versus 33 –44 × 5.5–8.0 μm) (Luo and Zhuang 2007). A comparison of the ITS and TUB2 nucleotides of C. eriocamporesiana and C. byssicola (CBS 364.78) shows 6/480 bp (1%) and 18/569 bp (3.1%) nucleotide differences. Another comparison of the ITS and TUB2 nucleotides of C. eriocamporesiana and C. wenpingii (HMAS 172156) shows 6/480 bp (1%) and 22/574 bp (3.8%) nucleotide differences. ITS and TUB2 nucleotide differences and significant morphological differences reveals that C. eriocamporesiana is a distinct taxon (Jeewon and Hyde 2016).
Clonostachys eriocamporesii R.H. Perera & K.D. Hyde, sp. nov.
Index Fungorum number: IF556897; Facesoffungi number: FoF 06964; Fig. 127
Clonostachys eriocamporesii (MFLU 18-2713, holotype). a Herbarium material. b Ascomata on host. c Section of ascoma. d Section of ostiole. e Section of peridium. f Face view of peridium. g–k Asci. l–q Ascospores. r Germinating ascospore. s, t Colony on PDA. Scale bars: b = 200 µm, c = 100 µm, d–k = 20 µm, l–q = 10 µm, r = 20 µm
Etymology: Named after the collector Erio Camporesi.
Holotype: MFLU 18-2718
Saprobic on stems of Pennisetum polystachion.Stromata absent. Sexual morphAscomata 90–150 × 80–170 μm (\( \bar{x} \) = 118 × 119 μm), perithecial, superficial, solitary to gregarious, globose to subglobose, not collapsing or sometimes slightly laterally pinched when dry, yellowish orange, KOH-, slightly papillate, surface warted. Ostiole periphysate. Perithecial warts 30 μm high, mostly developed in the upper part of the ascomata, off-white to yellowish white, cells angular to globose with unevenly thickened walls. Peridium 15–21 μm thick, composed of two regions: outer region 5–11 μm, 2–3 cells thick, cells angular to globose, with uniformly thickened walls, merging with the cells of the warts, vacuoles not observed; inner region 9–17 μm thick, composed of flat hyaline cells. Paraphyses absent. Asci 40–52.5 × 7.4–9.4 μm (\( \bar{x} \) = 44.8 × 8 μm, n = 20), 4-, 6- or 8-spored, unitunicate, cylindrical to narrowly clavate, apex flat with somewhat rounded edges, short pedicellate, apical ring J-. Ascospores 10–12.7 × 2.7–4.2 μm (\( \bar{x} \) = 11.3 × 3.5 μm, n = 30), 1–2-seriate, hyaline, median or submedian, 1-septate, equilateral or inequilateral, slightly constricted at the septum, ellipsoidal to occasionally subglobose, multi-guttulate, smooth-walled. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on PDA within 12 h. Colonies growing on PDA, reaching up to 20 cm in 7 days at 28 °C, surface effused, smooth, margin entire to undulate, aerial mycelium abundant, zonate, white, reverse white.
Material examined: THAILAND, Southern Thailand, on dead stems of Pennisetum polystachion (L.) Schult. (Poaceae), 11 November 2017, A. Karunarathne, Bion 78 (MFLU 18-2718, holotype), ex-type living culture MFLUCC 19-0486.
GenBank numbers: ITS = MN699133, LSU = MN699128.
Notes: Our strain MFLUCC 19-0486 clusters with the ex-type of Clonostachys epichloë Schroers (CBS 101037) and C. parva (Schroers) Rossman, L. Lombard & Crous (CBS 997.69) with high statistical support (100% ML, 1.00 BYPP; Fig. 128). DNA sequences of C. eriocamporesii and C. epichloë (CBS 101037) differ in 6 nucleotides of the ITS region (1.1%), and 4 nucleotides of LSU region (0.8%). DNA sequences of new fungus and ex-type of C. parva (CBS 997.69) differ in 8 nucleotides of the ITS region (1.6%), and 10 nucleotides of LSU region (1.2%). Clonostachys eriocamporesii is similar to C. epichloë and C. parva in having perithecial warts (Schroers 2001). However, it can be distinguished from C. epichloë by its larger ascospores (10–12.7 versus 9–10.8 μm) (Schroers 2001). Clonostachys eriocamporesii can be distinguished from C. parva in having smooth-walled, smaller ascospores (10–12.7 versus 12.2–13.4 μm) while C. parva has finely spinulose ascospores (Schroers 2001). We, therefore, introduce C. eriocamporesii as a novel taxon.
Phylogram generated from RAxML analysis based on combined ITS and TUB2 sequence data of Clonostachys isolates. Related sequences were obtained from GenBank. Seventy-one taxa are included in the analyses, which comprise 3352 characters including gaps. Single gene analyses were carried out and compared with each species, to compare the topology of the tree and clade stability. The tree is rooted to Nectria cinnabarina NRRL 20484 and Fusarium acutatum CBS 402.97. Tree topology of the ML analysis was similar to the BI. The best scoring RAxML tree with a final likelihood value of − 12301.505960 is presented. The matrix had 608 distinct alignment patterns, with 19.34% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.209639, C = 0.282711, G = 0.249015, T = 0.258636; substitution rates AC = 1.056089, AG = 2.708175, AT = 1.086912, CG = 0.685016, CT = 3.448496, GT = 1.000000; gamma distribution shape parameter α = 0.804098. Maximum likelihood bootstrap support values ≥ 75% (BT) and Bayesian posterior probabilities ≥ 0.99 (BYPP) are given at the nodes as ML/BYPP. The scale bar indicates 0.07 changes. The isolates obtained in this study are in blue and ex-types taxa are in black bold
Nectriaceae Tul. & C. Tul.
Notes: Nectriaceae (Sordariomycetes, Hypocreales) is a large family that contains 63 genera, based on sexual or asexual forms (Wijayawardene et al. 2018a; Index Fungorum 2020). The fungi of this family are found as soil-borne saprobes or obligatory or facultative plant pathogens. Some are fungicolous or insecticolous. Species are also reported as pathogenic human opportunists or producers of mycotoxins of medical concern (Lombard et al. 2015; Hyde et al. 2018a; Sun et al. 2019).
Mariannaea G. Arnaud ex Samson
Notes: Mariannaea, proposed initially by Arnaud in 1952 was recomposed by Samson (1974). Species of Mariannaea are characterised by their flask-shaped phialides, 1- or 2-celled conidia forming imbricate or straight chains. The genus accommodates species from soils, freshwater and wood (Hu et al. 2016) and contains 22 species (Index Fungorum 2020). A new species isolated from soil of Atlantic Forest, Pernambuco, Brazil is described based on morphological characters and phylogeny of the DNA sequences of the ITS and LSU regions.
Mariannaea atlantica A.L. Alves, A.C.S Santos & P.V. Tiago, sp. nov.
Index Fungorum number: IF556755; Facesoffungi number: FoF 06376; Fig. 129
Etymology: The specific epithet “atlantica” refers to forest type whose soil sample was collected and the fungus was isolated.
Holotype: URM 93044
Saprobic on soil. Sexual morph Undetermined. Asexual morph Hyphomycetous, amerosporous. Hyphae 2–8 μm wide, septate, hyaline, smooth, thin-walled, branched. Conidiophores up to 327.5 μm long, 3–5 μm wide at the base cell, macronematous, mononematous, erect, straight, smooth or verrucose, thin-walled, septate, hyaline, cylindrical, tapering with base cell wall slightly verrucouse, simple with 1 phialide or verticillate in the upper part. Phialides 12–20 × 2–3 μm length/width, flask-like, hyaline, smooth-walled, solitary or grouped with 2–7 in the same branch. Conidia 5–10 × 2–4 μm length/width, fusoid, hyaline, thin-walled, smooth, produced in imbricate chains. Chlamydospores single, terminal and intercalary, globose with 4–15 μm diam. and doliiform with 8–20 × 5–10 μm length/width, hyaline, thick-walled.
Culture characteristics: Colonies growing on PDA, circular, reaching 4–6 cm diam. in 2 weeks at 25 °C, white to ochreous closest to margins and rosy buff at centre; zonate; reverse ochreous close to margins to sepia at centre after 7 days, becoming rose at centre and wine to margins on verse and wine on reverse after 14 days.
Material examined: BRAZIL, Paudalho, Pernambuco state, on soil of Atlantic Forest, 20 July 2017, A.L. Alves, URM 93044 (holotype), ex-type living culture, URM 8146.
GenBank numbers: ITS = MN151372, LSU = MN151398.
Notes: ITS and LSU sequences are important markers for identification of Mariannaea species. Based on the current phylogenetic analysis, the new species Mariannaea atlantica forms a well-supported clade with M. terricola A.L. Alves et al., M. punicea (Samson) D.M. Hu & L. Cai and M. fusiformis D.M. Hu & L. Cai (Fig. 130), which present in common red–purple colonies after 14 days in PDA at 25 °C. Within this clade, M. atlantica is a distinct lineage, clustering with M. fusiformis. However, M. fusiformis differs in width and length of conidiophores, phialides, conidia and chlamydospores. In addition, M. fusiformis has fusiform to subglobose conidia and globose to subglobose chlamydospores, while M. atlantica has fusoid conidia and globose and doliform chlamydospores. Mariannaea atlantica was isolated from soil collected in the Brazilian Tropical Atlantic Forest, in the city of Paudalho, Pernambuco state.
Bayesian inference tree based on combined LSU and ITS sequence data, generated using MrBayes 3.2.6 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). Sequences of 18 strains were included in the combined analyses which comprised 1256 characters (566 characters for LSU and 690 characters for ITS) after alignment. Calonectria brassicae (CBS 111869) (Nectriaceae, Hypocreales) is used as the outgroup taxon. Estimated base frequencies were as follows: A = 21.77%, C = 22.12%, G = 24.64%, T/U = 22.10%, other: 9.37%; alpha shape parameter α = 0.10. Bayesian posterior probabilities greater than 0.50 and Bootstrap support values for ML greater than 50% are given near nodes. Bar represents substitutions expected per site. The tree is rooted with Calonectria brassicae (CBS 111869). Ex-type strains are in bold. The newly generated sequence was indicated in bold and blue
Subclass Savoryellomycetidae Hongsanan et al.
Conioscyphales Réblová & Seifert
Notes: Conioscyphales was introduced by Réblová et al. (2016c) based on a combined multi-gene (ITS, SSU, LSU, TUB2, RPB2 and MCM7) sequence dataset. The order was placed within Hypocreomycetidae in Réblová et al. (2016c). It clustered within the newly introduced subclass Savoryellomycetidae (Hongsanan et al. 2017). Conioscyphales is a monotypic order with Conioscyphaceae as the type family.
Conioscyphaceae Réblová & Seifert
Notes: Réblová et al. (2016c) established the family Conioscyphaceae to accommodate the holomorphic genus Conioscypha Höhn. Conioscyphascus Réblová & Seifert was introduced to accommodate sexual morphs which have Conioscypha asexual morphs (Réblová and Seifert 2004). Connections between Conioscypha and Conioscyphascus have been proven by cultural studies and with molecular evidence (Réblová and Seifert 2004; Zelski et al. 2015). Réblová et al. (2016b) recommended to protect Conioscypha over Conioscyphascus (Art. 59.1).
Conioscypha Höhn.
Notes: Von Höhnel (1904) introduced Conioscypha with C. lignicola Höhn. as the type species. Currently, 17 species are accepted in the genus including 14 asexual species (C. aquatica Z.L. Luo, K.D. Hyde & H.Y. Su, C. bambusicola Matsush., C. boutwelliae Hern.-Restr., C. dimorpha Matsush., C. fabiformis Matsush., C. hoehnelii P.M. Kirk, C. japonica Udagawa & Toyaz., C. lignicola, C. minutispora Hern.-Restr., Gené & Guarro, C. nakagirii Chuaseehar., Somrith., Suetrong & Boonyuen, C. pleiomorpha Hern.-Restr., C. submersa Z.L. Luo, K.D. Hyde & H.Y. Su, R.F. Castañeda & Gené, C. taiwaniana J.L. Chen & Tzean, C. tenebrosa N.G. Liu, K.D. Hyde & J.K. Liu), and one sexual morph (C. gracilis (Munk) Zelski, Raja, A.N. Mill. & Shearer) and two holomorphs (C. peruviana Zelski, Raja, A.N. Mill. & Shearer and C. varia Shearer). Conioscypha species are known from freshwater and terrestrial habitats on dead decaying wood, leaves or bamboo stems, some species are also isolated from soil or animal skin and hairs (Shearer 1973; Shearer and Motta 1973; Cole and Samson 1979; Goh and Hyde 1998a; Réblová and Seifert 2004; Crous et al. 2018a). Sexual morphs of Conioscypha are characterised by pale-coloured, immersed to superficial ascomata, papillate or elongated neck, cylindrical to clavate, unitunicate asci with a pronounced J-, apical ring, and narrowly fusiform to fusiform-navicular, hyaline ascospores, lacking a mucilaginous sheath or appendages (Réblová and Seifert 2004; Zelski et al. 2015; Réblová et al. 2016c). Asexual morphs of Conioscypha have unique conidiogenesis which occurs at inconspicuous loci along hyphae; a basipetal succession of blastically produced, aseptate, brown conidia leave behind hyaline, cup-like collarettes that are remnants of the initial outer wall of the conidia; these accumulate centripetally to form a multi-layered collarette appearing similar to annellations (Shearer and Motta 1973; Goh and Hyde 1998a).
Conioscypha verrucosa J. Yang & K.D. Hyde, sp. nov.
Index Fungorum number: IF556630; Facesoffungi number: FoF 06275; Fig. 131
Etymology: Referring to the verrucose conidia.
Holotype: MFLU 18-1503
Saprobic on twigs from freshwater habitats. Sexual morph Undetermined. Asexual morphColonies on natural substrate sporodochial, scattered, dark brown or black. Mycelium partly immersed, partly superficial, composed of septate, hyaline hyphae. Conidiophores up to 40 μm long, macronematous, mononematous, compact, flexuous, simple or branched, mostly moniliform, with globose to subglobose, ellipsoidal or clavate cells, hyaline, smooth-walled, sometimes reduced to conidiogenous cells. Conidiogenous cells 5.5–13 × 5–11.5 μm (\( \bar{x} \) = 9 × 7.6 μm, n = 30) monoblastic, integrated, terminal, globose to ellipsoidal, hyaline, smooth-walled. Conidia 12.5–23 × 10.5–20 μm (\( \bar{x} \) = 16.5 × 14.5 μm, n = 50), acrogenous, globose, subglobose, ellipsoidal or obovoid, aseptate, verrucose, guttulate, dark olivaceous to dark brown, with a central basal pore.
Culture characteristics: Conidia germinating on WA within 24 h and germ tubes produced from both ends. Colonies on PDA reaching 5–10 mm diam. at two weeks at 25 °C, in natural light, circular, wrinkled, with sparse dark brown mycelium in the middle, sparse pale brown mycelium on the outer ring with irregular margin; in reverse with a brown inner ring and a pale brown outer ring.
Material examined: CHINA, Guizhou Province, Dushan District, 25° 57.9′ N, 107° 39′ E, on decaying wood submerged in a freshwater stream, 26 August 2017, J. Yang, SG 2-1 (MFLU 18-1503, holotype; KUN-HKAS 102158, isotype), ex-type living culture MFLUCC 18-0419.
GenBank numbers: ITS = MN061350, LSU = MN061364, RPB2 = MN061668, SSU = MN061352.
Notes: In the phylogenetic analysis Conioscypha verrucosa clustered within Conioscypha as a sister taxon to C. pleiomorpha with strong support (100% ML, 91% MP, 1.00 BYPP; Fig. 135), based on a combined LSU, ITS, SSU and RPB2 sequence dataset comprising 60 taxa. Morphologically, Conioscypha verrucosa and C. pleiomorpha share similar conidial morphology having unicellular, globose, subglobose, ellipsoidal or obovoid conidia of similar size. But the conidia of C. verrucosa are olivaceous green to dark brown while those in C. pleiomorpha are mid brown. Conioscypha pleiomorpha has micronematous conidiophores, often reduced to conidogenous cells; conspicuous cupulate conidiogenous cells forming multi-layer, cupulate collarette after several percurrent proliferations and enteroblastic conidia. However, the unique generic feature of conidiogenesis was not observed in Conioscypha verrucosa. Conioscypha verrucosa produces macronematous, mononematous, septate or moliniform conidiophores, globose to ellipsoidal conidiogenous cells and holoblastic conidia.
Pleurotheciales Réblová & Seifert
Notes: Pleurotheciales was introduced by Réblová et al. (2016c) based on morphological characters and phylogenetic analyses. Members of Pleurotheciales are mostly saprobic on wood and rarely human pathogens causing keratomycosis (Réblová et al. 2016c). Guarro et al. (2000) reported that Phaeoisaria clematidis caused a corneal ulcer in a Brazilian man. An updated phylogenetic tree for Pleurotheciales is provided (Fig. 132) and Neomonodictys is introduced as a new genus.
Pleurotheciaceae Réblová & Seifert
Notes: Pleurotheciaceae was typified by Pleurothecium with Pleurothecium recurvatum (Morgan) Höhn. as the type species (Réblová et al. 2016c). Hyde et al. (2018a) introduced two new species of Pleurotheciella from Thailand and a new species of Phaeoisaria from China. We introduce a new genus Neomonodictys with Neomonodictys muriformis as the type species based on morphological characteristics and phylogenetic analyses.
Neomonodictys Y.Z. Lu, C.G. Lin & K.D. Hyde, gen. nov.
Index Fungorum number: IF556730; Facesoffungi number: FoF 06710
Etymology: Name reflects its morphological similarity to Monodictys.
Saprobic on decaying wood. Sexual morph Undetermined. Asexual morph Hyphomycetous, dictyosporous. Colonies on natural substratum superficial, scattered, black, glistening. Mycelium immersed in the substrate, composed of septate, branched, smooth, thin-walled, hyaline to pale brown hyphae. Conidiophores lacking. Conidiogenous cells holoblastic, monoblastic, integrated, terminal, determinate, brown. Conidia acrogenous, solitary, subglobose to globose, muriform, pale brown when immature, darkened to black when mature, sometimes with one basal cell paler than the others.
Type species: Neomonodictys muriformis Y.Z. Lu, C.G. Lin & K.D. Hyde
Notes: The monophyletic asexual genus Neomonodictys is established for a fungus collected from freshwater habitat from Thailand that is morphologically similar to members of Monodictys S. Hughes, but phylogenetically distinct (Fig. 132). Monodictys is characterised by monoblastic, hyaline to brown conidiogenous cells, solitary, dictyospores, subglobose, pyriform or clavate conidia (Ellis 1971; Seifert et al. 2011). Conidiophores of Monodictys are often reduced to conidiogenous cells (Ellis 1971; Seifert et al. 2011). Monodictys was placed in Dothideomycetes (Day et al. 2006; Seifert et al. 2011; Wijayawardene et al. 2017a, 2018a). Phylogenetic inference in this study showed that Neomonodictys muriformis formed a highly supported clade within Pleurotheciaceae (Pleurotheciales, Sordariomycetes) (Fig. 132), which is distant to Monodictys. Therefore, the new generic name, Neomonodictys, is introduced.
Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence data representing Pleurotheciales and the closely related orders. Eighty-five strains are included in the combined analyses which comprise 1871 characters (1194 characters for LSU, 677 characters for ITS) after alignment. Leotia tubrica (AFTOL-1) is used as the outgroup taxon. The best RAxML tree with a final likelihood values of − 18609.630764 is presented. The matrix had 1200 distinct alignment patterns, with 30.71% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.231344, C = 0.256609, G = 0.300675, T = 0.211372; substitution rates AC = 1.000000, AG = 3.698119, AT = 1.000000, CG = 1.000000, CT = 3.698119, GT = 1.000000; gamma distribution shape parameter α = 0.309377. Bootstrap values for maximum likelihood (ML) equal to or greater than 75% and clade credibility values greater than 0.90 from Bayesian-inference analysis labeled on the nodes. Ex-type strains are in bold and black, the new isolate is indicated in bold and red
Neomonodictys muriformis Y.Z. Lu, C.G. Lin & K.D. Hyde, sp. nov.
Index Fungorum number: IF556731; Facesoffungi number: FoF 06819; Fig. 133
Etymology: Name reflects its muriform conidia.
Holotype: MFLU 17-1178
Saprobic on decaying wood in a freshwater stream. Sexual morph Undetermined. Asexual morph Hyphomycetous, dictyosporous. Colonies on natural substratum superficial, scattered, black, glistening. Mycelium immersed in the substrate, composed of septate, branched, smooth, thin-walled, hyaline to pale brown, 2–3 μm wide hyphae. Conidiophores lacking. Conidiogenous cells holoblastic, monoblastic, integrated, terminal, determinate, brown. Conidia 20–30 × 15–25 μm (\( \bar{x} \) = 22 × 20 μm, n = 20), acrogenous, solitary, subglobose to globose, muriform, smooth-walled, pale brown when immature, darkened to black when mature, sometimes with one basal cell paler than the others.
Culture characteristics: Conidia germinating on WA and germ tubes produced from conidia within 36 h. Colonies growing on MEA, circular, with flat surface, edge entire, reaching 19 mm in 6 weeks at 28 °C, pale brown to brown in MEA medium. Mycelium superficial and partially immersed, branched, septate, hyaline to pale brown, smooth.
Material examined: THAILAND, Krabi Province, Plai Praya, Khao To, Ban Bang Thao Mae, on submerged decaying wood in a freshwater stream, 17 December 2015, S. Boonmee, BTM05-4 (MFLU 17-1178, holotype), ex-type living culture, MFLUCC 16-1136.
GenBank numbers: ITS = MN644509, LSU = MN644485, TEF1-α = MN646856.
Savoryellales Boonyuen et al.
Notes: Boonyuen et al. (2011) established the order Savoryellales in the subclass Hypocreomycetidae based on the phylogenetic analyses of a combined LSU, SSU, 5.8S rDNA, RPB1, RPB2 and TEF1-α sequence dataset. Hongsanan et al. (2017) raised Savoryellales to subclass Savoryellomycetidae based on both phylogenetic analysis and molecular clock analysis.
Savoryellaceae Jaklitsch & Réblová
Notes: The family Savoryellaceae includes three holomorph genera viz. the type genus Savoryella E.B.G. Jones & R.A. Eaton, Ascotaiwania Sivan. & H.S. Chang and Canalisporium Nawawi & Kuthub. (Sri-indrasutdhi et al. 2010; Sadowski et al. 2012; Réblová et al. 2016c; Yang et al. 2017; Zhang et al. 2019b). Two new genera Dematiosporium and Helicoascotaiwania Dayarathne, Maharachch. & K.D. Hyde were introduced to the family (Dayarathne et al. 2019; Luo et al. 2019).
Canalisporium Nawawi & Kuthub.
Notes: Canalisporium was established by Nawawi and Kuthubutheen (1989) to accommodate Berkleasmium caribense Hol.-Jech. & Mercado, B. pulchrum Hol.-Jech. & Mercado and a new species Canalisporium elegans Nawawi & Kuthub. So far, 15 species are known in the genus, which are common on submerged bamboo and wood in freshwater (Goh and Hyde 1998b; Ho et al. 2002). Ascothailandia grenadoidea Sri-indr. et al., the only known sexual morph in the genus, was introduced by Sri-indrasutdhi et al. (2010) with the Canalisporium asexual morph sporulating from in culture. Further molecular studies also supported the connection between Ascothailandia and Canalisporium (Sri-indrasutdhi et al. 2010; Jones et al. 2016; Réblová et al. 2016c; Hernández-Restrepo et al. 2017). Réblová et al. (2016b) recommended to protect Canalisporium over Ascothailandia (Art. 59.1). Canalisporium is morphologically similar to Berkleasmium in having muriform conidia born in sporodochia but conidia in Canalisporium are complanate and have ordered rows of cells (Bussaban et al. 2001; Cai et al. 2006). In this study a novel species, C. aquaticium is introduced from decaying wood submerged in a freshwater stream in Thailand.
Canalisporium aquaticium J. Yang & K.D. Hyde, sp. nov.
Index Fungorum number: IF556631; Facesoffungi number: FoF 06276; Fig. 134
Etymology: Referring to the aquatic habitat.
Holotype: MFLU 18-1504
Saprobic on twigs from freshwater habitats. Sexual morph Undetermined. Asexual morphColonies sporodochial, scattered, punctiform, pulvinate, dark brown to black, glistening. Mycelium immersed, consisting of branched, septate, smooth, thin-walled, pale to brown hyphae. Conidiophores micronematous or semi-macronematous, subhyaline or lightly pigmented, smooth, thin-walled, septate, simple or sparsely branched, loosely fasciculate, becoming vesiculate and disintegrating as conidia mature, up to 66 μm long, 3.5–6 μm wide. Conidiogenous cells monoblastic, integrated, terminal, determinate, hyaline or pale brown, cylindrical to ellipsoidal, giving rise to a single crop of conidia which mature synchronously. Conidia acrogenous, holoblastic, solitary, yellowish brown to mid brown, guttulate, muriform, with ordered arrangement of vertical and transverse septa, slightly constricted at the septa, complanate, smooth-walled, consisting of three vertical columns and 7–8 transverse rows of cells with a single cuneiform basal cell, 45–58 × 21–25.5 μm (\( \bar{x} \) = 51 × 23 μm, n = 30), thickened at the septa of middle column.
Culture characteristics: Conidia germinating on PDA within 24 h and germ tubes produced from several cells. Colonies on PDA slow-growing, reaching 5–10 mm diam. at a month at 25 °C, in natural light, circular, with sparse dark brown mycelium on the surface with entire margin, in reverse dark brown to black.
Material examined: THAILAND, Phang Nga Province, Bann Tom Thong Khang, on decaying wood submerged in a freshwater stream, 17 December 2015, J. Yang, Site 7-12-1 (MFLU 18-1504, holotype; KUN-HKAS 102159, isotype), ex-type living culture MFLUCC 16-0862.
GenBank numbers: ITS = MN061351, LSU = MN061365, SSU = MN061353.
Notes: Canalisporium aquaticium, C. kenyense Goh, W.H. Ho & K.D. Hyde and C. pulchrum (Hol.-Jech. & Mercado) Nawawi & Kuthub. have three columns of conidial cells, C. variabile Goh & K.D. Hyde rarely has three columns and mostly has two columns of conidial cells, while the remaining species produce conidia with two columns (Nawawi and Kuthubutheen 1989; Goh and Hyde 2000; Phookamsak et al. 2019). Canalisporium aquaticium differs from C. kenyense in having yellowish brown conidia with a single basal cell, while C. kenyense has reddish brown to dark brown conidia often with three basal cells (Goh et al. 1998; Phookamsak et al. 2019). Canalisporium aquaticium is distinguished from C. variabile in having regular muriform conidia which are slightly constricted at the septa, while C. variabile has variable-shaped conidia which are strongly constricted at the septa (Goh and Hyde 2000). In addition, conidia in C. aquaticium (45–58 × 21–25.5 μm) are wider than those in C. kenyense (34–56 × 24–34 μm) and larger than those in C. variabile (22–35 × 15–23 μm).
Canalisporium aquaticium mostly resembles C. pulchrum in having regular muriform conidia of a similar size with a single cuneiform basal cell. However, conidia of C. aquaticium (45–58 × 21–25.5 μm) often comprise 7–8 transverse rows of cells, while conidia of C. pulchrum (36–55 × 22–27 μm) have 5–7 rows (Nawawi and Kuthubutheen 1989). Conidiophores in C. aquaticium are hyaline and up to 66 μm long but those in C. pulchrum are mostly pale brown and up to 20 μm long. The combined four-gene phylogenetic analyses (Fig. 135) showed that C. aquaticium clustered close to C. elegans Nawawi & Kuthub., C. jinghongense L. Cai, K.D. Hyde & McKenzie and C. pulchrum with significant support (79% ML, 95% MP, 1.00 BYPP). Sequence data of C. pulchrum generated in Sri-indrasutdhi et al. (2010) with only one conidial photo which matches the conidial characters in the original diagnosis. However, sequence data for its ex-type is unavailable. Canalisporium pulchrum has a worldwide in distribution (Goh et al. 1998; Ferrer and Shearer 2005; Damyanti et al. 2015). Goh et al. (1998) showed several collections of the species but one collection from Brunei (HKU(M) 2911) shares the same conidial characters with C. aquaticium. We therefore assume that the previous collections of C. pulchrum may not be correctly identified for all collections. Thus, the epitypification of C. pulchrum is necessary to resolve the taxonomic problem (Hyde and Zhang 2008).
Maximum likelihood majority rule consensus tree for the analyzed Sayoryellales and Conioscyphales isolates based on a dataset of combined LSU, ITS, SSU and RPB2 sequence data. Bootstrap support values for maximum likelihood (ML) and maximum parsimony (MP) greater than 75% and Bayesian posterior probabilities greater than 0.95 are indicated above the nodes as ML/MP/BYPP. The scale bar represents the expected number of changes per site. The tree is rooted with Plagiascoma frondosum. The strain numbers are noted after the species names with ex-type strains and direct sequence from holotype indicated in bold. The new strains are in blue bold. Branches with 100% ML, 100% MP and 1.00 BYPP are shown as black nodes. Genera are indicated as coloured blocks
Subclass Sordariomycetidae O.E. Erikss & Winka
Coniochaetales Huhndorf et al.
Notes: Coniochaetaceae was shown to be independent of Sordariales and Coniochaetales was established to accommodate this family by Huhndorf et al. (2004). García et al. (2006) clarified the phylogenetic relationships of Coniochaetales based on SSU and LSU data. The MCC tree supports that Cordanales should be synonymised under Coniochaetales (77 MYA) (Hongsanan et al. 2017; Hyde et al. 2017). The order is characterised by glabrous or setose ascomata, aseptate ascospores with germ slit and phialidic conidiogenesis (Huhndorf et al. 2004; Phookamsak et al. 2019).
Coniochaetaceae Malloch & Cain
Notes: Coniochaetaceae was introduced by Malloch and Cain (1971) and is typified by Coniochaeta (Sacc.) Cooke. Maharachchikumbura et al. (2015) listed Barrina A.W. Ramaley, Coniochaeta (Sacc.) Cooke and Lecythophora Nannf in Coniochaetaceae based on multi-gene analysis. Khan et al. (2013) treated the asexual genus Lecythophora Nannf. as a synonym of Coniochaeta based on nomenclature study and this is in agreement of Maharachchikumbura et al. (2016) and Wijayawardene et al. (2018a). Species of Coniochaetaceae are characterised by brown to black ascomata, cylindrical to clavate asci and ascospores usually with elongated germ slits and ellipsoid to reniform conidia (Malloch and Cain 1971; Huhndorf et al. 2004; Khan et al. 2013; Maharachchikumbura et al. 2016; Wanasinghe et al. 2018).
Coniochaeta (Sacc.) Cooke
Notes: Coniochaeta, typified by Coniochaeta ligniaria (Grev.) Cooke, was established by Cooke (1887). Coniochaeta is characterised by pyriform to globose ascomata, cylindrical asci, ellipsoid to globose ascospores with an elongated germ slit and hyaline to orange, ellipsoid to reniform conidia (Huhndorf et al. 2004; García et al. 2006; Khan et al. 2013; Maharachchikumbura et al. 2016; Wanasinghe et al. 2018; Phookamsak et al. 2019). Based on morphological data and phylogenetic inferences, a new species Coniochaeta vineae is introduced.
Coniochaeta vineae S.K. Huang, T.C. Wen & K.D. Hyde, sp. nov.
Index Fungorum: IF556804; Facesoffungi number: FoF 06526; Fig. 136
Coniochaeta vineae (KUN-HKAS 99606, holotype). a Material. b Appearance of ascomata on the host. c Obpyriform ascoma. d Ascomata cross section. e Peridium. f Periphyses. g Asci with paraphyses (in Melzer’s reagent). h–j Asci (j stained in Melzer’s reagent). k–o Ascospores (l–o show germ slits). Scale bars: c = 200 μm, d = 100 μm, e–j = 50 μm, k–o = 5 μm
Etymology: The specific name “vineae” refers to the host vine, of which the holotype was collected.
Holotype: KUN-HKAS 99606
Saprobic on dead vine. Sexual morphAscomata 170–185 × 145–185 μm (\( \bar{x} \) = 180 × 170 μm, n = 5), perithecial, gregarious and scattered, semi-immersed to superficial, unilocular, obpyriform to subglobose, dark brown to black, roughened surface, the ostiole periphysate, covered by dark brown, inconspicuous, straight setae with rounded apex. Peridium 15–35 μm diam. near to ostiole, 10–20 μm diam. at base, membranaceous, outer layer composed of brown cells of textura angularis; inner layer composed of hyaline cells of textura prismatica. Paraphyes 1–3.5 μm wide, numerous, filiform, septate, branched. Asci 85–115 × 5–10 μm (\( \bar{x} \) = 98 × 7.5 μm, n = 20), 8-spored, unitunicate, cylindrical, with short pedicel, rounded at the apex, with a distinct, J-, apical ring. Ascospores 6.5–9.5 × 4–6 μm (\( \bar{x} \) = 8 × 5 μm, n = 50), 1-seriate, hyaline becoming dark brown, ovoid to broadly ellipsoid, aseptate, multi-guttulate, with a conspicuous, straight germ slit across the entire length, smooth, without sheath or appendages. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on PDA within one week at 23 °C. Colony on PDA reaching 2 cm diam., after 30 days at 23 °C. Mycelium superficial to semi-immersed, filamentous, branched, septate, hyaline. Colonies on PDA, initially white, becoming grayish-olivaceous, rough surface and raised elevation, with concentric zonations, smooth margins; reverse white to pale brown at the extremities.
Material examined: CHINA, Xinjiang, Zhaosu, a lake near by the snow mountains, on dead vine, 14 September 2017, J.W. Liu, XJ03 (KUN-HKAS 99606, holotype), ex-type living culture KUMCC 17-0322.
GenBank numbers: ITS = MN473469, LSU = MN473512, SSU = MN473471, TUB2 = MN485898, RPB2 = MN480811.
Notes: Based on LSU-ITS phylogenetic analyses, Coniochaeta vineae clusters with C. ligniaria (Grev.) Cooke and C. mutabilis (J.F.H. Beyma) Z.U. Khan, Gené & Guarro, with a significant support in the ML analysis (72% ML; Fig. 137). Coniochaeta vineae is compared across the ITS regions reveals 2% differences between C. ligniaria and 4% differences between C. mutabilis, and the LSU region reveals 2% differences between C. ligniaria and 7% differences between C. mutabilis. Coniochaeta mutabilis was reported as a human and plant pathogen which is only known in asexual morph (Mitteilung 1939; Drees et al. 2007; Ňancucheo and Johnson 2012). Coniochaeta vineae is characterised by inconspicuous ostioles, surrounded by setae and ovoid to broadly ellipsoid ascospores, while C. ligniaria has irregular black setae covering the ascomata and ellipsoid ascospores (Checa et al. 1988).
Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence data in Coniochaetaceae. Related sequences are taken from Samarakoon et al. (2018) and Phookamsak et al. (2019). Fifty strains are included in the combined genes sequence analyses which comprise total 1368 characters (852 characters for LSU, 516 characters for ITS) after alignment. Chaetosphaeria innumera (SMH2748) and C. pygmaea (MR 1365) in Chaetosphaeriaceae are used as outgroup taxa. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RAxML tree with a final likelihood values of − 6998.732264 is presented. The matrix had 1968 distinct alignment patterns, with 31.90% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.242211, C = 0.2460165, G = 0.2883425, T = 0.2234305; substitution rates AC = 1.1566885, AG = 1.7661595, AT = 0.9155275, CG = 1.246033, CT = 7.313758, GT = 1.000000; gamma distribution shape parameter α = 0.787081. Bootstrap values for maximum likelihood (ML) equal to or greater than 70% and clade credibility values greater than 0.90 (the rounding of values to 2 decimal proportions) from Bayesian-inference analysis labeled on the nodes. Ex-type strains are in bold and black, the new isolate is indicated in bold and blue
Pseudodactylariales Crous
Notes: Pseudodactylariales was introduced by Crous et al. (2017) based on two dactylaria-like species. Pseudodactylariales formed a distinct clade close to Chaetosphaeriales Huhndorf, A.N. Mill. & F.A. Fernández, Sordariales Chadef. ex D. Hawksw. & O.E. Erikss. and Vermiculariopsiellales Hern.-Restr., J. Mena, Gené & Crous in the phylogenetic analysis of Crous et al. (2017). Pseudodactylariales is a monotypic order with Pseudodactylariaceae as the type family and currently placed in the subclass Sordariomycetidae.
Pseudodactylariaceae Crous
Notes: The monotypic family Pseudodactylariaceae was introduced in a newly established order Pseudodactylariales by Crous et al. (2017). Only one asexual genus Pseudodactylaria is accommodated in Pseudodactylariaceae. The familial placement of Pseudodactylariaceae was confirmed by Lin et al. (2018).
Pseudodactylaria Crous
Notes: Pseudodactylaria was introduced by Crous et al. (2017) to accommodate a new species P. xanthorrhoeae Crous and a new combination P. hyalotunicata (C.K.M. Tsui, Goh & K.D. Hyde) Crous and typified by P. xanthorrhoeae. Pseudodactylaria is characterised by erect, hyaline conidiophores, integrated conidiogenous cells with a rachis and hyaline, fusoid-ellipsoid conidia surrounded by a thin mucilaginous sheath (Crous et al. 2017). Pseudodactylaria was distinguished from Dactylaria Sacc. by 1-septate conidia covered by a mucoid sheath (Crous et al. 2017), but lacking a mucoid sheath in P. brevis C.G. Lin, McKenzie & K.D. Hyde (Lin et al. 2018). Pseudodactylaria species are saprobic on submerged wood or dead wood collected from Australia (Crous et al. 2017), China (Tsui et al. 1997) and Thailand (Lin et al. 2018). We introduce the fourth species P. camporesiana, which was collected from submerged wood in Thailand. No sheath was found in P. camporesiana.
Pseudodactylaria camporesiana W. Dong, Doilom & K.D. Hyde, sp. nov.
Index Fungorum number: IF556924; Facesoffungi number: FoF 06344; Fig. 138
Pseudodactylaria camporesiana (MFLU 18-1535, holotype). a Colonies on submerged wood. b Conidiophore, conidiogenous cell and attached conidia. c–e Upper part of conidiophores and conidiogenous cells. f–i Conidia. j Colony on PDA (from front). k Colony on PDA (from reverse). Scale bars: b–e = 20 μm, f–i = 5 μm
Etymology: Named in honour of Mr. Erio Camporesi for his contributions to fungi.
Holotype: MFLU 18-1535
Saprobic on decaying, submerged wood in freshwater. Sexual morph Undetermined. Asexual morphColonies on natural substrate, superficial, effuse, gregarious, white. Mycelium partly immersed in natural substratum, composed of hyaline, branched, septate hyphae. Conidiophores 35–45 × 3.5–5 µm (\( \bar{x} \) = 40 × 4.2 µm, n = 5), macronematous, mononematous, unbranched, subcylindrical, robust at base, straight to flexuous, rough, brown at the base, hyaline at upper part, septate, constricted at septa, thin-walled. Conidiogenous cells 20–30 × 4–4.6 µm (\( \bar{x} \) = 26 × 4.2 µm, n = 5), holoblastic, polyblastic, integrated, terminal, sympodial, subcylindrical with apical taper, straight or flexuous, apical part forming a rachis with numerous, aggregated, cylindrical denticles, hyaline, rough-walled. Conidia 18–22 × 3.5–4.5 µm (\( \bar{x} \) = 20 × 3.8 µm, n = 20), solitary, aggregating in slimy mass, pleuroacrogenous, narrowly fusiform, hyaline, smooth, guttulate, 1-septate, slightly constricted at the septa, lacking a sheath.
Culture characteristics: On PDA, colony circular, reaching 15 mm in 10 days at 25 °C, white from above, pale yellow from below, surface smooth with sparse mycelium, dry, edge entire, mycelium mostly immersed in culture.
Material examined: THAILAND, Satun Province, on submerged wood in a stream, 10 May 2018, W. Dong, hat290 (MFLU 18-1535, holotype), ex-type living culture MFLUCC 18-1410; ibid., hat290-2 (HKAS 105053, isotype), ex-isotype living culture KUMCC 19-0074.
GenBank numbers: ITS = MN796325, LSU = MN796326, SSU = MN796327.
Notes: A megablast search using the LSU sequence data of Pseudodactylaria camporesiana (MFLUCC 18-1410) revealed closest affinities with members of Pseudodactylaria, i.e. Pseudodactylaria brevis (identities = 810/829 (98%), gaps = 2/829 (0%)), P. xanthorrhoeae (identities = 805/835 (96%), gaps = 2/835 (0%)) and P. hyalotunicata (identities = 805/837 (96%), gaps = 2/837 (0%)). In our phylogenetic analyses based on LSU and ITS genes (Fig. 139), P. camporesiana formed a well-supported clade with other Pseudodactylaria species in Pseudodactylariales (Sordariomycetes). The morphologically similar genus Dactylaria was shown to be polyphyletic and phylogenetically distant from Pseudodactylariales (Réblová 2009).
Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence data representing Pseudodactylariaceae and the closely related families in the subclass Sordariomycetidae. Related sequences are taken from Crous et al. (2017). Twenty-six strains are included in the combined analyses which comprise 1439 characters (844 characters for LSU, 595 characters for ITS) after alignment. Paracremonium binnewijzendii (CBS 143277) in Nectriaceae (Hypocreales) is used as the outgroup taxon. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RAxML tree with a final likelihood values of − 7304.691457 is presented. The matrix had 550 distinct alignment patterns, with 13.99% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.240180, C = 0.246116, G = 0.299689, T = 0.214015; substitution rates AC = 1.357550, AG = 1.592995, AT = 2.143251, CG = 0.789747, CT = 6.478576, GT = 1.000000; gamma distribution shape parameter α = 0.216986. Bootstrap values for maximum likelihood (ML) equal to or greater than 95% and clade credibility values greater than 0.95 (the rounding of values to 2 decimal proportions) from Bayesian-inference analysis labeled above or below the nodes. Ex-type strains are in bold and black, the new isolate is indicated in bold and blue
Pseudodactylaria xanthorrhoeae and P. hyalotunicata have somewhat longer or thinner conidia in comparison with P. camporesiana (Tsui et al. 1997; Crous et al. 2017), and both species have a hyaline gelatinous sheath surrounding the conidia, a feature that is lacking in P. camporesiana. Pseudodactylaria brevis is similar to P. camporesiana in having hyaline, 1-septate conidia without a sheath. However, they can be distinguished by conidial size (11.5–17.5 × 2.5–4 μm in P. brevis versus 18–22 × 3.5–4.5 µm in P. camporesiana) (Lin et al. 2018). The other three accepted species in the genus possess hyaline conidiophores, while they are brown at the base in P. camporesiana (Tsui et al. 1997; Crous et al. 2017; Lin et al. 2018). Thus, P. camporesiana sp. nov. is introduced here.
Chaetosphaeriales, generaincertae sedis
Notes: Chaetosphaeriales comprised of Chaetosphaeriaceae Réblová, M.E. Barr & Samuels, Helminthosphaeriaceae Samuels, Cand. & Magni, Leptosporellaceae S. Konta & K.D. Hyde and Linocarpaceae S. Konta & K.D. Hyde (Maharachchikumbura et al. 2016; Konta et al. 2017). In recent phylogenetic analyses, Caudatispora biapiculata Huhndorf & F.A. Fernández, Erythromada lanciospora Huhndorf et al., Lasiosphaeriella nitida Huhndorf & F.A. Fernández, Neoleptosporella clematidis, Neonawawia malaysiana and Rimaconus (R. coronatus and R. jamaicensis) appear to have a distant relationship from other certain families in Chaetosphaeriales (Huhndorf and Fernández 1999; Huhndorf and Miller 2011; Yang et al. 2018b; Hyde et al. 2019a; Phukhamsakda et al. in prep.). However, the statistical value is not support for the segregation of independent families and thus are placed in Chaetosphaeriales genera incertae sedis (Fig. 141).
Neoleptosporella Phukhams. & K.D. Hyde
Notes: Neoleptosporella species are characterised by subglobose to depressed globose ascomata, immersed beneath a small clypeus, asci with J- apical ring and, fusiform, fasciculate ascospores (Phukhamsakda et al. in prep.). We follow the latest treatments and updated accounts of Chaetosphaeriales in Phukhamsakda et al. (in prep.). The new species N. camporesiana is introduced in this study.
Neoleptosporella camporesiana R.H. Perera & K.D. Hyde, sp. nov.
Index Fungorum number: IF556898; Facesoffungi number: FoF 06962; Fig. 140
Neoleptosporella camporesiana (MFLU 19-0978, holotype). a Herbarium material. b Appearance of ascomata on host substrate. c Section through ascoma. d Section of peridium. e Paraphyses. f–j Asci (i in Melzer’s reagent, j upper view). k–n Ascospores. Scale bars: b = 500 µm, c = 200 µm, d, e = 20 µm, f–h = 50 µm, i = 20 µm, j = 10 µm, k–n = 20 µm
Etymology: Named after the collector Erio Camporesi.
Holotype: MFLU 19-0978
Saprobic on a plant stem. Sexual morph Appearing as shiny black, raised dome-shaped spots, with a central short papilla. Ascomata 210–300 μm high, 705–980 μm diam. (\( \bar{x} \) = 250 × 800 μm, n = 10), solitary or aggregated, immersed beneath small clypeus appearing as a disc around the neck, uni-loculate, subglobose to depressed globose, ostiolate. Peridium 75–125 μm diam. (\( \bar{x} \) = 96 μm, n = 10), outer cells merging with the host epidermal cells, composed of dark brown to black cells dome-shaped, of textura angularis. Paraphyses 3–6 µm diam. (\( \bar{x} \) = 4.5 μm, n = 10), hyaline, branched, septate. Asci 145–242 × 8–13 μm (\( \bar{x} \) = 187 × 10 μm, n = 20), 8-spored, unitunicate, cylindrical, long pedicellate, apex rounded with a wedge-shaped, J-, apical ring. Ascospores 100–156 × 2.5–4 μm (\( \bar{x} \) = 126 × 3 μm, n = 20), fasciculate, parallel becoming spiral at maturity, filiform, straight or curved, hyaline, aseptate, rounded at the apex, pointed at the base, smooth-walled, without appendages. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on PDA within 18 h. Colonies on MEA, reaching 10 cm diam. after 2 months at 25 °C, white at the edge, brown in the centre, outwardly with strong light brown radiations. After 30 days of incubation, colonies smooth, flat, margin entire.
Material examined: THAILAND, Chiang Rai Province, on dead branch of unidentified plant, 10 January 2016, R.H. Perera, RHP 132 (MFLU 19-0978, holotype), ex-type living culture, MFLUCC 15-1016.
GenBank numbers: ITS = MN699136, LSU = MN699131.
Notes: Phylogenetic analysis revealed our isolate is closely related to the ex-type of Neoleptosporella clematidis (MFLUCC 17-2074), Chaetosphaeriales genera incertae sedis (Fig. 141). It resembles Neoleptosporella in having subglobose to depressed globose ascomata, immersed beneath a small clypeus, asci with wedge-shaped, J- apical ring and, fusiform, fasciculate ascospores (Phukhamsakda et al., in prep.). Neoleptosporella camporesiana can be distinguished from N. clematidis by its longer asci (145–242 μm versus 60–86 μm) and ascospores (100–156 μm versus 32–50 μm) (Phukhamsakda et al. in prep.). A comparison of LSU sequences of N. camporesiana (MFLUCC 15-1016) and N. clematidis (MFLUCC 17-2074) revealed 82 different nucleotides (9.5%). Hence, we identify our fungus as a new lineage of Neoleptosporella (Jeewon and Hyde 2016). ITS data of N. clematidis is not available for the comparison.
Phylogram generated from RAxML analysis based on combined LSU and ITS sequence data of selected Chaetosphaeriales isolates. Related sequences were obtained from GenBank. Sixty-seven taxa are included in the analyses, which comprise 1655 characters including gaps. The tree is rooted to Gelasinospora tetrasperma CBS 178.33 and Neurospora crassa MUCL 19026. Tree topology of the ML analysis was similar to the BI. The best scoring RAxML tree with a final likelihood value of − 20560.409372 is presented. The matrix had 1025 distinct alignment patterns, with 29.72% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.229701, C = 0.267716, G = 0.312263, T = 0.190320; substitution rates AC = 1.260151, AG = 1.676101, AT = 1.305607, CG = 1.114083, CT = 5.846852, GT = 1.000000; gamma distribution shape parameter α = 0.497360. Maximum likelihood bootstrap support values ≥ 75% (BT) and Bayesian posterior probabilities ≥ 0.99 (BYPP) are given at the nodes as ML/BYPP. The scale bar indicates 0.1 changes. The isolates obtained in this study are in blue and ex-types taxa are in black bold
Subclass Xylariomycetidae O.E. Erikss & Winka
Amphisphaeriales D. Hawksw. & O.E. Erikss.
Notes: Hawksworth and Eriksson (1986) established the order Amphisphaeriales and currently the order comprises ten families viz. Amphisphaeriaceae G. Winter, Apiosporaceae K.D. Hyde et al., Beltraniaceae Nann., Clypeophysalosporaceae A. Giraldo & Crous, Hyponectriaceae Petr., Melogrammataceae G. Winter, Oxydothidaceae S. Konta & K.D. Hyde, Phlogicylindriaceae Senan. & K.D. Hyde, Pseudomassariaceae Senan., Maharachch. & K.D. Hyde, Sporocadaceae Corda and Vialaeaceae P.F. Cannon (Wijayawardene et al. 2018a).
Apiosporaceae K.D. Hyde et al.
Notes: Hyde et al. (1998) established the family Apiosporaceae to accommodate Apiospora Sacc., Appendicospora K.D. Hyde, Dictyoarthrinium S. Hughes, Endocalyx Berk. & Broome, Scyphospora L.A. Kantsch. and Spegazzinia Sacc. Crous and Groenewald (2013) showed Arthrinium Kunze as the asexual morph of Apiospora based on morphology and DNA sequence data and Apiospora was reduced to synonymy with Arthrinium. Currently, Apiosporaceae is established as a well-defined family in the order Amphisphaeriales, with six genera viz. Appendicospora, Arthrinium, Dictyoarthrinium, Endocalyx, Scyphospora and Spegazzinia (Wijayawardene et al. 2017a, 2018a).
Arthrinium Kunze
Notes: Arthrinium was introduced by Schmidt and Kunze (1817) and comprises approximately 64 species (Wijayawardene et al. 2017a; Jiang et al. 2019a). Arthrinium is a cosmopolitan fungal genus consisting of endophytes, pathogens or saprobes associated with a wide range of substrates (Crous and Groenewald 2013; Dai et al. 2016b; Thambugala et al. 2018).
Arthrinium marii Larrondo & Calvo, Mycologia 82 (3): 397 (1990)
Facesoffungi number: FoF 06384; Fig. 142
Holotype: SPAIN, from atmosphere, pharmaceutical excipients, home dust and beach sands, IMI 326872, ex-type culture, CBS 497.90.
Associated with dead stems of Ammophila arenaria. Sexual morph Undetermined. Asexual morphMycelium comprising 1–4 μm diam., smooth, hyaline, septate branched, hyphae. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 5–15 × 2–5 μm, basauxic, arising from hyphae and aggregated in clusters on hyphae, hyaline, smooth, ampulliform. Conidia 4–14 × 3–7 μm (\( \bar{x} \) = 7.2 × 5.2 μm, n = 60), brown to dark brown, smooth, globose to elongate in surface view, aseptate, lenticular in side view, with pale equatorial slit.
Culture characteristics: Colonies growing on PDA reaching 50 mm diam. after 6 days at 25 °C, circular, flat, dense, surface white to grayish, reverse smoke-grey to olivaceous-grey, white at the margin, smooth surface with edge entire to slightly curled.
Material examined: ITALY, Province of Ravenna, Lido di Dante, on dead aerial stems of Ammophila arenaria (L.) Link (Poaceae), 11 February 2015, E. Camporesi, IT 2376H (MFLU 15-1030), living culture MFLUCC 16-0282; ibid., IT 2376H2 (MFLU 18-0759), living culture MFLUCC 16-0283.
Known host and distribution: Beach sand, on stems of Phragmites australis (Cav.) Trin. ex Steud., on leaf of Beta vulgaris L., on culm of Arundinaria hindsii, on stems of Ammophila arenaria (L.) Link worldwide (Farr and Rossman 2020).
GenBank numbers: MFLUCC 16-0282: ITS = MH109526, TEF1-α = MH206166; MFLUCC 16-0283: ITS = MH109527, TEF1-α = MH220419.
Notes: In the present phylogenetic analysis (Fig. 143), MFLUCC 16-0283 clustered with other strains of A. marii including the ex-type strain (CBS 497.90). There is also no significant morphological divergence compared to the type (Crous and Groenewald 2013). Arthrinium marii was reported on several different hosts in Europe and Asia, while most of the records were from species of Poaceae (Farr and Rossman 2020). This is the first report of A. marii on stems of Ammophila arenaria (Poaceae) in Italy.
Phylogram generated from maximum likelihood analysis based on ITS sequence data representing Arthrinium. Related sequences are taken from Crous and Groenewald (2013), Dai et al. (2016b) and Thambugala et al. (2018). Sixty-four strains are included in the analyses which comprise 637 characters (ITS) after alignment. Seiridium phylicae (CBS 133587) in Pestalotiopsidaceae (Amphisphaeriales) is used as the outgroup taxon. The best RaxML tree with a final likelihood values of − 3089.572070 is presented. Estimated base frequencies were as follows: A = 0.259453, C = 0.230435, G = 0.223495, T = 0.286617; substitution rates AC = 0.775950, AG = 1.961909, AT = 0.980782, CG = 0.452485, CT = 3.571483, GT = 1.000000. Bootstrap values for maximum likelihood (ML) equal to or greater than 50% are labeled on the nodes. Ex-type strains are in bold and black, the new isolate is indicated in bold and blue
Pseudotruncatellaceae Crous
Notes: The family Pseudotruncatellaceae was established by Crous et al. (2019a) to accommodate Pseudotruncatella in Amphisphaeriales. It consists of three members and is phylogenetically affiliated to the family Hyponectriaceae. Morphologically, Pseudotruncatellaceae is similar to Truncatella Steyaert in Sporocadaceae, but they are phylogenetically distinct.
Pseudotruncatella R.H. Perera et al.
Notes: Pseudotruncatella was introduced by Perera et al. (2018b) as a monotypic genus typified by P. arezzoensis R.H. Perera et al. This species was found on Cytisus scoparius (L.) Link and Helichrysum italicum (Roth) G. Don fil. from Italy. Crous et al. (2019a) introduced a second species, P. bolusanthi Crous, collected on Bolusanthus speciosus (Bolus) Harms from South Africa. Pseudotruncatella is characterised by its truncatella-like, fusoid to clavate conidia, bearing 1 or 3 apical appendages and dark brown, thick-walled, median cells. The sexual morph is undetermined.
Pseudotruncatella camporesii Goonas. & Hyde, sp. nov.
Index Fungorum number: IF556928; Facesoffungi number: FoF 06822, Fig. 144
Pseudotruncatella camporesii (MFLU 15-0796, holotype). a Conidiomata on dead stem of Helichrysum sp. b A close-up of conidioma. c Cross section of conidioma. d Pycnidial wall. e Developing conidia from conidiogenous cells. f–h Conidia. i Germinating spore. Scale bars c = 50 µm, d, e, g, h = 10 µm, f, i = 20 µm
Etymology: Named after the collector Erio Camporesi.
Holotype: MFLU 15-0796
Saprobic on dead stem of Helichrysum sp. Sexual morph Undetermined. Asexual morphConidiomata black, shiny, raised or flattened, rounded, 170–250 μm high, 280–400 μm wide (\( \bar{x} \) = 200 × 340 μm, n = 10) acervular to pycnidioid, scattered or gregarious, uni-loculate, globose or oval with a flattened base, semi-immersed to superficial, sometimes erumpent. Pycnidial walls comprising several indistinct cell layers of golden brown to pale brown cells of textura angularis to textura prismatica, and towards the upper part of the fruiting body, more of textura prismatica and textura globulosa. Conidiophores arising from the basal and lateral cells of the conidioma cavity, cylindrical, reduced to conidiogenous cells, unbranched, smooth, hyaline. Conidiogenous cells 10–38 × 1.5–3 μm (\( \bar{x} \) = 26 × 2 μm, n = 20) cylindrical, hyaline, smooth, determinate, holoblastic, proliferating percurrently at apex, aseptate. Conidia 28–35 × 6–8.5 μm, (\( \bar{x} \) = 31 × 7.3 μm, n = 40), fusoid, straight to slightly curved, 2-septate, slightly constricted at second septum from base, basal cell 5.8–12 × 2.5–3.5 μm (\( \bar{x} \) = 9 × 2.8 μm), 15–30 × 1.5–2 μm, narrowly obconic, truncate at base, hyaline, smooth-walled, thin-walled, second and third cells from the base, together 18.5–26 × 5.5–9 μm (\( \bar{x} \) = 22 × 6.8 μm), dark brown, smooth-walled, thick-walled, slightly guttulate, fusoid, bearing three tubular apical appendages, 18.6–25.8 μm (\( \bar{x} \) = 23.2 μm) long, smooth, tubular, unbranched.
Culture characteristics: Colonies on PDA growing up to 20 mm in diam. at 16–18 °C after seven days. Colony irregular in shape, flat to effuse, surface smooth, with undulate margin, yellowish-white, reverse grey.
Material examined: ITALY, Province of Forlì-Cesena [FC], near Passo delle Forche-Galeata, on dead stem of Helichrysum sp. (Asteraceae), E. Camporesi, 16 July 2014, IT 2001 (MFLU 15-0796, holotype); MFLU 15-0797 = KUN-HKAS 96293, isotype, ex-type living culture, MFLUCC 15-0562.
GenBank numbers: ITS = MN648203.
Notes: Pseudotruncatella arezzoensis and P. camporesii are reported on Helichrysum sp. as saprobes. The two species can be distinguished by their conidiomata dimensions (230–275 μm high, 255–280 μm wide versus 170–250 μm high, 280–400 μm wide). Pseudotruncatella arezzoensis has pycnidioid, immersed to semi-immersed conidiomata, while in P. camporesii they are acervular to pycnidioid, semi-immersed to superficial (Perera et al. 2018b). Pseudotruncatella camporesii has longer conidiogenous cells and larger conidia (28–35 × 6–8.5 μm versus 20–25 × 5.4–6.5 μm, Perera et al. 2018b). Furthermore, P. arezzoensis produces 3-septate conidia with no constriction at the septum while the conidia of P. camporesii are 2-septate and slightly constricted at the septum. Likewise, P. bolusanthi has 2-septate conidia showing constriction at mid-septum (Crous et al. 2019a). However, the latter has smaller conidiomata of 150–200 μm diam., with 0–3-septate, branched conidiophores and shorter conidia (17–20 × 6.5–7 μm), with one apical appendage (Crous et al. 2019a). In the phylogenetic analysis our new isolate clustered with P. arezzoensis with high bootstrap support (96% ML, 1.00 BYPP; Fig. 145).
Phylogram generated from maximum likelihood analysis based on ITS sequence data representing Pseudotruncatellaceae and closely related families. Related sequences were obtained from Perera et al. (2018b), Crous et al. (2019a) and BLAST results. Thirty-six strains were included in the analysis which comprised of 659 characters after alignment. Eutypa lata (AFTOL-ID 929) in Diatrypaceae (Xylariales) was selected as the outgroup taxon. Tree topology of the maximum likelihood analysis was similar to the Bayesian analysis. The best RAxML tree with a final likelihood value of − 6953.648717 is presented. The matrix had 434 distinct alignment patterns, with 20.77% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.248803, C = 0.249335, G = 0.221058, T = 0.280804; substitution rates AC = 0.899580, AG = 1.933534, AT = 1.335762, CG = 0.977859, CT = 2.816070, GT = 1.000000; gamma distribution shape parameter α = 0.372495. Bootstrap values for maximum likelihood (ML) equal to or greater than 60% and clade credibility values greater than 0.90 (the rounding of values to 2 decimal proportions) from Bayesian-inference analysis are labeled at the nodes. Ex-type strains are in bold and black, the new isolate is indicated in bold and blue
Sporocadaceae Corda
Notes: Sporocadaceae was introduced by Corda (1842) designating Sporocadus Corda as the type genus. Species of Sporocadaceae are endophytic, plant pathogenic or saprobic, and associated with a wide range of host plants. There are 30 genera in this family (Liu et al. 2019).
Pseudopestalotiopsis Maharachch et al.
Notes: Pseudopestalotiopsis was introduced by Maharachchikumbura et al. (2014) based on Pseudopestalotiopsis theae (Sawada) Maharachch., K.D. Hyde & Crous. Pseudopestalotiopsis comprises species with dark concolourous median cells with knobbed apical appendages. There are 16 species in this genus (Index Fungorum 2020). Some species in this genus are plant pathogens and some are saprobes (Maharachchikumbura et al. 2014; Norphanphoun et al. 2019). Pseudopestalotiopsis theae is reported on living leaves of a palm from Guangdong, China for the first time.
Pseudopestalotiopsis theae (Sawada) Maharachch., K.D. Hyde & Crous, in Maharachchikumbura et al., Stud. Mycol. 79: 183 (2014)
Facesoffungi number: FoF 07204; Fig. 146
Mildly pathogenic on living leaves of palm forming spots. Sexual morph Undetermined. Asexual morphConidiomata 250–800 μm diam., scattered, immersed, acervuli, globose to oblate, black, coriaceous. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 6–14 × 1–3 μm (\( \bar{x} \) = 12 × 2.5 μm, n = 30), annellidic, ampulliform or subcylindrical, separate, hyaline, smooth-walled, straight to slightly curved. Conidia 18–32 × 5–8 μm (\( \bar{x} \) = 28 × 7 μm, n = 30), five-celled, clavate fusiform, ellipsoid to subcylindrical, slightly constricted at septa; basal cell 3–5 μm long obconical, hyaline, with or without a truncate base, thin walled, with a basal appendage 3–6 μm long, single, tubular, unbranched, knobbed, centric or excentric, attached to the basal cell; median cells 12–18 μm long, (1–)3, doliiform, brown to light brown, or olivaceous in colour, darker septate; apical cell 3–5 μm long, hyaline, cylindrical to subcylindrical, with 2–4 tubular, filiform, knobbed, 18–35 μm long apical appendages.
Culture characteristics: Colonies growing on PDA attaining 1 cm diam. within a week when incubated at 18 °C, circular, entire, flat, off to pinkish white, aerial mycelia appears as small clumps.
Material examined: CHINA, Guangdong, Shenzhen, Nanshan, Nanhai Avenue, Shenzhen University, on living leaves of palm, 28 August 2018, I.C. Senanayake, SI 33 (MFLU 20-0057), living culture, KUMCC 20-0034.
Known host and distribution: Human eye (India), Camellia sinensis (Thailand, Taiwan), Elaeis guineensis (Thailand), Pharus latifolius (USA), Macaranga sp. (Malaysia), mango leaf (Malaysia).
GenBank numbers: ITS = MN764320, LSU = MN764360, TEF1-α = MT080694, TUB2 = MT080693.
Notes: Phylogenetic analyses of a combined ITS, TUB2 and TEF1-α sequence dataset showed that our strain KUMCC 20-0034 clusters with Pseudopestalotiopsis theae (MAFF 752011, MFLUCC 12-0055 and PSHI2004) with high support (95% ML, 0.90 BYPP; Fig. 147) Pseudopestalotiopsis theae is the generic type of Pseudopestalotiopsis, which was based on Pestalotiopsis theae (Sawada) Steyaert (Maharachchikumbura et al. 2014). This is a common plant pathogen causing various plant diseases (Lu 1995; Chen et al. 2013b; Norphanphoun et al. 2019). This taxon has been reported from China several times. Mukhtar et al. (2019) reported this species from Fuzhou, China causing a leaf spot of Ixora chinensis Lam. A fruit spot in Eriobotrya japonica (Thunb.) Lindl., an important fruit and ornamental plant, has been reported by Chen et al. (2013b) from Jingxian, Anhui Province, China. We collected Pseudopestalotiopsis theae from Shenzhen, Guangdong Province, China from palm. However, we could not see any disease symptoms and this association appear to be saprobic. This is the first record of P. theae in Shenzhen.
Phylogram generated from maximum likelihood analysis based on combined ITS, TUB2 and TEF1-α sequence dataset. Bootstrap support values for ML greater than 50% and Bayesian posterior probabilities greater than 0.90 are given near nodes. The tree is rooted with Neopestalotiopsis saprophytica CBS 115452. Ex-type strains are in bold and black. The newly generated sequence is indicated in blue
Xylariales Nannf.
Notes: Xylariales is a large order in the subclass Xylariomycetidae. Maharachchikumbura et al. (2016) accepted 22 families in this order, while Hongsanan et al. (2017) transferred several families from Xylariales to Amphisphaeriales based on molecular clock evidence. Members of Xylariales have been revised in recent studies (Daranagama et al. 2018; Voglmayr et al. 2018; Wendt et al. 2018), of which three families Barrmaeliaceae Voglmayr & Jaklitsch, Graphostromataceae M.E. Barr, J.D. Rogers & Y.M. Ju and Hypoxylaceae DC. were included and revised.
Diatrypaceae Nitschke
Notes: We follow the latest treatment and updated accounts of Diatrypaceae in de Almeida et al. (2016), Shang et al. (2017, 2018), Senwanna et al. (2017), Hyde et al. (2019a), Phookamsak et al. (2019) and Konta et al. (2020). This family comprises 19 genera (Wijayawardene et al. 2018a).
Diatrypella (Ces. & De Not.) De Not.
Cesati and De Notaris (1863) introduced Diatrypella to accommodate members of stromatic Sphaeriales (Rao 1964). There are 144 epithets of Diatrypella listed in Index Fungorum (2020). Diatrypella is characterised by conical or truncate, cushion-like or discoid stromata, which are usually delimited by a black zone on host tissues, perithecial ascomata, umbilicate or sulcate ostioles, and numerous small ovoid to allantoid ascospores (Simmons 1948; Rao 1964; Vasilyeva and Stephenson 2005; Mehrabi et al. 2016). Diatrypella species have a libertella-like asexual morph (Kirk et al. 2008).
Diatrypella yunnanensis Brahmanage, Thyagaraja & K.D. Hyde, sp. nov.
Index Fungorum number: IF556926; Facesoffungi number: FoF 07205; Fig. 148
Etymology: Name refers to the geographical region “Yunnan” where the species was collected.
Holotype: JZBH3380001
Saprobic on unidentified dead wood. Sexual morphAscomata solitary and scattered, immersed, raising the substratum, or erumpent. Entostroma effuse, a faint black line between the groups sometimes present, wood hardened; a thin layer of white pulvinate fungal material sometimes present around the ascomal venter. Ascomal venter 360–440 µm high, 245–260 µm wide (\( \bar{x} \) = 400 × 255 μm, n = 20), at sides 20–30 μm (\( \bar{x} \) = 28 μm, n = 10), subglobose to broadly ellipsoidal, ostiolate, periphysate, comprising an outer layer of completely melanized cells, and an inner layer of less pigmented, elongate cells; singly erumpent, protruding above the level of the raised substratum; the outermost ascomata in an aggregate often bent towards the centre of the group. Peridium 45–55 μm wide (\( \bar{x} \) = 52 μm, n = 10), in transverse section with an outer, layer of small, thick-walled, melanized, rounded cells, and an inner layer of hyaline, elongate cells in textura angularis. Paraphyses up to 4–7 µm wide, hyaline, septate, deliquescent. Asci 105–210 × 15–30 μm (\( \bar{x} \) = 170 × 28 μm, n = 20), polysporous, clavate, truncate, pedicellate, with a refractive subapical, J-, ring at the base of an invagination of the ascus apex. Ascospores 18–22 × 3–4 μm (\( \bar{x} \) = 19 × 3 μm, n = 20), allantoid, slightly curved, pale yellow to pale brown. Asexual morph Undetermined.
Material examined: CHINA, Yunnan Province, Kunming Forest Park, on dead and dying twigs of an unidentified plant, 30 June 2018, V. Thyagaraja, VT 01 (JZBH3380001, holotype).
GenBank numbers: ITS = MN653008, TUB2 = MN887112.
Notes: Diatrypella yunnanensis is phylogenetically related to D. pulvinata Nitschke and D. favacea (Fr.) Ces. & De Not. and grouped within Diatrypellasensu lato (Fig. 149). Diatrypella yunnanensis can be easily distinguished from D. favacea and D. pulvinata by relatively bigger ascomta, asci and ascospores (Table 3). When comparing the ITS base pair differences of D. yunnanensis with D. favacea and D. pulvinata, there were 3.4% (18 bp out of 527 bp without gaps) and 1.3% (7 bp out of 527 bp without gaps), respectively. TUB2 base pair differences between D. yunnanensis and D. pulvinata are 5.3% (23 bp out of 435 bp without gaps). There are no TUB2 sequences generated from D. favacea. We could not obtain a culture for this species as the ascospores did not get germinate in any media used.
Maximum likelihood analysis with 1000 bootstrap replicates yielded a best tree with the likelihood value of − 8974.532790. The combined ITS and TUB2 sequence datasets comprised 96 strains with Xylaria hypoxylon and Kretzschmaria deusta (CBS 122620 and CBS 826.72) as the outgroup taxa. Tree topology of the ML analysis was similar to the BI. The matrix had 403 distinct alignment patterns, with 10.72% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.232805, C = 0.250707, G = 0.235491, T = 0.280998; substitution rates AC = 1.078239, AG = 3.114105, AT = 1.506841, CG = 0.917572, CT = 3.843879, GT = 1.000000; gamma distribution shape parameter α = 0.753063. Maximum likelihood bootstrap (ML) values > 65% and bayesian posterior probabilities (BYPP) > 0.80% are given above the nodes. The scale bar indicates 0.04 changes. The ex-type strains are in bold and new isolates in blue bold
Fasciatisporaceae S.N. Zhang, K.D. Hyde & J.K. Liu, fam. nov.
Index Fungorum number: IF557171; Facesoffungi number: FoF 07234
Saprobic on driftwood and palms or other monocotyledons, distributed in marine and terrestrial habitats. Sexual morphAscomata solitary or gregarious, immersed under a clypeus or very little stromatal structure, ostiolate, some with a papilla. Peridium composed of several layers of cells of textura angularis. Paraphyses hypha-like, septate. Asci 8-spored, unitunicate, cylindrical, with a short pedicel, and a J + (or J-), wedge-shaped subapical ring. Ascospores 1-seriate or overlapping 1-seriate, ellipsoidal, ovoid or rhomboid, unicellular, pale brown to brown, with a central pallid band, and with appendages at each end or surrounded by a mucilaginous sheath. Asexual morph Undetermined.
Type genus: Fasciatispora K.D. Hyde
Notes: We establish the family Fasciatisporaceae to accommodate the monotypic genus Fasciatispora K.D. Hyde based on its unique morphological features and distinct branch in the phylogeny. Fasciatispora was introduced by Hyde (1991) with F. nypae K.D. Hyde as the type species and was thought to be closely related to Xylariaceae and Amphisphaeriaceae. Liu et al. (2015) provided a reference sequence for F. nypae based on a collection from Nypa fruticans Wurmb. and showed that F. nypae is phylogenetically basal in Xylariaceae. Daranaragama et al. (2018) and Wijayawardene et al. (2018a) treated it as Amphisphaeriales genera incertae sedis based on phylogeny, but they also suggested more gene (RPB2 and TUB2) sequences were needed to determine the placement. In this study, we collected a new species Fasciatispora cocoes from Cocos nucifera L. (Arecaceae) and carried out multi-gene analyses based on ITS, LSU, RPB2, SSU, TEF1-α and TUB2 sequences. Our phylogeny agrees with Hyde et al. (2019a), which suggests Fasciatispora belongs in Xylariales. The character of unicellular ascospores with a central pallid band distinguish Fasciatispora from other members in Xylariales. Phylogenetically, Fasciatispora always forms a distinct branch in this and previous studies (Liu et al. 2015; Hyde et al. 2017, 2019a), and it is a distinct, well-supported clade in Xylariales in Hyde et al. (2019a) and this study (Fig. 153). Therefore, a new family Fasciatisporaceae in Xylariales is introduced herein.
Fasciatispora K.D. Hyde
Facesoffungi number: FoF 03041
Saprobic on driftwood, Pandanaceae and Arecaceae in marine and terrestrial habitats. Sexual morphAscomata immersed to semi-immersed beneath a clypeus, usually forming under a blackened region, solitary or gregarious, raised, in vertical section globose or subglobose, ostiolate, sometimes with papilla. Peridium composed of several layers of cells of textura angularis. Paraphyses hypha-like, septate, irregular, embedded in a gelatinous matrix. Asci 8-spored, unitunicate, cylindrical, short-pedicellate, apical rounded, with a J + (or J-), wedge-shaped subapical ring. Ascospores 1-seriate or overlapping 1-seriate, pale brown to brown, ellipsoidal, ovoid or rhomboid, unicellular, with a central pallid band, and with appendages at each end or surrounded by a mucilaginous sheath. Asexual morph Undetermined.
Type species: Fasciatispora nypae K.D. Hyde
Notes: Hyde (1991) reported Fasciatispora nypae from intertidal Nypa fruticans, and Alias et al. (1994) introduced the second marine species F. lignicola Alias, E.B.G. Jones & Kuthub. from a mangrove habitat. Currently, there are eleven Fasciatispora species (Hyde 1995; Hyde and Wong 1999; Hidayat et al. 2007; Hyde et al. 2017; Doilom et al. 2018) including the new species F. cocoes that we describe in this study. Five species have available sequence data and were included in our phylogenetic analyses. F. arengae Konta & K.D. Hyde, F. calami Konta & K.D. Hyde, F. cocoes and the type species F. nypae formed a well-supported clade in multi-gene analyses (Fig. 153), while F. petrakii (Mhaskar & V.G. Rao) K.D. Hyde (culture HKUCC 207) clustered with Vamsapriya species (data not shown) as in Hyde et al. (2017). Protein-coding genes (such as RPB2 and TUB2) are needed for this genus, which are important for introducing new taxa and for a better understanding the phylogenetic relationships in higher level ranking.
Fasciatispora cocoes S.N. Zhang, K.D. Hyde & J.K. Liu, sp. nov.
Index Fungorum number: IF557000; Facesoffungi number: FoF 06503; Fig. 150
Fasciatispora cocoes (MFLU 18-1589, holotype). a, b Ascomata on substrate surface. c Vertical section through an ascoma. d, e Ostiole with periphyses. f, g Peridium (g = at the base). h–j Asci. k Ascus in Melzer’s reagent, with a J + , subapical ring. l Paraphyses. m–o Ascospores. p Germinated ascospore. q Ascospore in India ink presents the clear mucilaginous sheath. r Colony on PDA. Scale bars: a = 500 μm, b = 200 μm, c = 50 μm, d–f, h–j = 20 μm, g, l, q = 10 μm, k, m–p = 5 μm
Etymology: The epithet refers to the host plant, on which the fungus was collected.
Holotype: MFLU 18-1589
Saprobic on rachides of Cocos nucifera. Sexual morphAscomata 150–330 μm diam., solitary or gregarious, immersed under a clypeus, visible as black dots on substrate, with an ostiolar or papilla, in vertical section 210–320 μm diam., 190–220 μm high, subglobose or globose, with a rounded or flattened base, coriaceous. Ostiole central, periphysate. Peridium 9–18 µm wide, consisting several layers of cells of textura angularis, outside layers brown cells and inter layers hyaline, thinner at the base. Paraphyses 2–3 µm wide, hyphae-like, irregular, septate, rarely branched. Asci 78–110 × 6–12 µm (\( \bar{x} \) = 94.2 × 9 µm, n = 20), 8-spored, unitunicate, cylindrical, pedicellate, thin-walled, with a J + , wedge-shaped, subapical ring, 2–2.5 µm diam., 1 µm high. Ascospores 10–15 × 4.5–6 µm, (\( \bar{x} \) = 12.7 × 5.4 µm, n = 50), 1-seriate, pale brown to brown, ellipsoidal, unicellular, with a central pallid band, smooth-walled, surrounded by a mucilaginous sheath. Asexual morph Undetermined.
Culture characteristics: Ascospores germinated on PDA within 24 h and transferred to MEA. Colonies growing on MEA, circular, with flat surface, edge entire or a little filamentous, creamy white, reached about a diam. of 20 mm after three weeks at 25–28 °C. Mycelium superficial, branched, septate, hyaline to pale brown, smooth.
Material examined: THAILAND, Trat Province, Koh Chang, on decaying rachides of Cocos nucifera, 27 April 2017, S.N. Zhang, SNT125 (MFLU 18-1589, holotype); ibid., KUN-HKAS105470, isotype, ex-type living culture MFLUCC 18-1445.
GenBank numbers: ITS = MN482680, LSU = MN482675, RPB2 = MN481517, SSU = MN482676, TEF1-α = MN481516, TUB2 = MN505104.
Notes: The new collection Fasciatispora cocoes fits well within Fasciatispora and is similar to F. petrakii (Hyde 1995). However, in multi-gene analyses, F. cocoes is distinct from F. petrakii but clustered with F. calami (Fig. 153). Fasciatispora cocoes is different from F. petrakii as the latter species has ovoid or almost rhomboid ascospores with a mucilaginous sheath. Fasciatispora cocoes also differs from F. calami in having a relatively thinner peridium (9–18 μm versus 17–36 μm) and larger asci in average size (94.2 × 9 μm versus 80 × 8 μm). The comparison of nucleotide difference between F. cocoes and F. calami showed that 4 nucleotide differences across the 843 nucleotides (0.47%) of LSU and a significant difference in TUB2 sequence data.
Xylariaceae Tul. & C. Tul.
Notes: Xylariaceae species are saprobic, pathogenic or endophytic in plant tissues, fruits, dung or insects (Maharachchikumbura et al. 2015, 2016), and also have been studied for secondary metabolites production (Whalley and Edwards 1995; Stadler and Hellwig 2005; Stadler 2011). The family is traditionally characterised by well-developed stromata, or sometimes with reduced or lack of stroma, 8-spored, unitunicate asci, with or without a J + , apical ring, pigmented ascospores with germ slits or pores, and geniculosporium-like, nodulisporium-like, or libertella-like asexual morphs (Rogers 2000; Daranaragama et al. 2018). In an updated classification of Xylariaceae 37 genera were accepted (Daranaragama et al. 2018). A new geographical record of Astrocystis bambusicola is introduced and illustrated herein.
Astrocystis Berk. & Broome
Notes: Astrocystis was introduced by Berkeley and Broome (1873), with A. mirabilis Berk. & Broome as the type species. Twenty-three species have been accommodated in the genus (Index Fungorum 2020). No asexual morph is known for the genus (Wijayawardene et al. 2017a, b). We follow the treatment and classification in Smith et al. (2001) and Hyde et al. (2017).
Astrocystis bambusicola R.H. Perera & K.D. Hyde, Fungal Diversity 87: 173 (2017)
Facesoffungi number: FoF 03592; Fig. 151
Astrocystis bambusicola (KUN-HKAS 101769). a Ascostromata on host surface. b Vertical section through ascostromata. c Peridium. d–g Asci. h Paraphyses. i–l Ascospores. m Germinating ascospore. n, o Culture characteristics (n = from above, o = from below). Scale bars: h = 30 μm, c–g = 20 μm, i–m = 10 μm
Holotype: THAILAND, Doi Mae Salong, on twigs of bamboo, February 2016, R.H. Perera, MM-1 (MFLU 17-0738), ex-type living culture, MFLUCC 17-0127.
Saprobic on dead bamboo culms, visible as black raised spots on the host. Sexual morphPseudostromata 200–730 μm diam., 130–200 μm high, solitary to gregarious, scattered, superficial, subgolobose to dome-shaped, blackened, carbonaceous, 1–3-loculate. Locules 150–250 μm diam., 140–180 μm high, subglobose, dark brown to black, immersed within pseudostroma. Peridium 25–35 μm thick, comprising several layers of dark brown to hyaline, compressed, pseudoparenchymatous cells arranged in textuta angularis, and black, fragile, carbonaceous tissue at outermost layer. Paraphyses 3.5–6 μm broad, septate, ribbon-like. Asci 96–120 × 5–10 μm (\( \bar{x} \) = 108 × 7.5 μm, n = 20), 8-spored, unitunicate, cylindric-clavate, with a short pedicel, apically rounded. Ascospores 14–16 × 6–8 μm (\( \bar{x} \) = 15 × 7 μm, n = 20), 1-seriate, ellipsoidal, aseptate, brown to dark brown, guttulate, smooth-walled, with an indistinct germ-slit. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on PDA within 24 h, at 23–28 °C. Colonies growing on PDA, reaching 40 mm in 10 days at 23–28 °C, cottony, circular, umbonate, edge irregular, white from above and below. Mycelium superficial to immersed in media, with hyaline, branched, septate, smooth hyphae.
Material examined: CHINA, Yunnan Province, Chuxiong Yi Autonomous Prefecture, Chuxiong City, Biji Village, on dead bamboo culms, 5 February 2019, H.B. Jiang, CX002 (KUN-HKAS 101769), living culture KUMCC 19-0245.
Known hosts and distribution: Bamboo (Yunnan, China; Thailand) (Hyde et al. 2017).
GenBank numbers: ITS = MN636698, LSU = MN636699, SSU = MN636700.
Notes: Astrocystis bambusicola R.H. Perera & K.D. Hyde was introduced in Hyde et al. (2017) based on a specimen collected from Thailand. Phylogenetic analyses of a combined ITS, LSU, RPB2, TUB2, SSU and TEF1-α sequence dataset showed that our strain KUMCC 19-0245 clades with the type strain of A. bambusicola (MFLUCC 17-0127) with high support (100% ML, 99% MP; Fig. 153). The new collection is morphologically close to A. bambusicola. Furthermore, our new collection has consistent base pairs with A. bambusicola (MFLUCC 17-0127) based on a comparison of ITS, LSU and SSU loci. Thus, we identify our new isolate as A. bambusicola, a species reported only in Thailand (Hyde et al. 2017); we report this species in China for the first time.
Xylariales , genera incertae sedis
Notes: Maharachchikumbura et al. (2016) listed 24 genera as incertae sedis in Xylariales. Several genera previously included in Xylariales incertae sedis have been ranked as families that belong to different orders (Konta et al. 2016, 2017; Hernández-Restrepo et al. 2017). However, more genera were added and the number of Xylariales genera incertae sedis increased to 44 in Wijayawardene et al. (2018a).
Melanographium Sacc.
Notes: The hyphomycetes genus Melanographium was erected based on M. spleniosporum Sacc. (Saccardo 1913). However, Ellis (1963) designated M. selenioides (Sacc. & Paoletti) M.B. Ellis (= M. spleniosporum Sacc.) as the type species, and recognized five species. Goh and Hyde (1997) accepted nine species, and provided a synopsis, as well as a key to Melanographium species. The genus was reviewed by Somrithipol and Jones (2005), and ten species were considered valid. Members of Melanographium are characterised by unbranched conidiophores generally arising in a group from immersed stromata, polyblastic conidiogenous cells with sympodial proliferation, and holoblastic, 1-celled conidia, frequently reniform and often with a hyaline germ slit (Ellis 1963, 1971). The length of conidiophores, pattern of conidiophore grouping (single, synnemata, loosely or dense fascicles), conidial shape (straight, curved, or strongly curved), and conidial size are morphological markers to distinguish species in this genus (Goh and Hyde 1997; Somrithipol and Jones 2005). However, the taxonomic placement of Melanographium is unresolved, because there was no sequence data available. Wijayawardene et al. (2018a) treated it as belonging to Sordariomycetes genera incertae sedis. We provide sequence data of Melanographium, and carried out multi-gene phylogeny which showed the genus nested in Xylariales. A new species M. phoenicis is introduced and illustrated herein.
Melanographium phoenicis S.N. Zhang, K.D. Hyde & J.K. Liu, sp. nov.
Index Fungorum number: IF557001; Facesoffungi number: FoF 06504; Fig. 152
Melanographium phoenicis (MFLU18-1587, holotype). a–c Colonies on natural substratum. d, e Conidiophores with conidiogenous cells bearing condia, note (e) conidiophores in loose flared fascicles. f Conidiophores in loosely fascicles. g–i Attached conidia at the proliferation of conidiophores that often at nodose. j Conidiogenous cells with conidiogenous scars. k, l Conidia. m Colony on PDA. n–p Conidia producing in culture. Scale bars: a–c = 100 μm, d–f = 50 μm, g, h, k, l = 10 μm, i, j = 20 μm, n–p = 5 μm
Etymology: The epithet refers to the genus of the host plant, Phoenix.
Holotype: MFLU18-1587
Saprobic on decaying rachides of palm (Phoenix paludosa). Sexual morph Undetermined. Asexual morphColonies on the substratum superficial, effuse, tufted, black. Mycelium immersed, composed of branched, septate, smooth, pale brown hyphae. Conidiophores 125–170 μm long (\( \bar{x} \) = 147.7 μm, n = 20), 3.5–6.5 μm wide (\( \bar{x} \) = 5.5 μm, n = 20), macronematous, 3–5 in loose flared fascicle or rarely single, unbranched, multi-septate, thick-walled, smooth, slightly geniculate near the apex, straight or flexuous, with percurrent proliferation that is often nodose, brown below, becoming pale brown above to subhyaline at the tip. Conidiogenous cells polyblastic, integrated, terminal and intercalary, cylindrical, cicatrized, intercalary conidiogenous loci often at the nodose proliferations, with conidiogenous scars. Conidia 11–18 μm long (\( \bar{x} \) = 14.4 μm, n = 30), 6–9 μm wide (\( \bar{x} \) = 7.9 μm, n = 50), holoblastic, solitary, aseptate, olivaceous to dark brown, reniform or obovoid, with a subhyaline longitudinal germ-slit on the convex side, guttulate, verruculose, smooth-walled. Conidial secession schizolytic.
Culture characteristics: Conidia germinated on PDA within 24 h and been transferred to MEA. Colonies growing on MEA, velvety, with flat surface, firstly whitish, and becoming olive to grey-green or light grey-green. Mycelium superficial and partially immersed, branched, septate, hyaline to pale brown, smooth. Conidia produced in culture smaller than that found in substrate, 8.2–14.5 μm long (\( \bar{x} \) = 11.7 μm, n = 10), 5.9–8.7 μm wide (\( \bar{x} \) = 6.9 μm, n = 10).
Material examined: THAILAND, Trat Province, Koh Chang, on decaying rachides of Phoenix paludosa Roxb. (Arecaceae), 27 April 2017, S.N. Zhang, SNT116 (MFLU 18-1587, holotype); KUN-HKAS102208, isotype, ex-type living culture MFLUCC 18-1481.
GenBank numbers: ITS = MN482677, LSU = MN482678, SSU = MN482679, TEF1-α = MN481518.
Notes: Most Melanographium species were found from palms (Ellis 1963; Goh and Hyde 1997; Somrithipol and Jones 2005) including M. phoenicis, which was collected from Phoenix paludosa (mangrove date palm). Morphological characters of M. phoenicis fit well with Melanographium. Conidiophores of M. phoenicis arise in a loose flared fascicle, which is similar to M. indicum Saikia & A.K. Sarbhoy (Saikia and Sarbhoy 1981), but it differs from the latter species because it has shorter conidiophores with obvious conidiogenous scars, and verrucose conidia. According to the key to Melanographium species in Somrithipol and Jones (2005), M. phoenicis is close to M. palmicola Goh & K.D. Hyde and M. spinulosum (Speg.) S. Hughes because of the verrucose conidia. However, M. phoenicis differs from M. palmicola as the conidiophores of the latter species are in widely divergent fascicles. The colour and size of conidia also distinguish M. phoenicis from M. spinulosum, M. phoenicis has olivaceous to dark brown conidia, while M. spinulosum is characterised by dark reddish-brown conidia. Multi-gene phylogenetic analyses in this study (Fig. 153) showed M. phoenicis belongs to Xylariales but is of uncertain familial placement. Therefore, we consider Melanographium belongs to Xylariales genus incertae sedis.
Phylogram generated from maximum likelihood analysis based on combined ITS, LSU, RPB2, TUB2, SSU and TEF1-α sequence data representing the order Xylariales. Related strains are referred to Hsieh et al. (2010), Daranagama et al. (2015), Liu et al. (2015), Becerra-Hernández et al. (2016), Maharachchikumbura et al. (2016), Dai et al. (2017), Hyde et al. (2017), Wendt et al. (2018), Voglmayr et al. (2018). Ninety-three strains are included in the combined analyses which comprise 6715 characters (828 characters for ITS, 885 characters for LSU, 1063 characters for RPB2, 1990 characters for TUB2, 1025 characters for SSU, 924 characters for TEF1-α) including gaps. Sordaaria fimicola (CBS 723.96) and Sordaria macrospora (Buck s.n.) are used as the outgroup taxa. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RAxML tree with a final likelihood values of − 69169.901135. The matrix had 3540 distinct alignment patterns, with 60.93% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.241739, C = 0.259560, G = 0.259228, T = 0.239474; substitution rates AC = 1.286285, AG = 3.197641, AT = 1.369896, CG = 1.063194, CT = 5.517353, GT = 1.000000; gamma distribution shape parameter α = 0.718586. Bootstrap values for maximum likelihood (ML) and maximum parsimony equal to or greater than 75%, and clade credibility values greater than 0.95 are indicated in bold branches. The new isolates are indicated in bold and blue, and the type strains are indicated in black bold
Xenoanthostomella Mapook & K.D. Hyde, gen. nov.
Index Fungorum number: IF556907; Facesoffungi number: FoF 06795
Etymology: Xeno = ξένος in Greek, distinct; Anthostomella = anthostomella-like.
Saprobic on dead stems. Sexual morphAscomata immersed beneath clypeus, unilocular, globose to obpyriform, coriaceous, solitary to scattered. Clypeus extending outwards around the ascomata, thicker around the papilla. Ostiole central, papillate. Peridium comprising several layers, inner layers comprising hyaline to brown cells of textura epidermoidea, outer layers comprising brown cells of textura intricata at side with textura angularis at upper part. Paraphyses cylindrical, septate with small guttules, hyaline, branching, embedded in a gelatinous matrix. Asci 8-spored, unitunicate, cylindrical to broadly filiform, pedicellate, straight or slightly curved, with inconspicuous apical ring. Ascospores 1-seriate, hyaline when immature, becoming brown to dark olivaceous-brown when mature, unicellular, inequilateral to broadly fusiform, tapering towards narrow ends, verruculose when immature with 1–2 large guttules when mature. Asexual morph Undetermined.
Type species: Xenoanthostomella chromolaenae Mapook & K.D. Hyde
Notes: A phylogenetic analyses based on combined dataset of ITS, LSU, RPB2, TUB2, SSU and TEF1-α sequence data showed that Xenoanthostomella chromolaenae clusters with Idriella lunata, Gyrothrix ramosa and Gyrothrix inops (Fig. 153). However, we could not compare the morphological characteristics of these genera as Xenoanthostomella is found as sexual morph in nature and asexual morph did not form in culture, while Idriella, Gyrothrix and Circinotrichum were found only as asexual morphs in nature. Therefore, Xenoanthostomella is described here as a new genus based on phylogeny, together with a comparison of the ITS (+5.8S) gene region of X. chromolaenae and Gyrothrix ramosa reveals 13 base pair differences (2.56%) across 507 nucleotides.
Xenoanthostomella chromolaenae Mapook & K.D. Hyde, sp. nov.
Index Fungorum number: IF556908; Facesoffungi number: FoF 06796; Fig. 154
Etymology: Name reflects the host genus Chromolaena, from which this species was isolated.
Holotype: MFLU 20-0048
Saprobic on dead stems of Chromolaena odorata. Sexual morphAscomata 190–220 µm high × 175–210 µm diam. (\( \bar{x} \) = 200 × 190 µm, n = 5), immersed beneath clypeus, unilocular, globose to obpyriform, coriaceous, solitary to scattered. Clypeus extending outwards around the ascomata, thicker around the papilla. Ostiole central, papillate. Peridium 15–25 µm wide, comprising several layers, inner layers comprising hyaline to brown cells of textura epidermoidea, outer layers comprising brown cells of textura intricata at side with textura angularis at upper part. Paraphyses 2.5–4 µm wide, cylindrical, septate with small guttules, hyaline, branching, embedded in a gelatinous matrix. Asci 60–98 × 5–7.5 µm (\( \bar{x} \) = 83.5 × 6 µm, n = 25), 8-spored, unitunicate, cylindrical to broadly filiform, pedicellate, straight or slightly curved, with inconspicuous apical ring. Ascospores 10.5–14 × 4–5.5 µm (\( \bar{x} \) = 12 × 5 µm, n = 20), 1-seriate, hyaline when immature, becoming brown to dark olivaceous-brown when mature, unicellular, inequilateral to broadly fusiform, tapering towards narrow ends, verruculose when immature with 1–2 large guttules when mature. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on MEA within 72 h at room temperature and germ tubes produced from both ends. Colonies on MEA irregular, mycelium umbonate and undulate margins, white aerial hyphae at the surface and brown in reverse with white-yellow at the margin.
Material examined: THAILAND, Lampang Province, Ngao, on dead stem of Chromolaena odorata (Asteraceae), 21 September 2016, A. Mapook, LP6 (MFLU 20-0048, holotype), ex-type culture MFLUCC 17-1484.
GenBank numbers: ITS = MN638863, LSU = MN638848, SSU = MN638852, RPB2 = MN648729, TEF1-α = MN648732.
Notes: In a BLASTn search of NCBI GenBank, the closest match of the ITS sequence of Xenoanthostomella chromolaenae (MFLUCC 17-1484, ex-holotype) is Idriella lunata P.E. Nelson & S. Wilh. with 98.65% similarity to strain MUCL 4103 (KC775734). The closest match of the LSU sequence with 99% similarity was to Circinotrichum papakurae S. Hughes & Piroz. (strain CBS 101373, KR611897). The closest matches of the SSU sequence with 99.09% similarity was to Circinotrichum sinense D.W. Li et al. (strain UAMH11913, KY994108), while the closest matches of the RPB2 sequence with 81.51% similarity was to Camillea tinctor (Berk.) Læssøe, J.D. Rogers & Whalley (strain CBS 203.56, KU684282).
Subphylum Saccharomycotina O.E. Erikss. & Winka
Class Saccharomycetes O.E. Erikss. & Winka
Notes: The class Saccharomycetes is the only class in the Saccharomycotina, the budding yeasts, and contains a single order, Saccharomycetales (Suh et al. 2006a). It is a monophyletic group that is economically and environmentally important, with many species that are symbionts, pathogens or parasites and have clinical importance (e.g. causing candidiasis in immunocompromised patients).
Saccharomycetales Luerss.
Notes:Saccharomycetales is a monophyletic taxon uniting all ascomycetous budding yeasts, which is substantiated by recent multi-locus phylogenetic and phylogenomic analyses (Dujon 2010; Kurtzman and Robnett 2013; Nagy et al. 2014; Riley et al. 2016). It is the only order of the budding yeast in the subphylum Saccharomycotina. The status of several families and the status and familial placement of several genera has not yet been unequivocally resolved (Shen et al. 2018). The order includes 95 genera (Wijayawardene et al. 2017a).
Diutina Khunnamw. et al.
Notes: Diutina was erected by Khunnamwong et al. (2015) to accommodate species previously assigned to Candida Berkh. without clear phylogenetic connection to known sexual morphs. Diutina species are globally distributed but relatively rarely recorded, known from isolates from clinical specimens, insects, mangrove habitats, rotting wood, dung, or spoiled food (Khunnamwong et al. 2015). The genus lacks familial placement.
Diutina bernali Haelew., Pfliegler, Horváth & Imre, sp. nov.
Mycobank number: MB828895; Facesoffungi number: FoF 06510; Fig. 156a, d, g, j
Etymology: The specific epithet “bernali” refers to Dr. Juan A. Bernal-Vega† (Museo de Peces de Agua Dulce e Invertebrados, Universidad Autónoma de Chiriquí, Panama), entomologist and close collaborator of D. Haelewaters.
Holotype: NCAIM Y.02230T
Isolated from the gut of Canthon angustatus Harold, 1867 scarab beetle (Coleoptera: Scarabaeidae) collected in a primary rainforest (Isthmian-Atlantic moist forest) on Barro Colorado Island, Panama. Colonies are cream yellowish, flat, smooth with entire margin on YPGA agar after one month of growth at 25 °C (Fig. 156a). Cells are elongated, 5.5–9.0 µm × 2.5–3.5 µm, occur singly or in pairs and propagate by multi-polar budding (Fig. 156g) when grown in the liquid medium YPGL at 25 °C for 3 days. In exponential-phase YPGL cultures, small pseudo-hyphae-like chains of unseparated yeast cells are occasionally formed with cells up to 30 µm long. Transition to pseudohyphal growth is detectable in thin films of YPGA sandwiched between glass slides with mostly rounded cells of 5–6.5 µm × 2.9–4.4 µm, and few elongated cells up to 28 µm long in the pseudohyphae (Fig. 156j). No sporulation on YPGA or V8 agar at 25 °C. No growth at 37 °C. The physiological characteristics (Table 4) show that the strain differs in numerous properties from other species of the genus, which allows their differentiation by conventional taxonomic tests. Karyotype: four bands of similar size (Fig. 155b).
a Haplotype network illustrating the nucleotide differences in the various ITS repeats found in D. bernali sp. nov. NCAIM Y.02230T (isotype DEBM179T) and closely related Diutina species. b Chromosomal profiles (karyotypes) of the new species, from left to right: D. bernali sp. nov. NCAIM Y.02230T; D. sipiczkii sp. nov. NCAIM Y.02231; D. sipiczkii sp. nov. NCAIM Y.02232T
Material examined: PANAMA, Panamá Oeste Province, Isla de Barro Colorado, Martin Moynihan Visitors Center, primary rainforest (Isthmian-Atlantic moist forest), isolated from the gut of Canthon angustatus (Coleoptera, Scarabaeidae), 27 July 2017, NCAIM Y.02230T (holotype), DEBM179T (isotype). Both are permanently preserved in a metabolically inactive state.
GenBank numbers: ITS = MN219635–MN219643, LSU = MN219652, SSU = MN219646, ACT1 = MN238673, TEF1-α = MN238676.
Notes: Differences in LSU sequences of our new species and Diutina scorzettiae Middelhoven & Kurtzman emend. Khunnamwong et al. are 14.6% (71 substitutions, 1 gap). Both SSU and LSU sequences have clear ambiguous bases after PCR amplification and sequencing and the ITS region was found to contain several heterozygous bases. The internal transcribed spacer region separates the genes coding for the SSU 18S and the LSU 26S genes in the ribosomal DNA (rDNA) units in yeasts and other fungi. These genes and the ITS region are organized into long tandem arrays in which the units evolve via concerted evolution, which homogenizes their sequences. Exceptions to this concerted evolution and homogenization of rDNA arrays have been published in certain groups of plants and fungi, such as species of the related yeast genus Metschnikowia (Sipiczki et al. 2018). Weak assimilation of sucrose, melibiose, as well as no assimilation of ethylamine and weak assimilation of nitrate and nitrite, growth on vitamin free medium, and weak acid production differentiate Diutina bernali from the previously described species in the genus. The growth of this new species was conspicuously filamentous on D-trehalose, arbutin, D-melibiose, and quinic acid carbon sources on agar medium. Sampling protocols for the collection of this yeast were authorized by Ministerio de Ambiente de Panamá (MiAmbiente, permit number SC/AH-1-17).
Diutina sipiczkii Pfliegler, Haelew., Horváth & Imre, sp. nov.
Mycobank number: MB828896; Facesoffungi number: FoF 06511; Fig. 156b, c, e, f, h, i, k, l
Microphotographs of new species of Diutina. a, d, g, jDiutina bernali sp.nov., NCAIM Y.02230T. b, e, h, kD. sipiczkii sp. nov. NCAIM Y.02231. c, f, h, l and D. sipiczkii NCAIM Y.02232T. a–c 10-day-old colonies on YPD agar. d–f Invasivity on YPD agar after washing off 10-day-old colonies. g–i Exponential phase cellular morphology in YPD liquid medium, bars = 20 µm. j–l Exponential phase pseudohyphal morphology on YPD sandwich plates, bars = 100 µm
Etymology: The specific epithet “sipiczkii” refers to Prof. Matthias Sipiczki (University of Debrecen, Hungary) for his contributions to yeast genetics, diversity, and taxonomy.
Holotype: NCAIM Y.02232T
Isolated from the gut of Canthon angustatus scarab beetle from Panama. Colonies are pastel pink, flat, rough (NCAIM Y.02232T = DEBM181T) or smooth (paratype: NCAIM Y.02231 = DEBM180) with pseudohyphal margin on YPGA agar after 1 month of growth at 25 °C (Fig. 156b–c). Cells are elongated, 7–15 µm × 3–3.5 µm, occur singly, in pairs or and propagate by multi-polar budding (Fig. 156h–i) in exponential phase cultures in YPGL at 25 °C. In exponential-phase YPGL cultures, pseudo-hyphae-like branched groups or unbranched chains of unseparated yeast cells are frequently formed (Fig. 156h–i). Transition to hyphal or pseudohyphal growth is prominent in thin films of YPGA sandwiched between glass slides, with long pseudohyphae, with up to 90 µm (NCAIM Y.02232T) cells or short pseudohyphae with up to 15 µm long (NCAIM Y.02231), predominantly round cells in the pseudohyphae (Fig. 156k–l). No sporulation on YPGA, potassium-acetate agar or V8 agar at 25 °C, neither in pure, nor in mixed cultures of the holotype and paratype cultures. No growth at 37 °C. The physiological characteristics (Table 4) show that the strain differs in numerous properties from other species of the genus, which allows their differentiation by conventional taxonomic tests, except for the closely related and physiologically indistinguishable Diutina bernali sp. nov. Karyotype: two bands (probably representing three chromosomes) of similar size, plus one band of smaller size (Fig. 155b).
Material examined: PANAMA, Panamá Oeste Province, Isla de Barro Colorado, Martin Moynihan Visitors Center, primary rainforest (Isthmian-Atlantic moist forest), isolated from the gut of Canthon angustatus (Coleoptera, Scarabaeidae), 27 July 2017, NCAIM Y.02232T (holotype), DEBM181T (isotype). Same data, isolated from the gut of the same Canthon angustatus scarab beetle, 27 July 2017, NCAIM Y.02231 (paratype) = DEBM180 (paratype).
GenBank numbers: NCAIM Y.02232T: ITS = MN219645, LSU = MN219654, SSU = MN219648, ACT1 = MN238671, TEF1-α = MN238674. NCAIM Y.02231: ITS = MN219644, LSU = MN216953, SSU = MN219647, ACT1 = MN238672, TEF1-α = MN238675.
Notes: The holotype NCAIM Y.02232T and paratype isolate NCAIM Y.02231 were isolated from the same beetle specimen (the same host species but a different specimen than the one for NCAIM Y.02230T, Diutina bernali sp. nov.). The morphology of the two D. sipiczkii sp. nov. isolates is remarkably different. However, none of the sequenced genes revealed any nucleotide differences between the isolates. NCAIM Y.02232T and NCAIM Y.02231 both differ from the isolate NCAIM Y.02230T, holotype of D. bernali sp. nov., by 67 substitutions and 1 gap in the D1/D2 LSU region (13.7% difference) and in 62 substitutions and 2 gaps (20.8% difference) to 65 substitutions and 4 gaps of the ITS1–5.8S–ITS region (22.5% difference) depending on D. bernali haplotype. Difference of the LSU sequence to D. scorzettiae is 8.5% (42 substitutions, 1 gap). Apparently, the phenomenon of variable ITS haplotypes does not exist in D. sipiczkii sp. nov. However, the number of substitutions, even if taking intra-strain variations into account (as represented on the ITS haplotype network of Fig. 155a), and substitutions in other genes is sufficient to separate the two new species and the related D. scorzettiae from each other.
Growth of Diutina sipiczkii sp. nov. is conspicuously filamentous on various carbon sources on agar medium (D-ribose D-xylose, L-arabinose, L-rhamnose, sucrose, D-trehalose, α-D-glucoside, D-salicin, arbutin, D-melibiose, lactose, D-raffinose, melesitose, inulin, starch, glycerol, erythtritol, ribitol, xylitol, L-arabitinol, D-sorbitol, galactitol, myo-inositol, D-glucono-1, 5-lactone, 2-keto-D-gluconate, sodium-D-glucoronate, galacturonic acid, butane-2,3-diol, quinic acid, calcium-D-saccharate in both strains, and on sodium-D-gluconate in the case of the holotype).
Barro Colorado Island, the collecting location for the two newly described Diutina species, has been investigated for arthropod-associated yeasts prior to our work. Several undescribed species have been reported within the following clades: Meyerozyma guilliermondii (Suh and Blackwell 2004), Candida membranifaciens (Suh et al. 2005), and Teunomyces kruisii (Suh et al. 2006b). These observations and the currently described Diutina spp. from the same limited geographical region, often from the same or related hosts, reflect the vast diversity of Neotropical insect-associated microfungi. Sampling protocols for the collection of this yeast were authorized by Ministerio de Ambiente de Panamá (MiAmbiente, permit number SC/AH-1-17).
Our multi-locus SSU + D1/D2 LSU + ACT + TEF1-α dataset for Diutina and related genera in the CTG clade (Fig. 157) consisted of 55 isolates and 4132 characters (1721 + 524 + 980 + 907), of which 2458 were constant, 346 parsimony-uninformative, and 1328 parsimony-informative (proportion 32.1%). LSU was the region with highest proportion of parsimony-informative over total characters (50.2%), followed by ACT (36.1%), TEF1-α (35.7%), and SSU (22.5%). The following substitution models were selected by AICc: TIM1 + I+G (SSU, -lnL = 11457.2635), TrN + I+G (D1/D2 LSU, -lnL = 8785.3555), GTR + G (ACT, -lnL = 9678.5135), GTR + I+G (TEF1-α, -lnL = 9959.7673). The ITS dataset consisted of 14 isolates and 307 characters, of which 124 were constant, 26 parsimony-uninformative, and 157 parsimony-informative. The LSU dataset consisted of 14 isolates and 524 characters, of which 286 were constant, 43 parsimony-uninformative, and 195 parsimony-informative. Substitution models selected through jModelTest2 (by AICc) were TrN + I+G for ITS (-lnL = 2720.8057) and TIM3 + I+G for LSU (-lnL = 3140.3513). Gene sequences used for generating ML inference are listed in Table 5.
Phylogenetic position of new species of Diutina, D. bernali sp. nov. NCAIM Y.02230T and D. sipiczkii sp. nov. NCAIM Y.02232T and NCAIM Y.02231. ML inference reconstructed from the SSU + D1/D2 LSU + ACT1 + TEF1-α dataset, showing relationships in the genus and among genera of the CTG clade. A total of 55 strains are included in the combined sequence analyses. Schizosaccharomyces pombe (CBS 356T) is used as the outgroup taxon. Tree topologies of the ML and Bayesian analyses were highly similar. The best-scoring ML tree (-lnL = − 41951.984) is presented. The dataset had 4132 characters. ML bootstrap support values equal to or greater than 60% are given above or below the nodes. New species are highlighted in blue. Note that the status of the families Metschnikowiaceae and Debaryomycetaceae are currently unresolved
Phylum Basidiomycota R.T. Moore
Notes: Phylum Basidiomycota is the second largest phylum of fungi after Ascomycota (He et al. 2019). It includes three subphyla Agaricomycotina, Pucciniomycotina and Ustilaginomycotina. The most diverse of these is Agaricomycotina and its representatives range from yeast-forming species to mushroom.
Subphylum Agaricomycotina Doweld
Class Agaricomycetes Doweld
Notes: Agaricomycetes is a diverse class best known for its mushroom-forming fungi. It includes thousands of species classified in more than 15 orders and over 1000 genera.
Subclass Agaricomycetidae Parmasto
Agaricales Underw.
Notes: Agaricales includes mainly mushrooms with gills but also fungi like puffballs and clavarioid fungi. The order contains more than 30 families and 400 genera. The order was divided in seven suborders by Dentinger et al. (2016). The species with brown and dark spores are placed in suborder Agaricineae.
Cortinariaceae R. Heim ex Pouzar
Notes: The limits of the family Cortinariaceae remain undetermined. Most species in this family are in Cortinarius, with a cosmopolitan distribution and over 2000 described species. Many genera formerly placed in the Cortinariaceae, e.g., Phaeocollybia R. Heim, Hebeloma (Fr.) P. Kumm., Galerina Earle, have been moved to other families in the Agaricales. Conversely, the sequestrate genera, Thaxterogaster Singer, Quadrispora Bougher & Castellano, Protoglossum Massee and Hymenogaster p.p. as well as Cuphocybe R. Heim, Rapacea E. Horak and species of Rozites P. Karst., once thought to pertain to the Cortinariaceae, are currently included in Cortinarius (Peintner et al. 2001, 2002).
Cortinarius (Pers.) Gray
Notes: The species of Cortinarius are important ectomycorrhizal fungi forming associations with different trees and shrubs, belonging to the families Fagaceae, Salicaceae, Caesalpiniaceae, Cistaceae, Dipterocarpaceae, Myrtaceae, Rhamnaceae, Rosaceae and Pinaceae, as well as some herbaceous plants in the Cyperaceae and Polygonaceae (Frøslev et al. 2006). The basidiocarps range from agaricoid to sequestrate, and many have poorly to well–developed veils. The basidiospores are typically ornamented and cinnamon brown in deposit. Hundreds of species are still undescribed and below we introduce six new species that have been discovered from United Kingdom.
Cortinarius ainsworthii Liimat. & Niskanen, sp. nov.
Index Fungorum number: IF556991; Facesoffungi number: FoF 07206; Fig. 158
Etymology: In honour of A. Martyn Ainsworth, a long-term mycologist who first found this species from Britain.
Holotype: A.M. Ainsworth 236875 (K).
Pileus 1.5–6.5 cm wide, at first convex, later plano-convex, brown, margin whitish fibrillose; hygrophanous, drying up in a zone at the centre to pale ochraceous brown. Lamellae medium spaced, adnexed to emarginate, at first pale brown, later brown to dark brown, edge paler at least when young. Stipe 3–8 cm long, 0.6–1.1 cm thick at apex, 0.8–1.3 cm wide at base, cylindrical to somewhat clavate, at first whitish silky fibrillose, becoming pale brownish with age, especially at the base. Context: in pileus brown, in stipe pale brown, marbled hygrophanous. Universal veil white, rather sparse or more abundant and forming some incomplete girdles on the stipe. Odour in lamellae indistinct. Basidiospores 8–9(–9.5) × 5–5.8 μm, av. = 8.7 × 5.3 µm, Q = 1.4–1.9, Qav. = 1.65, amygdaloid, moderately to strongly verrucose, moderately dextrinoid. Basidia 27–39 × 7–9 µm, clavate, with four sterigmata. Lamellar trama hyphae golden brown, smooth with a few spot-like encrustations. Pileipellis: Epicutis pale, consisting of parallel hyphae, 5.5–8.5 μm wide, smooth with a few spot-like incrustations. Hypoderm pale brown, elements 17–31 × 11–15.5, smooth with a few spot-like encrustations. ITS sequence (GenBank MK010965, holotype) distinct from other members of C. sect. Bovini and deviating from them by more than 13 substitutions and indels.
Ecology and distribution: In deciduous forests (Quercus, Corylus, Fagus) on calcareous ground, also in open, grazed areas, presumably with Helianthemum. From temperate to hemiboreal areas. Known from Central and northern Europe.
Material examined: BELGIUM. Brabant, Tervuren, in Fagus forest on calcareous ground, 23 September 1989, Lindström et al. CFP848 p.p. (mixed collection) (S). SWEDEN. Öland, Langlot, Ismantorps borg, open, grazed area, Quercus, Corylus, 15 September 2003, T. Niskanen 03-1528 (H). UNITED KINGDOM, England, West Sussex. Devil’s Dyke, TQ26511133, in chalk downland, on calcareous soil with H. nummularium but with young Quercus not far away, 30 September 2017, A.M. Ainsworth 236875 (K, holotype).
GenBank number: ITS =MK795702 (Lindström et al. CFP848 p.p.), MK795703 (T. Niskanen 03-1528), MK010965 (A.M. Ainsworth 236875).
Notes: Cortinarius ainsworthi is a medium-sized species of C. sect. Bovini that can be recognized by the combination of brown pileus, amygdaloid, medium-sized spores (av. = 8.7 × 5.3 µm), and habitat in deciduous forests on calcareous ground. Part of the C. rheubarbarinus collection illustrated in Brandrud et al. (1992) represents this species (the other part, that has the smell of parsley in lamellae and narrowly obovoid to dacryoid spores, is C. phaeosmus Rob. Henry (Niskanen and Kytövuori 2012). However, the type specimen of C. rheubarbarinus does not represent C. ainsworthii (unpublished data). A published ITS sequence (GenBank number MH784666), labelled as originating from the type specimen of C. hydrobivelus Rob. Henry & Reumaux, also matches with our species. However, our own unpublished ITS sequence of the same type specimen matches another species, C. armeniacus (Schaeff.) Fr. The protologue of C. hydrobivelus describes a species with a whitish context and growing with spruce, both typical for C. armeniacus. Therefore, we conclude that C. hydrobivelus is a later synonym of C. armeniacus and either the type specimen is a mixed collection or the ITS sequence MH784666 is a contamination. Thus, our species still lacks any valid name and we here describe it as new.
Cortinarius aurae Niskanen & Liimat., sp. nov.
Index Fungorum number: IF556995; Facesoffungi number: FoF 07207; Fig. 159
Etymology: Named after our newborn daughter Aura.
Holotype: T. Niskanen 200315 (K).
Pileus 0.5–3 cm wide, conical to somewhat convex, often with an umbo, red brown, hygrophanous. Lamellae medium spaced, yellowish brown. Stipe 1.5–6 cm long, 0.1–0.4 cm thick, cylindrical or rooting, at first covered with white fibrils, soon yellowish brown. Context: brown. Universal veil white, fairly sparse. Odour at the base of the stipe weak, iodoform-like, best observed when slightly dried; in lamellae indistinct. Basidiospores 6.5–8 × 4.5–5(–5.5) µm, av. = 7.0 × 4.7 µm, Q = 1.4–1.7, Qav. = 1.51, mostly ellipsoid with few amygdaloid, moderately verrucose, somewhat dextrinoid. Basidia 24–32 × 7–9.5 µm, clavate, with four sterigmata. Lamellar trama hyphae golden brown, strongly encrusted with zebra-striped encrustations. Pileipellis: Epicutis pale, consisting of more or less parallel hyphae 8–13 μm wide, densely encrusted with zebra-striped incrustations. Hypoderm pale orange-brown, elements 15.5–47 × 10–23 μm, densely encrusted with zebra-striped incrustations. ITS sequence (GenBank MK795701, holotype) distinct from other members of C. sect. Obtusisensu lato and deviating from them by more than 14 substitutions and indels.
Ecology and distribution: In mesic to damp coniferous forests but also known to occur with Castanea sativa. Widespread, known from Europe and North America.
Material examined: UNITED KINGDOM, England, West Kent, Mereworth Woods, in woodland, on mossy stump, Castanea sativa, 12 September 2010, A. Henrici 166960 (K); Scotland, Mid Perthshire, Blackwood of Rannoch, mesic to damp Pinus dominated forest with Betula, 24 August 2015, T. Niskanen 15-035, 200315 (K, holotype).
GenBank number: ITS = MK795700 (A. Henrici 166960), MK795701 (T. Niskanen 15-035).
Notes: Cortinarius aurae is a small brown species with sparse white universal veil, iodoform-like odour at the base of the stipe and ellipsoid, small spores (7–8 × 4.5–5 µm). It is most reminiscent of C. obtusus but the latter has somewhat larger (8–9 × 4.5–5 μm), amygdaloid spores. Cortinarius aurae formed a well-supported (100% ML; Fig. 164) clade within C. sect. Obtusi in our phylogenetic analysis and is here described as new.
Cortinarius britannicus Liimat. & Niskanen, sp. nov.
Index Fungorum number: IF556996; Facesoffungi number: FoF 07208; Fig. 160
Etymology: The species was first found from Britain.
Holotype: D. J. Savage 237998 (K).
Pileus 2–3 cm wide, low convex, greyish purple, margin whitish fibrillose; hygrophanous. Lamellae medium spaced, adnexed, brown, edge paler. Stipe 3–4 cm long, 0.35 cm thick at apex, cylindrical to somewhat tapering downwards, silky whitish fibrillose over a greyish purple base. Universal veil white, forming incomplete girdles on the stipe. Odour in lamellae indistinct. Basidiospores 8–9 × 5–6 µm, av. = 8.6 × 5.6 µm, Q = 1.3–2.0, Qav. = 1.55, amygdaloid, with a suprahilar depression, moderately verrucose, more strongly at the apex, moderately to strongly dextrinoid. Basidia (26–)30–35 × 7–10 µm, somewhat clavate, with four sterigmata. Lamellar trama hyphae rusty-brown, moderately encrusted with spot-like encrustations. Pileipellis: Epicutis pale, consisting of more or less parallel hyphae, 5–13 μm wide, with zebra-like incrustations. Lower hyphae rusty brown, 25–30 μm wide, with abundant zebra-like incrustations. ITS sequence (GenBank MK795699, holotype) distinct from other members of C. sect. Bicolores, with a sister group relationship to C. cystidiobicolor (GenBank KX964411, holotype) and deviating from it in the ITS region by more than 20 substitutions and indel positions.
Ecology and distribution: Under Fagus among mosses on clay soil. Currently only known from Britain.
Material examined: UNITED KINGDOM, Scotland, Caithness, Olrig Wood, in woodland, in fine grained clay soil, under Fagus sylvatica, 4 November 2017, D.J. Savage 222/17, 237998 (K, holotype).
GenBank number: ITS =MK795699.
Notes: Based on our phylogenetic analysis (Fig. 164) C. britannicus belongs to Cortinarius sect. Bicolores that contains otherwise similar but far bigger species. The combination of small-sized, purplish-coloured basidiomata and amygdaloid, medium-sized spores (av. = 8.6 × 5.6 µm) distinguish C. britannicus from the other known species of C. subgenus Telamonia. The species is known from a single collection from Britain but we sequenced it twice to confirm that the ITS sequence of the species is correct and unique.
Cortinarius heatherae Overall, sp. nov.
Index Fungorum number: IF556997; Facesoffungi number: FoF 07209; Fig. 161
Etymology: The species is named after Heather Overall.
Holotype: A.S.Overall 08.11.2018, 252508 (K).
Pileus 3–5.7 cm wide, at first convex, later plano-convex with a broad umbo, surface silky whitish fibrillose when young, later more apparent only in the margin, reddish brown, later darkening-blackening in large spots, hygrophanous. Lamellae adnate, medium spaced, fairly broad, at first light brown with paler edge, becoming rusty brown to dark brown at maturity. Stipe 6.3–8.2 cm long, 0.7–1.5 cm thick, more or less clavate, at first greyish white silky fibrillose, later pale greyish brown. Context: greyish brown, marbled hygrophanous. Universal veil cream-coloured, forming scattered patterns on the surface of the stipe. Odour not recorded. Basidiospores 9–11 × 5.5–6.5(–7) µm, av. = 10.0 × 6.2 µm, Q = 1.5–1.8, Qav. = 1.6, mostly amygdaloid, strongly verrucose, strongly dextrinoid. Basidia 33–46 × 7–11 µm, somewhat clavate, with four sterigmata. Lamellar trama hyphae smooth. Pileipellis: Epicutis pale, consisting of parallel hyphae, 6–8 μm wide, smooth to encrusted with spot-like encrustations. Hypoderm colourless, elements 17–50 × 11–19, smooth. ITS sequence (GenBank MK795704, holotype) distinct from other members of C. sect. Bovini. With a close relationship to C. subbrunneus and deviating from it by nine substitutions and indels.
Ecology and distribution: In temperate mixed forests of Quercus robur, Q. ilex and Salix on calcareous soil. Currently only known from Britain.
Material examined: UNITED KINGDOM, England, Middlesex, Heathrow, River Crane (near), in soil (near water), Quercus or Salix spp., 8 November 2018, A.S. Overall 08.11.2018, 252508 (K, holotype); loc. cit., 13 November 2018, A.S. Overall 13.11.2018 a (K); loc. cit., 13 November 2018, A.S. Overall 13.11.2018 b (K).
GenBank number: ITS = MK795704 (252508), MK795705 (A.S. Overall 13.11.2018 a), MK795706 (A.S. Overall 13.11.2018 b)
Notes: Cortinarius heatherae is a typical member of C. sect. Bovini. The basidiomata are medium-sized and brown, the pileus becomes dark spotted with age, the stipe is clavate and the spores are amygdaloid. The sister species C. subbrunneus Bidaud, Careteret & Remaux and C. subbrunneoideus Kytöv., Liimat. & Niskanen grow in coniferous forests. In addition the spores of C. subbrunneus are relatively narrower (Qav. = 1.75) and slightly larger (9.5–11.5 × 6–7 µm) whereas the spores of C. subbrunneoideus are smaller (av. = 9 × 6 µm) and only somewhat to moderately dextrinoid. Cortinarius heatherae formed a well-supported clade (100% ML; Fig. 164) in our phylogenetic analysis. Also morphological and ecological differences support its distinction from the sister species and therefore we here describe it as new.
Cortinarius scoticus Niskanen & Liimat., sp. nov.
Index Fungorum number: IF556998; Facesoffungi number: FoF 07210; Fig. 162
Etymology: The name refers to Scotland where this species was first found.
Holotype: T. Niskanen 15-034, 200314 (K).
Pileus 0.7–1.5 cm wide hemispherical, later low convex to almost plane, red brown to dark red brown, hygrophanous. Lamellae medium spaced, brown. Stipe 4–6 cm long, 0.2–0.3 cm thick, cylindrical, at first covered with silky, white fibrils, later yellowish brown. Context: brown. Universal veil white, fairly sparse. Odour at the base of the stipe weak, iodoform-like, best observed when slightly dried; in lamellae indistinct. Basidiospores 7.5–8.5(–8.8) × 4.5–5.5 µm, av. = 8.0 × 4.8 µm, Q = 1.5–1.9, Qav. = 1.67, ellipsoid to somewhat amygdaloid, moderately verrucose, somewhat dextrinoid. Basidia 25–34 × 7–10 µm, clavate, with four sterigmata. Lamellar trama hyphae yellowish brown, fairly strongly encrusted with zebra-striped encrustations. Pileipellis: Epicutis pale, consisting of more or less parallel hyphae, 5 μm wide, without encrustations. Hypoderm elements 19–44 × 10–20 μm, some hyphae encrusted with zebra-like incrustations. ITS sequence (GenBank MK795697, holotype) distinct from other members of C. sect. Obtusisensu lato and deviating from them by more than 14 substitutions and indels.
Ecology and distribution: In mesic and damp pine dominated forests. Known from central and northern Europe.
Material examined: UNITED KINGDOM, Scotland, Mid Perthshire, Blackwood of Rannoch, mesic to damp Pinus dominated forest with Betula, 24 August 2015, T. Niskanen 15-034, 200314 (K, holotype).
GenBank number: ITS = MK795697.
Specimen details of downloaded sequences: NORWAY. GenBank number: KC842421.
Notes: Cortinarius scoticus is easy to identify as a member of C. sect. Obtusi by the weak, iodoform-like, odour at the base of the stipe, white and sparse universal veil and small and brown basidiomata. In the field C. scoticus looks like C. trossingenensis, but the latter has small (4.5–5.5 × 4–4.5 μm), subglobose spores. In our phylogenetic analysis C. scoticus formed a well-supported clade (100% ML; Fig. 164) and based on the molecular and morphological data is described here as new.
Cortinarius subsaniosus Liimat. & Niskanen, sp. nov.
Index Fungorum number: IF556999; Facesoffungi number FoF 07211; Fig. 163
Etymology: The name refers to the affinity to Cortinarius saniosus.
Holotype: S. E. Evans 77587 (K).
Pileus 0.7–1.5 cm wide, at first conical to somewhat convex, later convex to planoconvex with an acute umbo, yellowish brown, often dark brown at the centre, hygrophanous. Lamellae medium spaced, brown. Stipe 2–4 cm long, 0.15–0.3 cm thick at the apex, cylindrical, yellowish brown. Context yellowish brown, dark brown at the base of the stipe. Universal veil yellow, forming distinct complete and incomplete girdles on the stipe. Odour in lamellae indistinct. Basidiospores 9.5–11 × 6–6.5(–7) µm, av. = 10.2 × 6.1 µm, Q = 1.5–1.8, Qav. = 1.67, ellipsoid to amygdaloid, strongly verrucose, especially at the apex, moderately dextrinoid. Basidia 25–35 × 9–11 µm, clavate, with four sterigmata. Lamellar trama hyphae golden brown, mainly smooth with a few spot-like encrustations. Pileipellis: Epicutis rusty brown, consisting of more or less parallel hyphae, 5–8 μm wide, densely encrusted with zebra-striped incrustations. ITS sequence (GenBank MK795698, holotype) distinct from other members of C. sect. Saniosi and deviating from them by more than 8 substitutions and indels.
Ecology and distribution: At least with Salix on sandy soil but might also be associated with other tree species. In northern and central Europe.
Material examined: UNITED KINGDOM, England, Westmorland, Barrow-in-Furness, Sandscale Haws National Nature Reserve, on sandy soil with Salix, 31 August 1999, S. E. Evans 77587 (K, holotype).
GenBank number: ITS = MK795698.
Specimen details of downloaded sequences: DENMARK, Tønder, Rømø, grey dune, Salix repens var. arenaria, E. & K. Bendiksen 88.0921 (O), GenBank number: DQ102657. ESTONIA, Saaremaa Island, Tagamoisa, ectomycorrhizal root tip, GenBank number: AJ893233. NORWAY, Vest-Agder, Farsund, Lista, Havik, sand dune depression, Salix repens, KH 408-78 (O 123908), GenBank number: DQ102658. SWEDEN, Jämtland, Håsjö, Singsjön, lakeshore, H. Lindström 01.001 (H. Lindström pers. herb.), GenBank number: DQ102659.
Notes: Cortinarius subsaniosus is a small yellow brown species with an acutely umbonate pileus, yellow universal veil and rather large (9.5–11 × 6–6.5(–7) µm), strongly verrucose spores. It is most reminiscent of C. saniosus but this has somewhat smaller spores 8.5–10 × 5–6.5 μm. Based on our phylogenetic analysis C. subsaniosus belongs to C. sect. Saniosi. It forms a well-supported (96% ML; Fig. 164), monophyletic group within the section.
Phylogram resulting from the RAxML (Stamatakis 2014) analysis of ITS regions. Bootstrap values greater than 50% are indicated above branches. The specimens in boldface represent the type specimens of the species
Subclass Auriculariomycetidae Jülich
Auriculariales J. Schröt.
Notes: Auriculariales includes species with resupinate, effused-reflexed, hydnoid, cerebriform, coraloid and pileate basidiomata; thin or thick-walled basidia globose to cylindrical, cystidia present or absent, and thin-walled basidiospores that germinate by tubes or producing conidia (Bodman 1952; Lowy 1971). The classification of this order has been amended through the revisions of types and newly collected material of species earlier unavailable for morphological and phylogenetic analysis, thus, old genera were restored, and new genera, combinations, and species were proposed (Malysheva and Spirin 2017; Malysheva et al. 2018; Alvarenga et al. 2019; Spirin et al. 2019).
Auriculariaceae Fr.
Notes: Auriculariaceae typified by Auricularia mesenterica (Dicks.) Pers., belongs to Auriculariales. The family includes species with resupinate, effused-reflexed, hydnoid, cerebriform and pileate basidiomata; thin or thick-walled basidia globose to cylindrical, cystidia present or absent, and thin-walled basidiospores that germinate by tubes or producing conidia (Martin 1952; Lowy 1971). The most common genera of Auriculariaceae are Auricularia Bull., Heterochaete Pat., Exidia Fr., Eichleriella Bres., and Exidiopsis (Bref.) A. Møller. Several genera have been described or delimited using morphological and DNA analyses, viz. Adustochaete Alvarenga & K.H. Larsson, Amphistereum Spirin & Malysheva, Heteroradulum Lloyd ex Spirin & V. Malysheva, Myxariellum Spirin & V. Malysheva, Proterochaete Spirin & V. Malysheva and Sclerotrema Spirin & Malysheva (Malysheva and Spirin 2017; Alvarenga et al. 2019; Spirin et al. 2019)
Adustochaete Alvarenga & K.H. Larss.
Notes: Adustochaete was described by Alvarenga et al. (2019) with Adustochaete rava Alvarenga & K.H. Larsson as the type species, and comprises two species A. rava and A. interrupta Spirin & V. Malysheva. The genus is characterised by resupinate, arid, soft basidiomata; spiny or tuberculate hymenophore; monomitic hyphal structure; hyphidia with crystal and cystidia-like cells present or absent; 4-celled basidia; hyaline, thin-walled, cylindrical to broadly cylindrical basidiospores. So far, the genus is found on dead hard wood branches in the Neotropics (Brazil and Mexico).
Adustochaete nivea Alvarenga, sp. nov.
MycoBank number: MB831902; Facesoffungi number: FoF 07233; Fig. 165
Etymology: ‘nivea’ (Lat., adj.)—snow-white.
Holotype: RLMA 531* (URM 93408)
Basidiomata annual, resupinate, farinaceous, first up to 0.2 cm in diam., later fusing, 0.1–0.2 mm thick, white 1A1 (Kornerup and Wanscher 1978). Margin adnate or slightly detaching, arachnoid to fimbriate. Hymenial surface covered by sharp-tipped, rather regularly arranged spines 45–200 × 35–45 µm, 4–6 per mm. Hyphal structure monomitic; hyphae clamped. Subicular hyphae hyaline, thin-walled, loosely arranged, interwoven or in subparallel bundles, 3–4 μm in diam. Subhymenial hyphae hyaline, thin-walled, ascending, 2–3 μm in diam. Basidiospores thin-walled, cylindrical, slightly or distinctly curved, (8.4–) 10.2–13.6 (–14.6) × (4.3–) 4.6–5.9 (–6.2) μm (n = 21/1), L = 11.8, W = 5.09, Q’ = (1.86–) 2.1–2.5 (–2.8), Q = 2.31. Basidia embedded, narrowly ovoid to obconical, 4-celled, (14.4–) 14.9–16.2 (–17.2) × (9.3–) 9.7–10.1 (–10.3) μm, mostly sessile but occasionally bearing an enucleate stalk up to 2–4 × 2–2.5 μm. Cystidia-like structures not observed. Hyphidia abundant in sterile spines and hymenial layer, variably branched, 1–1.5 μm in diam. at the apex, thick-walled, densely arranged and cemented by colourless amorphous matter.
Material examined: BRAZIL, Pernambuco: Recife, Universidade Federal de Pernambuco - Centro de Biociências, 2018 RLMA 531(URM 93408, holotype).
GenBank numbers: ITS = MN165954, LSU = MN165989.
Notes: Adustochaete nivea is easily recognized by the white and farinaceous basidiomata covering up to 5 cm. It differs from A. rava, the only species of the genus reported in Brazil, by the larger basidia and smaller basidiospores [(9.7–)10.8–15.2(–16.7) × (7.2–)7.3–10(–10.5) μm and, (10.1–)10.2–13.7(–13.8) × (3.7–)3.8–4.7(–5.1) μm, respectly in A. rava]. Adustochaete interrupta, the other species of the genus, also has white basidiomata, but differs by the tuberculate hymenophore, presence of cystidia-like structures, smaller basidia and basidiospores [(14.3–)15.1–24(–24.3) × (8.7–)9.1–11.8(–12.2) μm and (10.1–)11.3–14.3(–16.8) × (5.2–)5.7–6.2(–6.3) μm, respectly] (Alvarenga et al. 2019). Adustohaete nivea clustered in the Adustochaete clade with strong support (Fig. 166). ML and BI analyses returned trees with nearly congruent topologies, and our phylogenetic analyses were consistent with the recent work introducing the genus (Alvarenga et al. 2019).
References
Abbott SP, Currah RS (1997) The Hevellaceae: systematic revision and occurrence in northern and northwestern North America. Mycotaxon 62:1–125
Abdullah SK (1980) Two hyphomycetes on litter in stagnant water from Britain. Trans Br Mycol Soc 75:514–517
Adamčík S, Cai L, Chakraborty D, Chen XH, Cotter VTH, Dai DQ, Dai YC, Das K, Deng C, Ghobad-Nejhad M, Hyde KD, Langer E, Deepna LKP, Liu F, Liu SL, Liu TT, Wei LV, Shu-Xia LV, Machado AR, Pinho DB, Pereira OL, Prasher IB, Rosado AWC, Qin J, Qin WM, Verma RK, Wang Q, Yang ZL, Yu XD, Zhou LW, Buyck B (2015) Fungal biodiversity profiles 1–10. Cryptogr Mycol 36:121–166
Adams GC, Wingfield MJ, Common R, Roux J (2004) Phylogenetic relationships and morphology of Cytospora species and related teleomorphs (Ascomycota, Diaporthales, Valsaceae) from Eucalyptus. Stud Mycol 52:1–144
Adams GC, Roux J, Wingfield MJ (2006) Cytospora species (Ascomycota, Diaporthales, Valsaceae): introduced and native pathogens of trees in South Africa. Australas Plant Phytopathol 35:521–548
Aguirre-Hudson B, Kokubun T, Spooner BM, Tibell L (2007) Taxonomy of Calicium victorianum (F. Wilson) Tibell (Caliciaceae, Lecanorales), a lichenized ascomycete new to Europe. Lichenologist 39:401–407
Ahmed SA, van de Sande WWJ, Stevens DA, Fahal A, van Diepeningen AD, Menken SBJ, de Hoog GS (2014) Revision of agents of black-grain eumycetoma in the order Pleosporales. Persoonia 33:141–154
Ahmed SA, Desbois N, Quist D, Miossec C, Atoche C, Bonifaz A, de Hoog GS (2015) Phaeohyphomycosis caused by a novel species, Pseudochaetosphaeronema martinelli. J Clin Microbiol 53:2927–2934
Alias SA, Jones EBG, Kuthubutheen AJ (1994) Fasciatispora lignicola sp. nov. (Amphisphaeriaceae, Ascomycotina) from drift mangrove wood. Mycotaxon 52:77–82
Almeida DAC, Gusmão LFP, Miller AN (2017) A new species of Bertiella (Melanommataceae) from Brazil and a key to accepted species. Mycosphere 8:392–396
Alvarenga RLM, Spirin V, Malysheva V, Gibertoni TB, Larsson KH (2019) Two new genera and six other novelties in Heterochaetesensu lato (Auriculariales, Basidiomycota). Botany 97:439–451
Alves A, Linaldeddu BT, Deidda A, Scanu B, Phillips AJ (2014) The complex of Diplodia species associated with Fraxinus and some other woody hosts in Italy and Portugal. Fungal Divers 67:143–156
Ariyawansa HA, Tanaka K, Thambugala KM, Phookamsak R, Tian Q, Camporesi E, Hongsanan S, Monkai J, Wanasinghe DN, Mapook A, Chukeatirote E, Kang JC, Xu JC, McKenzie EHC, Jones EBG, Hyde KD (2014) A molecular phylogenetic reappraisal of the Didymosphaeriaceae (= Montagnulaceae). Fungal Divers 68:69–104
Ariyawansa HA, Hyde KD, Jayasiri SC, Buyck B, Chethana KWT, Dai DQ, Dai YC, Daranagama DA, Jayawardena RS, Lücking R, Ghobad-Nejhad M, Niskanen T, Thambugala KM, Voigt K, Zhao RL, Li GJ, Doilom M, Boonmee S, Yang ZL, Cai Q, Cui YY, Bahkali AH, Chen J, Cui BK, Chen JJ, Dayarathne MC, Dissanayake AJ, Ekanayaka AH, Hashimoto A, Hongsanan S, Jones EBG, Larsson E, Li WJ, Li QR, Liu JK, Luo ZL, Maharachchikumbura SSN, Mapook A, McKenzie EHC, Norphanphoun C, Konta S, Pang KL, Perera RH, Phookamsak R, Phukhamsakda C, Pinruan U, Randrianjohany E, Singtripop C, Tanaka K, Tian CM, Tibpromma S, Abdel-Wahab MA, Wanasinghe DN, Wijayawardene NN, Zhang JF, Zhang H, Abdel-Aziz FA, Wedin M, Westberg M, Ammirati JF, Bulgakov TS, Lima DX, Callaghan TM, Callac P, Chang CH, Coca LF, Dal Forno M, Dollhofer V, Fliegerová K, Greiner K, Griffith GW, Ho HM, Hofstetter V, Jeewon R, Kang JC, Wen TC, Kirk PM, Kytövuori I, Lawrey JD, Xing J, Li H, Liu ZY, Liu XZ, Liimatainen K, Lumbsch HT, Matsumura M, Moncada B, Nuankaew S, Parnmen S, de Azevedo Santiago ALCM, Sommai S, Song Y, de Souza CAF, de Souza-Motta CM, Su HY, Suetrong S, Wang Y, Wei SF, Wen TC, Yuan HS, Zhou LW, Réblová M, Fournier J, Camporesi E, Luangsa-ard JJ, Tasanathai K, Khonsanit A, Thanakitpipattana D, Somrithipol S, Diederich P, Millanes AM, Common RS, Stadler M, Yan JY, Li XH, Lee HW, Nguyen TTT, Lee HB, Battistin E, Marsico O, Vizzini A, Vila J, Ercole E, Eberhardt U, Simonini G, Wen HA, Chen XH (2015a) Fungal diversity notes 111–252 taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 75:1–248
Ariyawansa HA, Phukhamsakda C, Thambugala KM, Bulgakov TS, Wanasinghe DN, Perera RH, Mapook A, Camporesi E, Kang JC, Jones EG, Bahkali AH (2015b) Revision and phylogeny of Leptosphaeriaceae. Fungal Divers 74(1):19–51
Ariyawansa HA, Phillips AJL, Chuang WY, Tsai I (2018) Tzeananiaceae, a new pleosporalean family associated with Ophiocordyceps macroacicularis fruiting bodies in Taiwan. MycoKeys 37:1–17
Årsvoll K (1965) Acrothecium carotae n. sp., a New Pathogen on Daucus carota L. Acta Agric Scand B 15:101–114
Arzanlou M, Groenewald JZ, Gams W, Braun U, Shin HD, Crous PW (2007) Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Stud Mycol 58:57–93
Aveskamp MM, de Gruyter J, Woudenberg JHC, Verkley GJM, Crous PW (2010) Highlights of the Didymellaceae: a polyphasic approach to characterise Phoma and related pleosporalean genera. Stud Mycol 65:1–60
Awasthi DD (1982) Pyxine in India. Phytomorphology 30:59–379
Bai JN, Wang XM, Shi YJ, Duan CX (2016) Occurrence and identification of Nothophoma quercina causing brown spot of jujube in China. Can J Plant Pathol 38:527–532
Bakhshi M, Arzanlou M, Groenewald JZ, Quaedvlieg W, Crous PW (2019) Parastagonosporella fallopiae gen. et sp. nov. (Phaeosphaeriaceae) on Fallopia convolvulus from Iran. Mycol Prog 18:203–214
Bandara AR, Rapior S, Mortimer PE, Kakumyan P, Hyde KD, Xu J (2019) A review of the polysaccharide, protein and selected nutrient content of Auricularia, and their potential pharmacological value. Mycosphere 10:579–607
Barr ME (1979) A classification of Loculoascomycetes. Mycologia 71:935–995
Barr ME (1984) Herpotrichia and its segregates. Mycotaxon 20:1–38
Barr ME (1996) Planistromellaceae, a new family in the Dothideales. Mycotaxon 60:433–442
Becerra-Hernández CI, González D, De Luna E, Mena-Portales J (2016) First report of pleoanamorphy in Gyrothrix verticiclada with an Idriella-like synanamorph. Cryptogr Mycol 37:241–253
Berkeley MJ, Broome CE (1850) Notices of British fungi (438–501). Ann Mag Nat Hist 5:455–466
Berkeley MJ, Broome CE (1873) Enumeration of the fungi of Ceylon. Part II. Bot J Linn Soc 14:29–141
Berlese AN (1896) Icones Fungorum. Pyrenomycetes 2:1–216
Bessey CE (1907) A synopsis of plant phyla. Univ Neb Stud 7:275–373
Bezerra JDP, Oliveira RJV, Paiva LM, Silva GA, Groenewald JZ, Crous PW, Souza-Motta CM (2016) Bezerromycetales and Wiesneriomycetales ord. nov. (class Dothideomycetes), with two novel genera to accommodate endophytic fungi from Brazilian cactus. Mycol Prog 16:297–309
Bodman MC (1952) A taxonomic study of the genus Heterochaete. Lloydia 15:193–233
Boehm EWA, Mugambi GK, Miller AN, Huhndorf SM, Marincowitz SL, Spatafora JW, Schoch CL (2009) A molecular phylogenetic reappraisal of the Hysteriaceae, Mytilinidiaceae and Gloniaceae (Pleosporomycetidae, Dothideomycetes) with keys to world species. Stud Mycol 64:49–84
Boonmee S, Zhang Y, Chomnunti P, Chukeatirote E, Tsui CKM, Bahkali AH, Hyde KD (2011) Revision of lignicolous Tubeufiaceae based on morphological reexamination and phylogenetic analysis. Fungal Divers 51:63–102
Boonmee S, Bhat JD, Maharachchikumbura SSN, Hyde KD (2014a) Clavatispora thailandica gen. et sp. nov., a novel taxon of Venturiales (Dothideomycetes) from Thailand. Phytotaxa 176:92–101
Boonmee S, Rossman AY, Liu JK, Li WJ, Dai DQ, Bhat JD, Jones EG, McKenzie EH, Xu JC, Hyde KD (2014b) Tubeufiales, ord. nov., integrating sexual and asexual generic names. Fungal Divers 68:239–298
Boonmee S, Dsouza MJ, Luo ZL, Pinruan U, Tanaka K, Su HY, Bhat DJ, McKenzie EHC, Jones EBG, Taylor JE (2016) Dictyosporiaceae fam. nov. Fungal Divers 80:457–482
Boonyuen N, Chuaseeharonnachai C, Suetrong S, Sri-indrasutdhi V, Sivichai S, Jones EBG, Pang KL (2011) Savoryellales (Hypocreomycetidae, Sordariomycetes): a novel lineage of aquatic ascomycetes inferred from multiple-gene phylogenies of the genera Ascotaiwania, Ascothailandia, and Savoryella. Mycologia 103:1351–1371
Brahmanage RS, Lu YZ, Bhat DJ, Wanasinghe DN, Yan JY, Hyde KD, Boonmee S (2017) Phylogenetic investigations on freshwater fungi in Tubeufiaceae (Tubeufiales) reveals the new genus Dictyospora and new species Chlamydotubeufia aquatica and Helicosporium flavum. Mycosphere 8:917–933
Brandrud TE, Lindström H, Marklund H, Melot J, Muskos S (1992) Cortinarius: Flora Photographica II. Cortinarius HB, Matfors, Sweden, p 40
Braun U (1994) Miscellaneous notes on phytopathogenic hyphomycetes. Mycotaxon 51:37–68
Braun U, Crous PW, Nakashima C (2014) Cercosporoid fungi (Mycosphaerellaceae) 2. Species on monocots (Acoraceae to Xyridaceae, excluding Poaceae). IMA Fungus 5:203–390
Braun U, Crous PW, Nakashima C (2015) Cercosporoid fungi (Mycosphaerellaceae) 4. Species on dicots (Acanthaceae to Amaranthaceae). IMA Fungus 6:373–469
Braun U, Crous PW, Nakashima C (2016) Cercosporoid fungi (Mycosphaerellaceae) 5. Species on dicots (Anacardiaceae to Annonaceae). IMA Fungus 7:161–216
Bussaban B, Lumyong S, Lumyong P, McKenzie EHC, Hyde KD (2001) A synopsis of the genus Berkleasmium with two new species and new records of Canalisporium caribense from Zingiberaceae in Thailand. Fungal Divers 8:73–85
Buyck et al (2017) Fungal Biodiversity Profiles 21-30. Cryptogr Mycol 38:101–146
Cai L, Hyde KD, Tsui CKM (2006) Genera of freshwater fungi. Fungal Divers Res Ser 18:1–261
Cai L, Hyde KD, Taylor PWJ, Weir BS, Waller JM, Abang MM, Zhang JZ, Yang YL, Phoulivong S, Liu ZY, Prihastuti H, Shivas RG, McKenzie EHC, Johnston PR (2009) A polyphasic approach for studying Colletotrichum. Fungal Divers 39:183–204
Câmara MP, Ramaley AW, Castlebury LA, Palm ME (2003) Neophaeosphaeria and Phaeosphaeriopsis, segregates of Paraphaeosphaeria. Mycol Res 107:516–522
Cannon PF, Damm U, Johnston PR, Weir BS (2012) Colletotrichum current status and future directions. Stud Mycol 73:181–213
Carpenter SE (1988) Leotiales, a name to replace Helotiales (Ascomycotina). Mycologia 80:127–130
Carris LM (1989) Vaccinium fungi: Helicoma vaccinii sp.nov. Mycotaxon. 36(1):29–34
Castañeda RRF (1985) Deuteromycotina de Cuba. Hyphomycetes III. Instituto de Investigaciones Fundamentales en Agricultura Tropical Alejandro de Humboldt Habana. 42 pp
Castlebury LA, Rossman AY, Jaklitsch WJ, Vasilyeva LN (2002) A preliminary overview of the Diaporthales based on large subunit nuclear ribosomal DNA sequences. Mycologia 94:1017–1031
Cesati V, De Notaris G (1863) Schema di classificazione degle sferiacei italici aschigeri piu’ o meno appartenenti al genere Sphaeria nell’antico significato attribuitoglide Persono. Comment Soc Crittogamol Ital 1:177–420
Chadefaud M (1960) Les végétaux non vasculaires (Cryptogamie). In: Chadefaud M, Emberger L (eds) Traité de botanique systématique. Tome I. Masson et Cie, Paris
Chaiwan N, Lu YZ, Tibpromma S, Bhat DJ, Hyde KD, Boonmee S (2017) Neotubeufia gen. nov. and Tubeufia guangxiensis sp. nov. (Tubeufiaceae) from freshwater habitats. Mycosphere 8:1443–1456
Checa J, Barrasa JM, Moreno G, Fort F, Guarro J (1988) The genus Coniochaeta Sacc. Cooke Coniochaetaceae Ascomycotina in Spain. Cryptogr Mycol 9:1–34
Chen SF, Wingfield MJ, Roux J (2013a) Diversimorbus metrosiderotis gen. et sp. nov. and three new species of Holocryphia (Cryphonectriaceae) associated with cankers on native Metrosideros angustifolia trees in South Africa. Fungal Biol 117:289–310
Chen Y, Wang WX, Zhang AF, Yang X, Xu YL (2013b) First Report of Pestalotiopsis theae on Loquat (Eriobotrya japonica) in Anhui Province of China. The American Phytopathological Society 97:558–568
Chen Q, Jiang JR, Zhang GZ, Cai L, Crous PW (2015) Resolving the Phoma enigma. Stud Mycol 82:137–217
Chen SF, Wingfield MJ, Li GQ, Liu FF (2016) Corticimorbus sinomyrti gen. et sp. nov. (Cryphonectriaceae) pathogenic to native Rhodomyrtus tomentosa (Myrtaceae) in South China. Plant Pathol 65:1254–1266
Chen Q, Hou LW, Duan WJ, Crous PW, Cai L (2017) Didymellaceae revisited. Stud Mycol 87:105–159
Chesters CG, Bell AE (1970) Pseudothecium development in the Lophiostomataceae. Trans Br Mycol Soc 54:27–34
Chethana KWT, Li XH, Wei ZH, Hyde KD, Yan J (2016) Trail of decryption of molecular research on Botryosphaeriaceae in woody plants. Phytopathol Mediterr 55:147–171
Chethana KWT, Jayawardene RS, Zhang W, Zhou YY, Liu M, Hyde KD, Li XH, Wang J, Zhang KC, Yan JY (2019) Molecular characterization and pathogenicity of fungal taxa associated with cherry leaf spot disease. Mycosphere 10:490–530
Chevallier FF (1826) Flore Générale des Environs de Paris. 1:1–674
Chilvers MI, Rogers JD, Dugan FM, Stewart JE, Chen W, Peever TL (2009) Didymella pisi sp. nov., the teleomorph of Ascochyta pisi. Mycol Res 113:391–400
Chomnunti P, Schoch CL, Aguirre-Hudson B, Ko-Ko TW, Hongsanan S, Jones EG, Kodsueb R, Phookamsak R, Chukeatirote E, Bahkali AH, Hyde KD (2011) Capnodiaceae. Fungal Divers 51:103–134
Chomnunti P, Hongsanan S, Aguirre-Hudson B, Tian Q, Peršoh D, Dhami MK, Alias AS, Xu J, Liu X, Stadler M, Hyde KD (2014) The sooty moulds. Fungal Divers 66:1–36
Cole GT, Samson RA (1979) Patterns of development in conidial fungi. London, U.K
Cooke MC (1887) Synopsis pyrenomycetum. Grevillea 16:16–19
Cooke WB (1983) Coniothyriaceae. Rev Biol (Lisbon) 12:289
Corda ACJ (1831) Deutschlands Flora, Abt. III. Die Pilze Deutschlands. 3:33–64
Corda ACJ (1836) Mykologische Beobachtungen. Weitenweber’s Beitrage zur gesammtem Natur- und Heilwissenschaften Prague
Corda ACJ (1839) Pracht - Flora. Europaeischer Schimmel-Bildungen
Corda ACJ (1842) Anleitung zum Studium der Mykologie, nebst Kritischer Beschreibung aller Bekannten Gattungen und Einer Kurzen Beschichte der Systematik. Bei Friedrich Ehrlich. pp 12–33
Corlett MA (1981) Taxonomic survey of some species of Didymella and Didymella-like species. Can J Bot 59:2016–2042
Crespo A, Blanco O, Llimona X, Hawksworth DL (2004) Coscinocladium, an overlooked endemic and monotypic mediterranean lichen genus of Physciaceae, reinstated by molecular phylogenetic analysis. Taxon 53:405–414
Crous PW, Groenewald JZ (2013) A phylogenetic re-evaluation of Arthrinium. IMA Fungus 4:133–154
Crous PW, Groenewald JZ (2017) The Genera of Fungi—G 4: Camarosporium and Dothiora. IMA Fungus 8:131–152
Crous PW, Mohammed C, Glen M, Verkley GJM, Groenewald JZ (2007a) Eucalyptus microfungi from culture. 3. Eucasphaeria and Sympoventuria genera nova, and new species of Furcaspora, Harknessia, Heteroconium and Phacidiella. Fungal Divers 25:19–36
Crous PW, Summerell BA, Carnegie AJ, Mohammed C, Himaman W, Groenewald JZ (2007b) Foliicolous Mycosphaerella spp. and their anamorphs on Corymbia and Eucalyptus. Fungal Divers 26:143–185
Crous PW, Schoch CL, Hyde KD, Wood AR, Gueidan C, De Hoog GS, Groenewald JZ (2009) Phylogenetic lineages in the Capnodiales. Stud Mycol 64:17–47
Crous PW, Groenewald JZ, Shivas RG, Edwards J, Seifert KA, Alfenas AC, Alfenas RF, Burgess TI, Carnegie AJ, Hardy GESJ, Hiscock N, Hüberli D, Jung T, Louis-Seize G, Okada G, Pereira OL, Stukely MJC, Wang W, White GP, Young AJ, McTaggart AR, Pascoe IG, Porter IJ, Quaedvlieg W (2011a) Fungal planet description sheets: 69–91. Persoonia 26:108–156
Crous PW, Summerell BA, Shivas RG, Romberg M, Mel’nik VA, Verkley GJM, Groenewald JZ (2011b) Fungal planet description sheets: 92–106. Persoonia 27:130–162
Crous PW, Summerell BA, Swart L, Denman S, Taylor JE, Bezuidenhout CM, Palm ME, Marincowitz S, Groenewald JZ (2011c) Fungal pathogens of Proteaceae. Persoonia 27:20–45
Crous PW, Summerell BA, Shivas RG, Burgess TI, Decock CA, Dreyer LL, Granke LL, Guest DI, Hardy GESJ, Hausbeck MK, Hüberli D, Jung T, Koukol O, Lennox CL, Liew ECY, Lombard L, McTaggart AR, Pryke JS, Roets F, Saude C, Shuttleworth LA, Stukely MJC, Vánky K, Webster BJ, Windstam ST, Groenewald JZ (2012) Fungal Planet description sheets: 107–127. Persoonia 28:138–182
Crous PW, Braun U, Hunter GC, Wingfield MJ, Verkley GJ, Shin HD, Nakashima C, Groenewald JZ (2013) Phylogenetic lineages in Pseudocercospora. Stud Mycol 75:37–114
Crous PW, Carris LM, Giraldo A, Groenewald JZ, Hawksworth DL, Hernández-Restrepo M, Jaklitsch WM, Lebrun MH, Schumacher RK, Stielow JB (2015a) The genera of fungi-fixing the application of the type species of generic names-G 2: Allantophomopsis, Latorua, Macrodiplodiopsis, Macrohilum, Milospium, Protostegia, Pyricularia, Robillarda, Rotula, Septoriella, Torula, and Wojnowicia. IMA Fungus 6:163–198
Crous PW, Schumacher RK, Wingfield MJ, Lombard L, Giraldo A, Christensen M, Gardiennet A, Nakashima C, Pereira O, Smith AJ, Groenewald JZ (2015b) FUSE 1. Sydowia 67:81–118
Crous PW, Wingfield MJ, Guarro J, Hernández-Restrepo M, Sutton DA, Acharya K, Barber PA, Boekhout T, Dimitrov RA, Dueñas M, Dutta AK, Gené J, Gouliamova DE, Groenewald M, Lombard L, Morozova OV, Sarkar J, Smith MTH, Stchigel AM, Wiederhold NP, Alexandrova AV, Antelmi I, Armengol J, Barnes I, Cano-Lira JF, Ruiz RF, Castañeda Contu M, Courtecuisse PR, da Silveira AL, Decock CA, de Goes A, Edathodu J, Ercole E, Firmino AC, Fourie A, Fournier J, Furtado EL, Geering ADW, Gershenzon J, Giraldo A, Gramaje D, Hammerbacher A, He XL, Haryadi D, Khemmuk W, Kovalenko AE, Krawczynski R, Laich F, Lechat C, Lopes UP, Madrid H, Malysheva EF, Marín-Felix Y, Martín MP, Mostert L, Nigro F, Pereira OL, Picillo B, Pinho DB, Popov ES, Peláez CA, Rodas Rooney-Latham S, Sandoval-Denis M, Shivas RG, Silva V, Stoilova-Disheva MM, Telleria MT, Ullah C, Unsicker SB, van der Merwe NA, Vizzini A, Wagner HG, Wong PTW, Wood AR, Groenewald JZ (2015c) Fungal Planet description sheets: 320–370. Persoonia. 34:167–266
Crous PW, Wingfield MJ, Burgess TI, Hardy GESJ, Crane C, Barrett S, Cano-Lira JF, Le Roux JJ, Thangavel R, Guarro J, Stchigel AM, Martín MP, Alfredo DS, Barber PA, Barreto RW, Baseia IG, Cano-Canals J, Cheewangkoon R, Ferreira RJ, Gené J, Lechat C, Moreno G, Roets F, Shivas RG, Sousa JO, Tan YP, Wiederhold NP, Abell SE, Accioly T, Albizu JL, Alves JL, Antoniolli ZI, Aplin N, Araújo J, Arzanlou M, Bezerra JDP, Bouchara J-P, Carlavilla JR, Castillo A, Castroagudín VL, Ceresini PC, Claridge GF, Coelho G, Coimbra VRM, Costa LA, da Cunha KC, da Silva SS, Daniel R, de Beer ZW, Dueñas M, Edwards J, Enwistle P, Fiuza PO, Fournier J, García D, Gibertoni TB, Giraud S, Guevara-Suárez M, Gusmão LFP, Haituk S, Heykoop M, Hirooka Y, Hofmann TA, Houbraken J, Hughes DP, Kautmanová I, Koppel O, Koukol O, Larsson E, Latha KPD, Lee DH, Lisboa DO, Lisboa WS, López-Villalba Á, Maciel JLN, Manimohan P, Manjón JL, Marincowitz S, Marney TS, Meijer M, Miller AN, Olariaga I, Paiva LM, Piepenbring M, Poveda-Molero JC, Raj KNA, Raja HA, Rougeron A, Salcedo I, Samadi R, Santos TAB, Scarlett K, Seifert KA, Shuttleworth LA, Silva GA, Silva M, Siqueira JPZ, Souza-Motta CM, Stephenson SL, Sutton DA, Tamakeaw N, Telleria MT, Valenzuela-Lopez N, Viljoen A, Visagie CM, Vizzini A, Wartchow F, Wingfield BD, Yurchenko E, Zamora JC, Groenewald JZ (2016a) Fungal planet description sheets: 469–557. Persoonia 37:218–403
Crous PW, Wingfield MJ, Richardson DM, Le Roux JJ, Strasberg D, Edwards J, Roets F, Hubka V, Taylor PWJ, Heykoop M, Martín MP, Moreno G, Sutton DA, Wiederhold NP, Barnes CW, Carlavilla JR, Gené J, Giraldo A, Guarnaccia V, Guarro J, Hernandez-Restrepo M, Kolařík M, Manjón JL, Pascoe IG, Popov ES, Sandoval-Denis M, Woudenberg JHC, Acharya K, Alexandrova AV, Alvarado P, Barbosa RN, Baseia IG, Blanchette RA, Boekhout T, Burgess TI, Cano-Lira JF, Čmoková A, Dimitrov RA, Dyakov M, Yu Dueñas M, Dutta AK, Esteve-Raventós F, Fedosova AG, Fournier J, Gamboa P, Gouliamova DE, Grebenc T, Groenewald M, Hanse B, Hardy GES, Held BW, Jurjević Ž, Kaewgrajang T, Latha KPD, Lombard L, Luangsa-ard JJ, Lysková P, Mallátová N, Manimohan P, Miller AN, Mirabolfathy M, Morozova OV, Obodai M, Oliveira NT, Ordóñez ME, Otto EC, Paloi S, Peterson SW, Phosri C, Roux J, Salazar WA, Sánchez A, Sarria GA, Shin HD, Silva BDB, Silva GA, Smith MTh, Souza-Motta CM, Stchigel AM, Stoilova-Disheva MM, Sulzbacher MA, Telleria MT, Toapanta C, Traba JM, Valenzuela-Lopez N, Watling R, Groenewald JZ (2016b) Fungal planet description sheets: 400–468. Persoonia 36:316–458
Crous PW, Wingfield MJ, Burgess TI, Carnegie AJ, Hardy GESJ, Smith D, Summerell BA, Cano-Lira JF, Guarro J, Houbraken J, Lombard L, Martin MP, Sandoval-Denis M, Alexandrova AV, Barnes CW, Baseia IG, Bezerra JDP, Guarnaccia V, May TW, Hernandez-Restrepo M, Stchigel AM, Miller AN, Ordonez ME, Abreu VP, Accioly T, Agnello C, Agustin Colman A, Albuquerque CC, Alfredo DS, Alvarado P, Araujo-Magalhaes GR, Arauzo S, Atkinson T, Barili A, Barreto RW, Bezerra JL, Cabral TS, Camello Rodriguez F, Cruz RHSF, Daniels PP, da Silva BDB, de Almeida DAC, de Carvalho Junior AA, Decock CA, Delgat L, Denman S, Dimitrov RA, Edwards J, Fedosova AG, Ferreira RJ, Firmino AL, Flores JA, Garcia D, Gene J, Giraldo A, Gois JS, Gomes AAM, Goncalves CM, Gouliamova DE, Groenewald M, Gueorguiev BV, Guevara-Suarez M, Gusmao LFP, Hosaka K, Hubka V, Huhndorf SM, Jadan M, Jurjevic Z, Kraak B, Kucera V, Kumar TKA, Kusan I, Lacerda SR, Lamlertthon S, Lisboa WS, Loizides M, Luangsa-ard JJ, Lyskova P, Mac Cormack WP, Macedo DM, Machado AR, Malysheva EF, Marinho P, Matocec N, Meijer M, Mesic A, Mongkolsamrit S, Moreira KA, Morozova OV, Nair KU, Nakamura N, Noisripoom W, Olariaga I, Oliveira RJV, Paiva LM, Pawar P, Pereira OL, Peterson SW, Prieto M, Rodriguez-Andrade E, Rojo De Blas C, Roy M, Santos ES, Sharma R, Silva GA, Souza-Motta CM, Takeuchi-Kaneko Y, Tanaka C, Thakur A, Smith MT, Tkalcec Z, Valenzuela-Lopez N, van der Kleij P, Verbeken A, Viana MG, Wang XW, Groenewald JZ (2017) Fungal planet description sheets: 625–715. Persoonia 39:270–467
Crous PW, Luangsa-Ard JJ, Wingfield MJ, Carnegie AJ, Hernández-Restrepo M, Lombard L, Roux J, Barreto RW, Baseia IG, Cano-Lira JF, Martín MP, Morozova OV, Stchigel AM, Summerell BA, Brandrud TE, Dima B, García D, Giraldo A, Guarro J, Gusmão LFP, Khamsuntorn P, Noordeloos ME, Nuankaew S, Pinruan U, Rodríguez-Andrade E, Souza-Motta CM, Thangavel R, van Iperen AL, Abreu VP, Accioly T, Alves JL, Andrade JP, Bahram M, Baral HO, Barbier E, Barnes CW, Bendiksen E, Bernard E, Bezerra JDP, Bezerra JL, Bizio E, Blair JE, Bulyonkova TM, Cabral TS, Caiafa MV, Cantillo T, Colmán AA, Conceição LB, Cruz S, Cunha AOB, Darveaux BA, da Silva AL, da Silva GA, da Silva GM, da Silva RMF, de Oliveira RJV, Oliveira RL, De Souza JT, Dueñas M, Evans HC, Epifani F, Felipe MTC, Fernández-López J, Ferreira BW, Figueiredo CN, Filippova NV, Flores JA, Gené J, Ghorbani G, Gibertoni TB, Glushakova AM, Healy R, Huhndorf SM, Iturrieta-González I, Javan-Nikkhah M, Juciano RF, Jurjević Ž, Kachalkin AV, Keochanpheng K, Krisai-Greilhuber I, Li YC, Lima AA, Machado AR, Madrid H, Magalhães OMC, Marbach PAS, Melanda GCS, Miller AN, Mongkolsamrit S, Nascimento RP, Oliveira TGL, Ordoñez ME, Orzes R, Palma MA, Pearce CJ, Pereira OL, Perrone G, Peterson SW, Pham THG, Piontelli E, Pordel A, Quijada L, Raja HA, Rosas de Paz E, Ryvarden L, Saitta A, Salcedo SS, Sandoval-Denis M, Santos TAB, Seifert KA, Silva BDB, Smith ME, Soares AM, Sommai S, Sousa JO, Suetrong S, Susca A, Tedersoo L, Telleria MT, Thanakitpipattana D, Valenzuela-Lopez N, Visagie CM, Zapata M, Groenewald JZ (2018a) Fungal Planet description sheets: 785–867. Persoonia 41:238–417
Crous PW, Schumacher RK, Wingfield MJ, Akulov A, Denman S, Roux J, Braun U, Burgess TI, Carnegie AJ, Váczy KZ (2018b) New and interesting fungi 1. FUSE 1:169–215
Crous PW, Carnegie AJ, Wingfield MJ, Sharma R, Mughini G, Noordeloos ME, Santini A, Shouche YS, Bezerra JDP, Dima B, Guarnaccia V, Imrefi I, Jurjević Ž, Knapp DG, Kovács GM, Magistà D, Perrone G, Rämä T, Rebriev YA, Shivas RG, Singh SM, Souza-Motta CM, Thangavel R, Adhapure NN, Alexandrova AV, Alfenas AC, Alfenas RF, Alvarado P, Alves AL, Andrade DA, Andrade JP, Barbosa RN, Barili A, Barnes CW, Baseia IG, Bellanger J-M, Berlanas C, Bessette AE, Bessette AR, Biketova AY, Bomfim FS, Brandrud TE, Bransgrove K, Brito ACQ, Cano-Lira JF, Cantillo T, Cavalcanti AD, Cheewangkoon R, Chikowski RS, Conforto C, Cordeiro TRL, Craine JD, Cruz R, Damm U, de Oliveira RJV, de Souza JT, de Souza HG, Dearnaley JDW, Dimitrov RA, Dovana F, Erhard A, Esteve-Raventós F, Félix CR, Ferisin G, Fernandes RA, Ferreira RJ, Ferro LO, Figueiredo CN, Frank JL, Freire KTLS, García D, Gené J, Gęsiorska A, Gibertoni TB, Gondra RAG, Gouliamova DE, Gramaje D, Guard F, Gusmão LFP, Haitook S, Hirooka Y, Houbraken J, Hubka V, Inamdar A, Iturriaga T, Iturrieta-González I, Jadan M, Jiang N, Justo A, Kachalkin AV, Kapitonov VI, Karadelev M, Karakehian J, Kasuya T, Kautmanová I, Kruse J, Kušan I, Kuznetsova TA, Landell MF, Larsson K-H, Lee HB, Lima DX, Lira CRS, Machado AR, Madrid H, Magalhães OMC, Majerova H, Malysheva EF, Mapperson RR, Marbach PAS, Martín MP, Martín-Sanz A, Matočec N, McTaggart AR, Mello JF, Melo RFR, Mešić A, Michereff SJ, Miller AN, Minoshima A, Molinero-Ruiz L, Morozova OV, Mosoh D, Nabe M, Naik R, Nara K, Nascimento SS, Neves RP, Olariaga I, Oliveira RL, Oliveira TGL, Ono T, Ordoñez ME, Ottoni AM, Paiva LM, Pancorbo F, Pant B, Pawłowska J, Peterson SW, Raudabaugh DB, Rodríguez-Andrade E, Rubio E, Rusevska K, Santiago ALCMA, Santos ACS, Santos C, Sazanova NA, Shah S, Sharma J, Silva BDB, Siquier JL, Sonawane MS, Stchigel AM, Svetasheva T, Tamakeaw N, Telleria MT, Tiago PV, Tian CM, Tkalčec Z, Tomashevskaya MA, Truong HH, Vecherskii MV, Visagie CM, Vizzini A, Yilmaz N, Zmitrovich IV, Zvyagina EA, Boekhout T, Kehlet T, Læssøe T, Groenewald JZ (2019a) Fungal Planet description sheets: 868–950. Persoonia 42:291–473
Crous PW, Schumacher RK, Akulov A, Thangavel R, Hernández-Restrepo M, Carnegie AJ, Cheewangkoon R, Wingfield MJ, Summerell BA, Quaedvlieg W, Coutinho TA, Roux J, Wood AR, Giraldo A, Groenewald JZ (2019b) New and interesting fungi 2. Fungal Syst Evol 3:57–134
Dai YC, Cui BK, Jing Si J, He SH, Hyde KD, Hai-Sheng Yuan HS, Xiao-Yong Liu XY, Zhou LW (2015) Dynamics of the worldwide number of fungi with emphasis on fungal diversity in China. Mycol Prog 14:62
Dai DQ, Bahkali AH, Ariyawansa HA, Li WJ, Bhat DJ, Hyde KD (2016a) Neokalmusia didymospora (Didymosphaeriaceae), a new species from bamboo. Sydowia 68:17–25
Dai DQ, Jiang HB, Tang LZ, Bhat DJ (2016b) Two new species of Arthrinium (Apiosporaceae, Xylariales) associated with bamboo from Yunnan, China. Mycosphere 7:1332–1345
Dai DQ, Phookamsak R, Wijayawardene NN, Li WJ, Bhat DJ, Xu JC, Taylor JE, Hyde KD, Chukeatirote E (2017) Bambusicolous fungi. Fungal Divers 82:1–105
Damm U, Cannon PF, Liu F, Barreto RW, Guatimosim E, Crous PW (2013) The Colletotrichum orbiculare species complex: Important pathogens of field crops and weeds. Fungal Divers 61:29–59
Damm U, Sato T, Alizadeh A, Groenewald JZ, Crous PW (2019) The Colletotrichum dracaenophilum, C. ámagnum and C. áorchidearum species complexes. Stud Mycol 92:1–46
Damyanti R, Patil KB, Borse KN (2015) Submerged aquatic Hyphomycetes Canalisporium from Madhya Pradesh. Int J Life Sci A3(Special Issue):144–146
Daranagama DA, Camporesi E, Tian Q, Liu X, Chamyuang S, Stadler M, Hyde KD (2015) Anthostomella is polyphyletic comprising several genera in Xylariaceae. Fungal Divers 73:203–238
Daranagama DA, Hyde KD, Sir EB, Thambugala KM, Tian Q, Samarakoon MC, McKenzie EHC, Jayasiri SC, Tibpromma S, Bhat JD, Liu XZ, Stadler M (2018) Towards a natural classification and backbone tree for Graphostromataceae, Hypoxylaceae, Lopadostomataceae and Xylariaceae. Fungal Divers 88:1–165
Day MJ, Gibas CF, Fujimura KE, Egger KN, Currah RS (2006) Monodictys arctica, a new hyphomycete from the roots of Saxifraga oppositifolia collected in the Canadian High Arctic. Mycotaxon 98:261–272
Dayarathne MC, Boonmee S, Braun U, Crous PW, Daranagama DA, Dissanayake AJ, Ekanayaka H, Jayawardena R, Jones EBG, Maharachchikumbura SSN, Perera RH, Phillips AJL, Stadler M, Thambugala KM, Wanasinghe DN, Zhao Q, Hyde KD, Jeewon R (2016) Taxonomic utility of old names in current fungal classification and nomenclature: conflicts, confusion & clarifications. Mycosphere 7:1622–1648
Dayarathne MC, Maharachchikumbura SSN, Jones EBG, Wei D, Devadatha B, Yang J, Ekanayake H, De Silva W, Sarma VV, AlSadi AM, Khongphinitbunjong K, Hyde KD, Zhao RL (2019) Phylogenetic revision of Savoryellaceae and evidence for its ranking as a subclass. Front Microbiol 10:840
De Almeida DAC, Gusmão LFP, Miller AN (2016) Taxonomy and molecular phylogeny of Diatrypaceae (Ascomycota, Xylariales) species from the Brazilian semi-arid region, including four new species. Mycol Prog 15:1–27
De Gruyter J, Aveskamp MM, Woudenberg JHC, Verkley GJM, Groenewald JZ, Crous PW (2009) Molecular phylogeny of Phoma and allied anamorph genera: Towards a reclassification of the Phoma complex. Mycol Res 113:508–519
De Gruyter J, Woudenberg JHC, Aveskamp MM, Verkley GJM, Groenewald JZ, Crous PW (2010) Systematic reappraisal of species in Phoma section Paraphoma, Pyrenochaeta and Pleurophoma. Mycologia 102:1066–1081
De Gruyter J, Woudenberg JH, Aveskamp MM, Verkley GJ, Groenewald JZ, Crous PW (2013) Redisposition of phoma-like anamorphs in Pleosporales. Stud Mycol 75:1–36
De Hoog GS, van Oorschot CAN (1985) Taxonomy of the Dactylaria complex. VI. Key to the genera and checklist of epithets. Stud Mycol 26:97–122
De Silva DD, Rapior S, Fons F, Bahkali AH, Hyde KD (2012a) Medicinal mushrooms in supportive cancer therapies: an approach to anti-cancer effects and putative mechanisms of action. Fungal Divers 55:1–35
De Silva DD, Rapior S, Hyde KD, Bahkali AH (2012b) Medicinal mushrooms in prevention and control of diabetes mellitus. Fungal Divers 56:1–29
Deighton FC (1973) Studies on Cercospora and allied genera. IV. Cercosporella Sacc., Pseudocercosporella gen. nov. and Pseudocercosporidium gen. nov. Mycol Pap 133:62
Delgado G, Koukol O, Cáceres O, Piepenbring M (2017) The phylogenetic placement of Ernakulamia cochinensis within Pleosporales (Dothideomycetes, Ascomycota). Cryptogr Mycol 38:435–452
Dennis RWG (1968) British ascomycetes. J. Cramer, Lehre
Dentinger BTM, Gaya E, O’Brien H, Suz LM, Lachlan R, Diaz-Valderrama JR, Koch A, Aime MC (2016) Tales from the crypt: genome mining from fungarium specimens improves resolution of the mushroom tree of life. Biol J Linn Soc 117:11–32
Dickinson CH (1968) Gliomastix Guéguen. Mycol Pap 115:1–24
Dissanayake AJ, Phillips AJL, Li XH, Hyde KD (2016) Botryosphaeriaceae: Current status of genera and species. Mycosphere 7:1001–1073
Dissanayake AJ, Camporesi E, Hyde KD, Yan JY, Li XH (2017a) Saprobic Botryosphaeriaceae, including Dothiorella italica sp. nov., associated with urban and forest trees in Italy. Mycosphere 8:1157–1176
Dissanayake AJ, Phillips AJL, Hyde KD, Yan JY, Li XH (2017b) The current status of species in Diaporthe. Mycosphere 8:1106–1156
Dissing H (1966) The genus Helvella in Europe, with special emphasis on the species found in Norden. Dansk Botanisk Arkiv 25:1–172
Doilom M, Dissanayake AJ, Wanasinghe DN, Boonmee S, Liu JK, Bhat DJ, Taylor JE, Bahkali AH, McKenzie EH, Hyde KD (2017) Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Divers 82:107–182
Doilom M, Hyde KD, Phookamsak R, Dai DQ, Tang LZ, Hongsanan S, Chomnunti P, Boonmee S, Dayarathne MC, Li WJ, Thambugala KM, Perera RH, Daranagama DA, Norphanphoun C, Konta S, Dong W, Ertz D, Phillips AJL, McKenzie EHC, Vinit K, Ariyawansa HA, Jones EBG, Mortimer PE, Xu JC, Promputtha I (2018) Mycosphere notes 225–274: types and other specimens of some genera of Ascomycota. Mycosphere 9:647–754
Drees M, Wickes B, Gupta M, Hadley S (2007) Lecythophora mutabilis prosthetic valve endocarditis in a diabetic patient. Med Mycol 45:463–467
Duarte S, Bärlocher F, Pascoal C, Cássio F (2016) Biogeography of aquatic hyphomycetes Current knowledge and future perspectives. Fungal Ecol 19:169–181
Dudka IA (1966) New and rare species of fungi imperfecti from the basins of the southern part of Kiev Polessye. Ukr Bot J 23:91–95
Dujon B (2010) Yeast evolutionary genomics. Nat Rev Genet 11:512–524
Ehrenberg CG (1818) Sylvae mycologicae Berolinenses: 1–32
Ekanayaka AH, Ariyawansa HA, Hyde KD, Jones EB, Daranagama DA, Phillips AJ, Hongsanan S, Jayasiri SC, Zhao Q (2017) DISCOMYCETES: the apothecial representatives of the phylum Ascomycota. Fungal Divers 87:237–298
Ekanayaka AH, Hyde KD, Jones EBG, Zhao Q (2018) Taxonomy and phylogeny of operculate discomycetes: Pezizomycetes. Fungal Divers 90:161–243
Ekanayaka AH, Hyde KD, Gentekaki E, McKenzie EHC, Zhao Q, Bulgakov TS, Camporesi E (2019) Preliminary classification of Leotiomycetes. Mycosphere 10:310–489
Elix JA (2009) Physciacae. 3. Pyxine. In: McCarthy PM (ed) Flora of Australia Volume 57. Lchens 5. ABRS & CSIRO Publishing, Canberra, pp 517–533
Ellis MB (1963) Dematiaceous hyphomycetes. V. Mycol Pap 93:1–33
Ellis MB (1971) Dematiaceous Hyphomycetes. Commonwealth Mycological Institute, Kew
Ellis MB (1976) More Dematiaceous Hyphomycetes. Commonwealth Mycological Institute, Kew, pp 1–507
Engler HGA, Prantl KAE (1897) Die Natürlichen Pflanzenfamilien nebst ihren Gattungen und wichtigeren Arten. 1:1–513
Eriksson OE (1982) Outline of the Ascomycetes. Mycotaxon 15:203–248
Eriksson OE, Hawksworth DL (1991) Outline of Ascomycetes—1990. Syst Ascom 9:39–257
Eriksson OE, Hawksworth DL (1993) Outline of the ascomycetes. Syst Ascom 12:51–257
Eriksson OE, Winka W (1997) Supraordinal taxa of ascomycota. Myconet 1:1–16
Fan XL, Bezerra JDP, Tian CM, Crous PW (2018) Families and genera of diaporthalean fungi associated with canker and dieback of tree hosts. Persoonia 40:119–134
Fan XL, Bezerra JD, Tian CM, Crous PW (2019) Cytospora (Diaporthales) in China. Persoonia 45:1–45
Farr DF, Rossman AY (2020) Fungal Databases, Systematic Mycology and Microbiology Laboratory, ARS, USDA. http://nt.ars-grin.gov/fungaldatabases/
Feng Y, Zhang SN, Liu ZY (2019) Tremateia murispora sp. nov. (Didymosphaeriaceae, Pleosporales) from Guizhou, China. Phytotaxa 416:79–87
Ferreira MA, de Oliveira ME, Silva GA, Mathioni SM, Mafia RG (2019) Capillaureum caryovora gen. sp. nov. (Cryphonectriaceae) pathogenic to pequi (Caryocar brasiliense) in Brazil. Mycol Prog 18:385–403
Ferrer A, Shearer CA (2005) New records and a new species of Canalisporium from aquatic habitats in Panama. Mycotaxon 93:179–188
Fournier J, Lechat C (2010) Phomatospora luteotingens sp. nov., a new aquatic species of Phomatospora from France and Spain. Mycosphere 1:39–43
Fries EM (1822) Systema Mycologicum 2:1–275
Fries EM (1825) Systema Orbis Vegetabilis 1:1–374
Frøslev TG, Jeppesen TS, Læssøe T (2006) Seven new calochroid and fulvoid species of Cortinarius. Mycol Res 110:1046–1058
Gams W (1971) Cephalosporium-artige Schimmelpilze (Hyphomycetes). G. Fischer, Stuttgart
García D, Stchigel AM, Cano J, Calduch M, Hawksworth DL, Guarro J (2006) Molecular phylogeny of Coniochaetales. Mycol Res 110:1271–1289
Gaya E, Högnabba F, Holguin A, Molnar K, Fernández-Brime S, Stenroos S, Arup U, Søchting U, den Boom PV, Lücking R, Sipman HJM, Lutzoni F (2012) Implementing a cumulative supermatrix approach for a comprehensive phylogenetic study of the Teloschistales (Pezizomycotina, Ascomycota). Mol Phylogenet Evol 63:374–387
Goh TK, Hyde KD (1997) Melanographium palmicolum sp. nov. from Hong Kong, and a key to the genus. Mycol Res 101:1097–1100
Goh TK, Hyde KD (1998a) A new hyphomycete genus, Conioscyphopsis, from wood submerged in a freshwater stream and a review of Conioscypha. Mycol Res 102:308–312
Goh TK, Hyde KD (1998b) Fungi on submerged wood in Lake Barrine, north Queensland, Australia. Mycol Res 102:739–749
Goh TK, Hyde KD (2000) A new species of Canalisporium from Australia. Mycologia 92:589–592
Goh TK, Ho WH, Hyde KD, Whitton SR, Umali TE (1998) New records and species of Canalisporium (Hyphomycetes), with a revision of the genus. Can J Bot 76:142–152
Goh TK, Hyde KD, Ho WH (1999) A revision of the genus Dictyosporium, with descriptions of three new species. Fungal Divers 2:65–100
Goncalves VN, Vaz ABM, Rosa CA, Rosa LH (2012) Diversity and distribution of fungal communities in lakes of Antarctica. FEMS Microbiol Ecol 82:459–471
Gönczöl J, Révay Á (2003) Treehole fungal communities: aquatic, aero-aquatic and dematiaceous hyphomycetes. Fungal Divers 12:19–34
Gönczöl J, Révay Á (2004) Fungal spores in rainwater: stemflow, throughfall and gutter conidial assemblages. Fungal Divers 16:67–86
Goodwin SB (2002) The barley scald pathogen Rhynchosporium secalis is closely related to the discomycetes Tapesia and Pyrenopeziza. Mycol Res 106:645–654
Gramaje D, Mostert L, Armengol J (2011) Characterization of Cadophora luteo-olivacea and C. melinii isolates obtained from grapevines and environmental samples from grapevine nurseries in Spain. Phytopathol Mediterr 50:S112–S126
Gryzenhout M, Myburg H, van der Merwe NA, Wingfield BD, Wingfield MJ (2004) Chrysoporthe, a new genus to accommodate Cryphonectria cubensis. Stud Mycol 50:119–141
Gryzenhout M, Glen HF, Wingfield BD, Wingfield MJ (2005a) Amphilogia gen. nov. for Cryphonectria-like fungi from Elaeocarpus spp. in New Zealand and Sri Lanka. Mycotaxon 54:1009–1021
Gryzenhout M, Myburg H, Wingfield BD, Montenegro F, Wingfield MJ (2005b) Rostraureum tropicale gen. [et] sp. nov. (Diaporthales) associated with dying Terminalia ivorensis in Ecuador. Mycol Res 109:1029–1044
Gryzenhout M, Myburg H, Wingfield BD, Wingfield MJ (2006) Cryphonectriaceae (Diaporthales), a new family including Cryphonectria, Chrysoporthe, Endothia and allied genera. Mycologia 98:239–249
Guarro J, Vieira LA, De Freitas D, Gené J, Zaror L, Hofling-Lima AL, Fischman O, Zorat-Yu C, Figueras MJ (2000) Phaeoisaria clematidis as a cause of keratomycosis. J Clin Microbiol 38:2434–2437
Hansen K, Perry BA, Dranginis AW, Pfister DH (2013) A phylogeny of the highly diverse cup-fungus family Pyronemataceae (Pezizomycetes, Ascomycota) clarifies relationships and evolution of selected life history traits. Mol Phylogenet Evol 67:311–335
Harkness HW (1884) New species of Californian fungi. Bull Calif Acad Sci 1:29–47
Hashimoto A, Hirayama K, Takahashi H, Matsumura M, Okada G, Chen CY, Huang JW, Kakishima M, Ono T, Tanaka K (2018) Resolving the Lophiostoma bipolare complex: generic delimitations within Lophiostomataceae. Stud Mycol 90:161–189
Hatakeyama S, Tanaka K, Harada Y (2005) Bambusicolous fungi in Japan (5): three species of Tetraploa. Mycoscience 46:196–200
Hawksworth DL (1981) The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol Res 105:1422–1432
Hawksworth DL (2011) A new dawn for the naming of fungi: impacts of decisions made in Melbourne in July 2011 on the future publication and regulation of fungal names. IMA Fungus 2:155–162
Hawksworth DL, Eriksson OE (1986) The names of accepted orders of Ascomycetes. Syst Ascomycetum 5:175–184
He MQ, Zhao RL, Hyde KD, Begerow D, Kemler M, Yurkov A, McKenzie EHC, Raspé O, Kakishima M, Sánchez-Ramírez S, Vellinga EC, Halling R, Papp V, Zmitrovich IV, Buyck B, Ertz D, Wijayawardene NN, Cui BK, Schoutteten N, Liu XZ, Li TH, Yao YJ, Zhu XY, Liu AQ, Li GJ, Zhang MZ, Ling ZL, Cao B, Antonín V, Boekhout T, da Silva BDB, Crop ED, Decock C, Dima B, Dutta AK, Fell JW, Geml J, Ghobad-Nejhad M, Giachini AJ, Gibertoni TB, Gorjón SP, Haelewaters D, He SH, Hodkinson BP, Horak E, Hoshino T, Justo A, Lim YW, Menolli N Jr, Mešić A, Moncalvo JM, Mueller GM, Nagy LG, Nilsson RH, Noordeloos M, Nuytinck J, Orihara T, Ratchadawan C, Rajchenberg M, Silva-Filho AGS, Sulzbacher MA, Tkalčec Z, Valenzuela R, Verbeken A, Vizzini A, Wartchow F, Wei TZ, Weiß M, Zhao CL, Kirk PM (2019) Notes, outline and divergence times of Basidiomycota. Fungal Divers 99:105–367
Hernandez-Restrepo M, Schumacher RK, Wingfield MJ, Ahmad I, Cai L, Duong TA, Edwards J, Gené J, Groenewald JZ, Jabeen S, Khalid AN, Lombard L, Madrid H, Marin-Felix Y, Marincowitz S, Miller AN, Rajeshkumar KC, Rashid A, Sarwar S, Stchigel AM, Taylor PWJ, Zhou N, Crous PW (2016) Fungal systematics and evolution: FUSE 2. Sydowia 68:193–230
Hernández-Restrepo M, Gené J, Castañeda-Ruiz RF, Mena-Portales JM, Crous PW, Guarro J (2017) Phylogeny of saprobic microfungi from Southern Europe. Stud Mycol 86:53–97
Hernández-Restrepo M, Bezerra JDP, Tan YP, Wiederhold N, Crous PW, Guarro J, Gené J (2019) Re-evaluation of Mycoleptodiscus species and morphologically similar fungi. Persoonia 42:205–227
Hidayat I, Meeboon J, To-anun C (2007) Anthostomella and Fasciatispora species (Xylariaceae) from palms including F. ujungkulonensis sp. nov. Mycotaxon 102:347–354
Hildebrand AA (1935) Root rot of ginseng in Ontario caused by members of the genus Ramularia. Can J Res 12:82–114
Ho WH, Hyde KD, Hodgkiss IJ (2002) Seasonality and sequential occurrence of fungi on wood submerged in Tai Po Kau Forest Stream, Hong Kong. Fungal Divers 10:21–43
Hongsanan S, Hyde KD, Bahkali AH, Camporesi E, Chomnunti P, Ekanayaka H, Gomes AA, Hofstetter V, Jones EG, Pinho DB, Pereira OL (2015) Fungal biodiversity profiles 11–20. Cryptogr Mycol 36:355–381
Hongsanan S, Sánchez-Ramírez S, Crous PW, Ariyawansa HA, Zhao RL, Hyde KD (2016) The evolution of fungal epiphytes. Mycosphere 7:1690–1712
Hongsanan S, Maharachchikumbura SSN, Hyde KD, Samarakoon MC, Jeewon R, Zhao Q, Al-Sadi AM, Bahkali AH (2017) An updated phylogeny of Sordariomycetes based on phylogenetic and molecular clock evidence. Fungal Divers 84:25–41
Hongsanan S, Hyde KD, Phookamsak R, Wanasinghe DN, Sarma VV, Boonmee S, Lücking R, Pem D, Liu NG, Tennakoon DS, Karunarathna A, Jones EBG, Phillips AJL, Tibpromma S, Jayawardena RS, Wijayawardene NN, Chathumini S, Ekanayaka AH, Manawasinghe IS, Sandamali D, Jeewon R, Ariyawansa HA, Dai DQ, Dissanayake AJ, Lu YZ, Luo ZL, Zeng XY, Pérez-Ortega S, Thambugala KM, Chethana KWT, Phukhamsakda C, Doilom M, Tian Q, Ertz D, Senwanna C, Konta S, Jiang HB, Zhang SN, Zhang JF, Thiyagaraja V, Bezerra JDP, Zhang H, Miranda-Gonzáles R, Aptroot A, Harishchandra D, Aluthmuhandiram JVS, Abeywickrama PD, Bao DF, Mapook A, Devadatha B, Wu HX, Chaiwan N, Wijesinghe NS, Bundhun D, Monkai J, Samarakoon MC, Suetrong S, Xu JC, Bahkali AH, Xie N (2020) Refined families of Dothideomycetes. Fungal Divers (in press)
Hoog GS, de Oorschot CAN van, Hijwegen T (1983) Taxonomy of the Dactylaria complex II: Dissoconium gen. nov. and Cordana Preuss. In Proceedings of the van de Koninklijke Nederlandse Akademie van Wetenschappen Section C, vol 86, pp 197–206
Hsieh HM, Lin CR, Fang MJ, Rogers JD, Fournier J, Lechat C, Ju YM (2010) Phylogenetic status of Xylaria subgenus Pseudoxylaria among taxa of the subfamily Xylarioideae (Xylariaceae) and phylogeny of the taxa involved in the subfamily. Mol Phylogenet Evol 54:957–969
Hu DM, Wang M, Cai L (2016) Phylogenetic assessment and taxonomic revision of Mariannaea. Mycol Prog 16:271–283
Huang F, Chen GQ, Hou X, Fu YS, Cai L, Hyde KD, Li HY (2014) Colletotrichum species associated with cultivated citrus in China. Fungal Divers 61:61–74
Huelsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogeny. Bioinformatics 17:754–755
Hughes SJ (1951) Studies on micro-fungi. Ill. Mastigosporium, Camposporium, and Ceratophorum. Mycol Pap 36:1–43
Hughes SJ (1958) Revisiones hyphomycetum aliquot cum appendice de nominibus rejiciendis. Can J Bot 36:727–836
Huhndorf SM, Fernández FA (1999) Neotropical ascomycetes 8. New species of Lasiosphaeriella. Mycologia 91:544–552
Huhndorf SM, Miller AN (2011) A molecular re-appraisal of taxa in the Sordariomycetidae and a new species of Rimaconus from New Zealand. Stud Mycol 68:203–210
Huhndorf SM, Miller AN, Fernández FA (2004) Molecular systematics of the Sordariales: the order and the family Lasiosphaeriaceae redefined. Mycologia 96:368–387
Hyde KD (1991) A new amphisphaeriaceous fungus from interdial fronds of Nypa fruticans. Trans Mycol Soc Jpn 32:265–271
Hyde KD (1992) Intertidal mangrove fungi from the west coast of Mexico, including one new genus and two new species. Mycol Res 96:25–30
Hyde KD (1993) Fungi from palms. V. Phomatospora nypae sp. nov. and notes on marine fungi from Nypa fruticans in Malaysia. Sydowia 45:199–203
Hyde KD (1995) Fungi from palms. XVII. The genus Fasciatispora and notes on Amphisphaerella. Nova Hedwigia 61:249–268
Hyde KD, Alcorn JL (1993) Some disease-associated microorganisms on plants of Cape York Peninsula and Torres Strait islands. Australas Plant Path 22:73–83
Hyde KD, Jones EBG (1986) Marine fungi from Seychelles. II. Lanspora coronata gen. et sp. nov. from driftwood. Can J Bot 64:1581–1585
Hyde KD, Philemon E (1994) Some disease-associated microorgansims on plants in the Western Province of Papua New Guinea. Australas Plant Path 23:69–76
Hyde KD, Wong SW (1999) An ultrastructural study of the asci and banded ascospores of Fasciatispora petrakii. Fungal Divers 2:129–134
Hyde KD, Zhang Y (2008) Epitypification: should we epitypify? Journal of Zhejiang University Science B 9:842–846
Hyde KD, Fröhlich J, Taylor JE (1998) Fungi from palms. XXXVI. Reflections on unitunicate ascomycetes with apiospores. Sydowia 50:21–80
Hyde K, Cai L, McKenzie EHC, Yang YL, Zhang JZ, Prihastuti H (2009) Colletotrichum: a catalogue of confusion. Fungal Divers 39:1–17
Hyde KD, Abd-Elsalam K, Cai L (2011) Morphology: still essential in a molecular world. Mycotaxon 114:439–451
Hyde KD, Jones EBG, Liu JK, Ariyawansa HA, Boehm E, Boonmee S, Braun U, Chomnunti P, Crous PW, Dai DQ, Diederich P, Dissanayake AJ, Doilom M, Doveri F, Hongsanan S, Jayawardena R, Lawrey JD, Li YM, Liu YX, Lücking R, Monkai J, Muggia L, Nelsen MP, Pang KL, Phookamsak R, Senanayake I, Shearer CA, Seutrong S, Tanaka K, Thambugala KM, Wijayawardene NN, Wikee S, Wu HX, Zhang Y, Aguirre-Hudson B, Alias SA, Aptroot A, Bahkali AH, Bezerra JL, Bhat JD, Camporesi E, Chukeatirote E, Gueidan C, Hawksworth DL, Hirayama K, De Hoog S, Kang JC, Knudsen K, Li WJ, Li X, Liu ZY, Mapook A, McKenzie EHC, Miller AN, Mortimer PE, Phillips AJL, Raja HA, Scheuer C, Schumm F, Taylor JE, Tian Q, Tibpromma S, Wanasinghe DN, Wang Y, Xu JC, Yan J, Yacharoen S, Zhang M (2013) Families of Dothideomycetes. Fungal Divers 63:1–313
Hyde KD, Nilsson RH, Alias SA, Ariyawansa HA, Blair JE, Cai L, de Cock AWAM, Dissanayake AJ, Glockling SL, Goonasekara ID, Gorczak M, Hahn M, Jayawardena RS, van Kan JAL, Laurence MH, André Lévesque C, Li XH, Liu JK, Maharachchikumbura SSN, Manamgoda DS, Martin FN, Mckenzie EHC, McTaggart AR, Mortimer PE, Nair PVR, Pawlowska J, Rintoul TL, Shivas RG, Spies CFJ, Summerell BA, Taylor PWJ, Terhem RB, Udayanga D, Vaghefi N, Walther G, Wilk M, Wrzosek M, Xu JC, Yan JY, Zhou N (2014) One stop shop: backbones tree for important phytopathogenic genera: I. Fungal Divers 67:21–125
Hyde KD, Hongsanan S, Jeewon R, Bhat DJ, McKenzie EHC, Jones EBG, Phookamsak R, Ariyawansa HA, Boonmee S, Zhao Q, Abdel-Aziz FA, Abdel-Wahab MA, Banmai S, Chomnunti P, Cui BK, Daranagama DA, Das K, Dayarathne MC, de Silva NI, Dissanayake AJ, Doilom M, Ekanayaka AH, Gibertoni TB, Góes-Neto A, Huang SK, Jayasiri SC, Jayawardena RS, Konta S, Lee HB, Li WJ, Lin CG, Liu JK, Lu YZ, Luo ZL, Manawasinghe IS, Manimohan P, Mapook A, Niskanen T, Norphanphoun C, Papizadeh M, Perera RH, Phukhamsakda C, Richter C, Santiago ALCMA, Drechsler-Santos ER, Senanayake I, Tanaka K, Tennakoon TMDS, Thambugala KM, Tian Q, Tibpromma S, Thongbai B, Vizzini A, Wanasinghe DN, Wijayawardene NN, Wu HX, Ynag J, Zeng XY, Zhang H, Zhang JF, Bulgakov TS, Camporesi E, Bahkali AH, Amoozegar MA, Araujo-Neta LS, Ammirati JF, Baghela A, Bhatt RP, Bojantchev D, Buyck B, da Silva GA, de Lima CLF, de Oliveira RJV, de Souza CAF, Dai YC, Dima B, Duong TT, Ercole E, Mafalda-Freire F, Ghosh A, Hashimoto A, Kamolhan S, Kang JC, Karunarathna SC, Kirk PM, Kytövuori I, Lantieri A, Liimatainen K, Liu ZY, Liu XZ, Lücking R, Medardi G, Mortimer PE, Nguyen TTT, Promputtha I, Raj KNA, Reck MA, Lumyong S, Shahzadeh-Fazeli SA, Stadler M, Soudi MR, Su HY, Takahashi T, Tangthirasunun N, Uniyal P, Wang Y, Wen TC, Xu JC, Zhang ZK, Zhao YC, Zhou JL, Zhu L (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 80:1–270
Hyde KD, Norphanphoun C, Abreu VP, Bazzicalupo A, Chethana KWT, Clericuzio M, Dayarathne MC, Dissanayake AJ, Ekanayaka AH, He MQ, Hongsanan S, Huang SK, Jayasiri SC, Jayawardena RS, Karunarathna A, Konta S, Kusˇan I, Lee H, Li J, Lin CG, Liu NG, Lu YZ, Luo ZL, Manawasinghe IS, Mapook A, Perera RH, Phookamsak R, Phukhamsakda C, Siedlecki I, Soares AM, Tennakoon DS, Tian Q, Tibpromma S, Wanasinghe DN, Xiao YP, Yang J, Zeng XY, Abdel-Aziz FA, Li WJ, Senanayake IC, Shang QJ, Daranagama DA, De Silva NI, Thambugala KM, Abdel-Wahab MA, Bahkali AH, Berbee ML, Boonmee S, Bhat DJ, Bulgakov TS, Buyck B, Camporesi E, Castañeda-Ruiz RF, Chomnunti P, Doilom M, Dovana F, Gibertoni TB, Jadan M, Jeewon R, Jones GEB, Kang JC, Karunarathna SC, Lim YW, Liu JK, Liu ZY, Plautz HL Jr, Lumyong S, Maharachchikumbura SSN, Matočec N, Mckenzie EHC, Meśič A, Miller D, Pawłowska J, Pereira OL, Promputtha I, Romero AI, Ryvarden L, Su HY, Suetrong S, Tkalčec Z, Vizzini A, Wen TC, Wisitrassameewong K, Wrzosek M, Xu JC, Zhao Q, Zhao RL, Mortimer PE (2017) Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Divers 87:1–235
Hyde KD, Al-Hatmi AM, Andersen B, Boekhout T, Buzina W, Dawson TL, Eastwood DC, Jones EG, de Hoog S, Kang Y, Longcore JE (2018a) The world’s ten most feared fungi. Fungal Divers 93:161–194
Hyde KD, Chaiwan N, Norphanphoun C, Boonmee S, Camporesi E, Chethana KWT, Dayarathne MC, de Silva NI, Dissanayake AJ, Ekanayaka AH, Hongsanan S, Huang SK, Jayasiri SC, Jayawardena RS, Jiang HB, Karunarathna A, Lin CG, Liu JK, Liu NG, Lu YZ, Luo ZL, Maharachchimbura SSN, Manawasinghe IS, Pem D, Perera RH, Phukhamsakda C, Samarakoon MC, Senwanna C, Shang Q, Tennakoon DS, Thambugala KM, Tibpromma S, Wanasinghe DN, Xiao YP, Yang J, Zeng XY, Zhang JF, Zhang SN, Bulgakov TS, Bhat DJ, Cheewangkoon R, Goh TK, Jones EBG, Kang JC, Jeewon R, Liu ZY, Lumyong S, Kuo CH, McKenzie EHC, Wen TC, Yan JY, Zhao Q (2018b) Mycosphere notes 169–224. Mycosphere 9:271–430
Hyde KD, Norphanphoun C, Chen J, Dissanayake AJ, Doilom M, Hongsanan S, Jayawardena RS, Jeewon R, Perera RH, Thongbai B, Wanasinghe DN, Wisitrassameewong K, Tibpromma S, Stadler M (2018c) Thailand’s amazing diversity: up to 96% of fungi in northern Thailand are novel. Fungal Divers 93:215–239
Hyde KD, Danushka S, Tennakoon DS, Jeewon R, Bhat DJ, Maharachchikumbura SSN, Rossi W, Leonardi M, Lee HB, Mun HY, Houbraken J, Nguyen TTT, Jeon SJ, Frisvad JC, Dhanushka N, Wanasinghe DN, Luücking R, Aptroot A, Cáceres MES, Karunarathna SC, Hongsanan S, Phookamsak R, de Silva NI, Thambugala KM, Jayawardena RS, Senanayake IC, Boonmee S, Chen J, Luo ZL, Phukhamsakda C, Pereira OL, Abreu VP, Rosado AWC, Bart B, Randrianjohany E, Hofstetter V, Gibertoni TB, da Silva Soares AM, Plautz HL Jr, Sotão HMP, Xavier WKS, Bezerra JDP, de Oliveira TGL, de Souza-Motta CM, Magalhães OMC, Bundhun D, Harishchandra D, Manawasinghe IS, Dong W, Zhang SN, Bao DF, Samarakoon MC, Pem D, Karunarathna A, Lin CG, Yang J, Perera RH, Kumar V, Huang SK, Dayarathne MC, Ekanayaka AH, Jayasiri SC, Xiao YP, Konta S, Niskanen T, Liimatainen K, Dai YC, Ji XH, Tian XM, Mešić A, Singh SK, Phutthacharoen K, Cai L, Sorvongxay T, Thiyagaraja V, Norphanphoun C, Chaiwan N, Lu YZ, Jiang HB, Zhang JF, Abeywickrama PD, Aluthmuhandiram JVS, Brahmanage RS, Zeng M, Chethana T, Wei DP, Réblová M, Fournier J, Nekvindová J, do Nascimento Barbosa R, dos Santos JEF, de Oliveira NT, Li GJ, Ertz D, Shang QJ, Phillips AJL, Kuo CH, Camporesi E, Bulgakov TS, Lumyong S, Jones EBG, Chomnunti P, Gentekaki E, Bungartz F, Zeng XY, Fryar S, Tkalčec Z, Liang J, Li GS, Wen TC, Singh PN, Gafforov Y, Promputtha I, Yasanthika E, Goonasekara ID, Zhao RL, Zhao Q, Kirk PM, Liu JK, Yan JY, Mortimer PE, Xu JC (2019a) Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers 96:1–242
Hyde KD, Xu JC, Rapior S, Jeewon R, Lumyong S, Niego AGT, Abeywickrama PD, Aluthmuhandiram JVS, Brahamanage RS, Brooks S, Chaiyasen A, Chethana KWT, Chomnunti P, Chepkirui C, Chuankid B, de Silva NI, Doilom M, Faulds C, Gentekaki E, Gopalan V, Kakumyan P, Harishchandra D, Hemachandran H, Hongsanan S, Karunarathna A, Karunarathna SC, Khan S, Kumla J, Jayawardena RS, Liu JK, Liu N, Luangharn T, Macabeo APG, Marasinghe DS, Meeks D, Mortimer PE, Mueller P, Nadir S, Nataraja KN, Nontachaiyapoom S, O’Brien M, Penkhrue W, Phukhamsakda C, Ramanan US, Rathnayaka AR, Sadaba RB, Sandargo B, Samarakoon BC, Tennakoon DS, Siva R, Sriprom W, Suryanarayanan TS, Sujarit K, Suwannarach N, Suwunwong T, Thongbai B, Thongklang N, Wei DP, Wijesinghe SN, Winiski J, Yan J, Yasanthika E, Stadler M (2019b) The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers 97:1–136
Hyde KD, de Silva NI, Jeewon R, Liu NG, Chaiwan N, Tennakoon DS, Boonmee S, Maharachchimbura SSN, Samarakoon MC, Norphanphoun C, Jayasiri SC, Jayawardena RS, Lin CG, Phookamsak R, Jiang HB, Karunarathna A, Manawasinghe IS, Pem D, Zeng XY, Li J, Luo ZL, Doilom M, Abeywickrama PD, Wijesinghe NS, Bandarupalli D, Brahamanage RS, Yang EF, Wanasinghe DN, Senanayake IC, Goonasekara ID, Wei DP, Aluthmuhandiram JVS, Dayarathne MC, Marasinghe DS, Li WJ, Huanraluek N, Sysouphanthong P, Dissanayake LS, Dong W, Lumyong S, Karunarathna SC, Al-Sadi AM, Harishchandra D, Sarma VV (2020a) AJOM new records and collections of fungi: 1–100. Asian Journal of Mycology 3
Hyde KD, Norphanphoun C, Maharachchikumbura SSN, Bhat DJ, Jones EBG, Bundhun D, Chen YJ, Bao DF, Boonmee S, Calabon MS, Chaiwan N, Chethana KWT, Dai DQ, Dayarathne MC, Devadatha B, Dissanayake AJ, Dissanayake LS, Doilom M, Dong W, Fan XL, Goonasekara ID, Hongsanan S, Huang SK, Jayawardena RS, Jeewon R, Karunarathna A, Konta S, Kumar V, Lin CG, Liu JK, Liu N, Luangsa-ard J, Lumyong S, Luo ZL, Marasinghe DS, McKenzie EHC, Niego AGT, Niranjan M, Perera RH, Phukhamsakda C, Rathnayaka AR, Samarakoon MC, Samarakoon SMBC, Sarma VV, Senanayake IC, Shang QJ, Stadler M, Tibpromma S, Wei DP, Wijayawardene NN, Xiao YP, Xiang MM, Yang J, Zeng XY, Zhang SN (2020b) Refined families of Sordariomycetes. Mycosphere 11
Ichinoe M (1971) Camposporium species from Japan. Trans Mycol Soc Jpn 12:79–88
Inderbitzin P, Gareth Jones EB, Vrijmoed LLP (2000) A new species of Leptosphaerulina from decaying mangrove wood from Hong Kong. Mycoscience 41:233–237
Index Fungorum (2020). http://www.indexfungorum.org/Names/Names.asp
Iturrieta-González I, Gené J, Guarro J, Castañeda-Ruiz RF, García D (2018) Neodendryphiella, a novel genus of the Dictyosporiaceae (Pleosporales). MycoKeys 37:19–38
Jaklitsch WM, Voglmayr H (2017) Three former taxa of Cucurbitaria and considerations on Petrakia in the Melanommataceae. Sydowia 69:81–95
Jaklitsch W, Baral HO, Lucking R, Lumbsch HT (2016) Ascomycota. In: Frey W (ed) Syllabus of plant families-Adolf Engler’s Syllabus der Pflanzenfamilien. Borntraeger, Stuttgart
Jayasiri SC, Hyde KD, Jones EBG, Jeewon R, Ariyawansa HA, Bhat JD, Camporesi E, Kang JC (2017) Taxonomy and multigene phylogenetic evaluation of novel species in Boeremia and Epicoccum with new records of Ascochyta and Didymella (Didymellaceae). Mycosphere 8:1080–1101
Jayasiri SC, Hyde KD, Jones EBG, Peršoh D, Camporesi E, Kang JC (2018) Taxonomic novelties of hysteriform dothideomycetes. Mycosphere 9:803–837
Jayasiri SC, Hyde KD, Jones EBG, McKenzie EHC, Jeewon R, Phillips AJL, Bhat DJ, Wanasinghe DN, Liu JK, Lu YZ, Kang JC, Xu J, Karunarathna SC (2019) Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 10:1–186
Jayawardena RS, Hyde KD, Damm U, Cai L, Liu M, Li XH, Zhang W, Zhao WS, Yan JY (2016) Notes on currently accepted species of Colletotrichum. Mycosphere 7:1192–1260
Jayawardena RS, Hyde KD, Jeewon R, Ghobad-Nejhad M, Wanas-inghe DN, Liu NG, Phillips AJL, Oliveira-Filho JRC, da Silva GA, Gibertoni TB, Abeywikrama P, Carris LM, Chethana KWT, Dissanayake AJ, Hongsanan S, Jayasiri SC, McTaggart AR, Perera RH, Phutthacharoen K, Savchenko KG, Shivas RG, Thongklang N, Dong W, Wei DP, Wijayawardena NN, Kang JC (2019a) One stop shop II: taxonomic update with molecular phylogeny for important phytopathogenic genera: 26–50. Fungal Divers 94:41–129
Jayawardena RS, Hyde KD, McKenzie EHC, Jeewon R, Phillips AJL, Perera RH, de Silva NI, Maharachchikumburua SSN, Samarakoon MC, Ekanayake AH, Tennakoon DS, Dissanayake AJ, Norphanphoun C, Lin C, Manawasinghe IS, Tian Q, Brahmanage R, Chomnunti P, Hongsanan S, Jayasiri SC, Halleen F, Bhunjun CS, Karunarathna A, Wang Y (2019b) One stop shop III: taxonomic update with molecular phylogeny for important phytopathogenic genera: 51–75 (2019b). Fungal Divers 97:1–84
Jayawardena RS, McKenzie EH, Chen YJ, Phillips AJL, Hongsanan S, Norphanphoun C, Abeywikrama P, Maharachchikumbura SSN, Manawasinghe IS, Hyde KD (2019c) https://onestopshopfungi.org/, a database to enhance identification of phytopathogenic genera. Asian Journal of Mycology 2: 281–286
Jeewon R, Hyde KD (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7:1669–1677
Jiang HB, Hyde KD, Doilom M, Karunarathna SC, Xu JC, Phookamsak R (2019a) Arthrinium setostromum (Apiosporaceae, Xylariales), a novel species associated with dead bamboo from Yunnan, China. AJOM 2:254–268
Jiang N, Fan XL, Tian CM (2019b) Identification and pathogenicity of Cryphonectriaceae species associated with chestnut canker in China. Plant Pathol 68:1132–1145
Jiao WL, Zhou RJ, Fu JF, Xu HJ, Hao N (2017) First report of Nothophoma quercina causing leaf spot disease of Phellodendron amurense in China. Plant Dis 101:1820
Jie CY, Zhou QX, Zhao WS, Jiang YL, Hyde KD, McKenzie EHC, Wang Y (2013) A new Myrmecridium species from Guizhou, China. Mycotaxon 124:1–8
Johnston PR, Quijada L, Smith CA, Baral HO, Hosoya T, Baschien C, Pärtel K, Zhuang WY, Haelewaters D, Park D, Carl S, López-Giráldez F, Wang Z, Townsend JP (2019) A multigene phylogeny toward a new phylogenetic classification of Leotiomycetes. IMA Fungus 10:1
Jones EBG, Sakayaroj J, Suetrong S, Somrithipol S, Pang KL (2009) Classification of marine Ascomycota, anamorphic taxa and Basidiomycota. Fungal Divers 35:1–187
Jones EBG, To-anun C, Suetrong S, Boonyuen N (2016) Mycosphere Essays 12. Progress in the classification of the water–cooling tower ascomycete Savoryella and a tribute to John Savory: a review. Mycosphere 7:570–581
Jones EBG, Devadatha B, Abdel-Wahab M, Dayarathne M, Zhang SN, Hyde K, Liu JK, Bahkali A, Sarma V, Tibell S, Tibell L, Wang MM, Liu F, Cai L (2019a) Phylogeny of new marine Dothideomycetes and Sordariomycetes from mangroves and deep-sea sediments. Bot Mar. https://doi.org/10.1515/bot-2019-0014
Jones EBG, Pang KL, Abdel-Wahab MA, Scholz B, Hyde KD, Boekhout T, Ebel R, Rateb ME, Henderson L, Sakayaroj J, Suetrong S, Dayarathne MC, Kumar V, Raghukumar S, Sridhar KR, Bahkali AH, Gleason FH, Norphanphoun C (2019b) An online resource for marine fungi. Fungal Divers 96:347–433
Jumbam B, Haelewaters D, Koch RA, Dentinger BTM, Henkel TW, Aime MC (2019) A new and unusual species of Hericium (Basidiomycota: Russulales, Hericiaceae) from the Dja Biosphere Reserve, Cameroon. Mycol Prog 18:1253–1262
Kalb K (1987) Brasilianische Flechten. 1. Die Gattung Pyxine. J. Cramer, Berlin-Stuttgart, pp 5–82
Kalb K (2004) New or otherwise interesting lichens II. Bibl Lichenol 88:301–329
Kendrick WB (1961) The Leptographium complex. Phialocephala gen. nov. Can J Bot 39:1079–1085
Khan Z, Gené J, Ahmad S, Cano J, Al-Sweih N, Joseph L, Guarro J (2013) Coniochaeta polymorpha, a new species from endotracheal aspirate of a preterm neonate, and transfer of Lecythophora species to Coniochaeta. Antonie Van Leeuwenhoek 104:243–252
Khunnamwong P, Lertwattanasakul N, Jindamorakot S, Limtong S, Lachance M-A (2015) Description of Diutina gen. nov., Diutina siamensis, f.a. sp. nov., and reassignment of Candida catenulata, Candida mesorugosa, Candida neorugosa, Candida pseudorugosa, Candida ranongensis, Candida rugosa and Candida scorzettiae to the genus Diutina. Int J Syst Evol Microbiol 65:4701–4709
King KM, West JS, Brunner PC, Dyer PS, Fitt BDL (2013) Evolutionary relationships between Rhynchosporium lolii sp nov and other Rhynchosporium species on grasses. PLoS ONE 8:e72536
Kirk PM, Cannon PF, Minter DW, Stalpers JA (2008) Dictionary of the Fungi, 10th edn. Wallingford, CAB International, p 547
Kirk PM, Stalpers JA, Braun U, Crous PW et al (2013) A without-prejudice list of generic names of fungi for protection under the International Code of Nomenclature for algae, fungi, and plants. IMA Fungus 4:381–443
Kirschstein W (1924) Beiträge zur Kenntnis der Ascomyceten. Verhandlungen des Botanischen Vereins der Provinz Brandenburg 66:23–29
Kohlmeyer J, Volkmann-Kohlmeyer B (1993) Atrotorquata and Loratospora: new ascomycete genera on Juncus roemerianus. Syst Ascom 12:7–22
Kohlmeyer J, Volkmann-Kohlmeyer B, Eriksson OE (1995) Fungi on Juncus roemerianus. 2. New dictyosporous ascomycetes. Bot Mar 38:165–174
Kohlmeyer J, Volkmann-Kohlmeyer B, Eriksson OE (1996) Fungi on Juncus roemerianus. 8. New bitunicate ascomycetes. Can J Bot 74:1830–1840
Konta S, Hongsanan S, Tibpromma S, Thongbai B, Maharachchikumbura SSN, Bahkali AH, Hyde KD, Boonmee S (2016) An advance in the endophyte story: Oxydothidaceae fam. nov. with six new species of Oxydothis. Mycosphere 7:1425–1446
Konta S, Hongsanan S, Eungwanichayapant PD, Liu JK, Jeewon R, Hyde KD, Maharachchikumbura SSN, Boonmee S (2017) Leptosporella (Leptosporellaceae fam. nov.) and Linocarpon and Neolinocarpon (Linocarpaceae fam. nov.) are accommodated in Chaetosphaeriales. Mycosphere 8:1943–1974
Konta S, Maharachchikumbura SSN, Senanayake IC, McKenzie EHC, Stadler M, Boonmee S, Phookamsak R, Jayawardena RS, Senwanna C, Hyde KD, Elgorban AM, Eungwanichayapant PD (2020) A new genus Allodiatrype, five new species and a new host record of diatrypaceous fungi from palms (Arecaceae). Mycosphere 11:239–268
Korf RP (1973) Discomycetes and Tuberales. In: Ainsworth GC, Sparrow FK, Sussman AS (eds) The fungi: an advanced treatise, vol IVA. Academic Press, New York
Korf RP (1990) Discomycete systematics today: a look at some unanswered questions in a group of unitunicate ascomycetes. Mycosystema 3:19–27
Korf RP, Lizon P (2000) Validation of Nannfeldt’s ordinal name Helotiales. Mycotaxon 75:501–502
Korf RP, Lizon P (2001) The status of the ordinal name Leotiales. Czech Mycol 52:255–257
Kornerup A, Wanscher JH (1978) Methuen handbook of colour, 3rd edn. Eyre Methuen Ltd., London
Kraichak E, Huang JP, Nelsen M, Leavitt SD, Lumbsch HT (2018) A revised classification of orders and families in the two major subclasses of Lecanoromycetes (Ascomycota) based on a temporal approach. J Linn Soc Bot 188:233–249
Kuhnert E, Sir EB, Lambert C, Hyde KD, Hladki AI, Romero AI, Rohde M, Stadler M (2017) Phylogenetic and chemotaxonomic resolution of the genus Annulohypoxylon (Xylariaceae) including four new species. Fungal Divers 85:1–43
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Kumar V, Cheewangkoon R, Thambugala KM, Jones EBG, Brahmanage RS, Doilom M, Jeewon R, Hyde KD (2019) Rhytidhysteron mangrovei (Hysteriaceae), a new species from mangroves in Phetchaburi Province, Thailand. Phytotaxa 401:166–178
Kurtzman C, Robnett CJ (2013) Relationships among genera of the Saccharomycotina (Ascomycota) from multigene phylogenetic analysis of type species. FEMS Yeast Res 13:22–33
Lantz H, Johnston PR, Park D, Minter DW (2011) Molecular phylogeny reveals a core clade of Rhytismatales. Mycologia 103:57–74
Lawrence DP, Holland LA, Nouri MT, Travadon R, Abramians A, Michailides TJ, Trouillas FP (2018) Molecular phylogeny of Cytospora species associated with canker diseases of fruit and nut crops in California, with the descriptions of ten new species and one new combination. IMA Fungus 9:333–369
Leuchtmann A (1987) Species of Heterospaeria (Discomycetes) and their anamorphs. Mycotaxon 28:261–284
Li WJ, McKenzie EHC, Liu JK(Jack), Bhat DJ, Dai DQ, Camporesi E, Tian Q, Maharachchikumbura SSN, Luo ZL, Shang QJ, Zhang FJ, Tangthirasunun N, Karunarathna SC, Xu JC, Hyde KD (2020) Taxonomy and phylogeny of hyaline-spored coelomycetes. Fungal Divers. https://doi.org/10.1007/s13225-020-00440-y
Li WJ, Bhat DJ, Camporesi E, Tian Q, Wijayawardene NN, Dai DQ, Phookamsak R, Chomnunti P, Bahkali AH, Hyde KD (2015) New asexual morph taxa in Phaeosphaeriaceae. Mycosphere 6:681–708
Li GJ, Hyde KD, Zhao RN, Hongsanan S, Abdel-Aziz FA, Abdel-Wahab MA, Alvarado P, Alves-Silva G, Ammirati JF, Ariyawansa HA, Baghela A, Bahkali AH, Beug M, Bhat DJ, Bojantchev D, Boonpratuang T, Bulgakov TS, Camporesi E, Boro MC, Ceska O, Chakraborty D, Chen JJ, Chethana KWT, Chomnunti P, Consiglio G, Cui BK, Dai DQ, Dai YC, Daranagama DA, Das K, Dayarathne MC, Crop ED, De Oliveira RJV, de Souza CAF, de Souza JI, Dentinger BTM, Dissanayake AJ, Doilom M, Drechsler-Santos ER, Ghobad-Nejhad M, Gilmore SP, Góes-Neto A, Gorczak M, Haitjema GH, Hapuarachchi KK, Hashimoto A, He MQ, Henske JK, Hirayama K, Iribarren MJ, Jayasiri SC, Jayawardena RS, Jeon SJ, Jerônimo GH, Jesus AL, Jones EBG, Kang JC, Karunarathna SC, Kirk PM, Konta S, Kuhnert E, Langer E, Lee HS, Lee HB, Li WJ, Li XH, Liimatainen K, Lima DX, Lin CG, Liu JK, Liu XZ, Liu ZY, Luangsa-ard JJ, Lücking R, Lumbsch HT, Lumyong S, Leanõ EM, Marano AV, Matsumura M, McKenzie EHC, Mongkolsamrit S, Mortimer PE, Nguyen TTT, Niskanen T, Norphanphoun C, O’Malley MA, Parnmen S, Pawłowska J, Perera RH, Phookamsak R, Phukhamsakda C, Pires-Zottarelli CLA, Raspé O, Reck MA, Rocha SCO, de Santiago ALCMA, Senanayake IC, Setti L, Shang QJ, Singh SK, Sir EB, Solomon KV, Song J, Srikitikulchai P, Stadler M, Suetrong S, Takahashi H, Takahashi T, Tanaka K, Tang LP, Thambugala KM, Thanakitpipattana D, Theodorou MK, Thongbai B, Thummarukcharoen T, Tian Q, Tibpromma S, Verbeken A, Vizzini A, Vlasák J, Voigt K, Wanasinghe DN, Wang Y, Weerakoon G, Wen HA, Wen TC, Wijayawardene NN, Wongkanoun S, Wrzosek M, Xiao YP, Xu JC, Yan JY, Yang J, Yang SD, Hu Y, Zhang JF, Zhao J, Zhou LW, Persoh D, Phillips AJL, Maharachchikumbura SSN (2016a) Fungal divers notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 78:1–237
Li JF, Phookamsak R, Mapook A, Boonmee S, Bhat JD, Hyde KD, Lumyong S (2016b) Seifertia shangrilaensis sp. nov. (Melanommataceae), a new species from Southwest China. Phytotaxa 273:34–42
Li JF, Jeewon R, Luo ZL, Phookamsak R, Bhat DJ, Mapook A, Phukhamsakda C, Camporesi E, Lumyong S, Hyde KD (2017a) Morphological characterization and DNA based taxonomy of Fusiconidium gen. nov. with two novel taxa within Melanommataceae (Pleosporales). Phytotaxa 308:206–218
Li JF, Phookamsak R, Jeewon R, Bhat DJ, Mapook A, Camporesi E, Shang QJ, Chukeatirote E, Bahkali AH, Hyde KD (2017b) Molecular taxonomy and morphological characterization reveal new species and new host records of Torula species (Torulaceae, Pleosporales). Mycol Prog 16:447–461
Libert MA (1830) Plantae Cryptogamae, quas in Arduenna collegit. Fasc 1:100
Liew ECY, Aptroot A, Hyde KD (2000) Phylogenetic significance of the pseudoparaphyses in Loculoascomycete taxonomy. Mol Phyl Evol 16:392–402
Lin CG, Mckenzie EH, Bhat DJ, Liu JK, Hyde KD, Lumyong S (2018) Pseudodactylaria brevis sp. nov. from Thailand confirms the status of Pseudodactylariaceae. Phytotaxa 369:241–250
Linnaeus C (1753) Species Plantarum, exhibentes plantas rite cognitas, ad genera relatas, cum differentiis specificis, nominibus trivialibus, synonymis selectis, locis natalibus secundum systema sexual digestas. Tomus II. Holmiae: 561–1200
Liu F, Hu DM, Cai L (2012) Conlarium duplumascospora gen. et. sp. nov. and Jobellisia guangdongensis sp. nov. from freshwater habitats in China. Mycologia 104:1178–1186
Liu JK, Hyde KD, Gareth Jones EB, Ariyawansa HA, Bhat DJ, Boonmee S, Maharachchikumbura SSN, McKenzie EHC, Phookamsak R, Phukhamsakda C, Shenoy BD, Abdel-Wahab MA, Buyck B, Chen J, Chethana KWT, Singtripop C, Dai DQ, Dai YC, Daranagama DA, Dissanayake AJ, Doilom M, D’souza MJ, Fan XL, Goonasekara ID, Hirayama K, Hongsanan S, Jayasiri SC, Jayawardena RS, Karunarathna SC, Li WJ, Mapook A, Norphanphoun C, Pang KL, Perera RH, Peršoh D, Pinruan U, Senanayake IC, Somrithipol S, Suetrong S, Tanaka K, Thambugala KM, Tian Q, Tibpromma S, Udayanga D, Wijayawardene NN, Wanasinghe DN, Wisitrassameewong K, Zeng XY, Abdel-Aziz FA, Adamčík S, Bahkali AH, Boonyuen N, Bulgakov T, Callac P, Chomnunti P, Greiner K, Hashimoto A, Hofstetter V, Kang JC, Lewis D, Li XH, Liu XZ, Liu ZY, Matsumura M, Mortimer PE, Rambold G, Randrianjohany E, Sato G, Sri-Indrasutdhi V, Tian CM, Verbeken A, von Brackel W, Wang Y, Wen TC, Xu JC, Yan JY, Zhao RL, Camporesi E (2015) Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Divers 72:1–197
Liu JK, Hyde KD, Jeewon R, Phillips AJ, Maharachchikumbura SS, Ryberg M, Liu ZY, Zhao Q (2017a) Ranking higher taxa using divergence times: a case study in Dothideomycetes. Fungal Divers 84:75–99
Liu NG, Hongsanan S, Yang J, Bhat DJ, Liu J, Jumpathong J, Liu Z (2017b) Periconia thailandica (Periconiaceae), a new species from Thailand. Phytotaxa 323:253–263
Liu NG, Hongsanan S, Yang J, Lin CG, Bhat DJ, Liu JK, Jumpathong J, Boonmee S, Hyde KD, Liu ZY (2017c) Dendryphiella fasciculata sp. nov. and notes on other Dendryphiella species. Mycosphere 8:1575–1586
Liu JK, Lu YZ, Cheewangkoon R, To-Anun C (2018a) Phylogeny and morphology of Helicotubeufia gen. nov., with three new species in Tubeufiaceae from aquatic habitats. Mycosphere 9:495–509
Liu M, Zhang W, Manawasinghe IS, Zhou Y, Xing QK, Li XH, Yan JY, Wang S (2018b) First report of Nothophoma quercina causing trunk canker on Crabapple (Malus micromalus) in China. Plant Dis 102:1462
Liu F, Bonthond G, Groenewald JZ, Cai L, Crous PW (2019) Sporocadaceae, a family of coelomycetous fungi with appendage-bearing conidia. Stud Mycol 92:287–415
Lombard L, van der Merwe NA, Groenewald JZ, Crous PW (2015) Generic concepts in Nectriaceae. Stud Mycol 80:189–245
Lowy B (1971) Tremellales. Flora Neotrop 6:1–153
Lu JY (1995) Diagnosis of plant diseases. In: Lu JY et al (eds) Pestalotiopsis. China Agriculture Press, Beijing, pp 1–194
Lu YZ, Boonmee S, Bhat DJ, Hyde KD, Kang JC (2017a) Helicosporium luteosporum sp. nov. and Acanthohelicospora aurea (Tubeufiaceae, Tubeufiales) from terrestrial habitats. Phytotaxa 319:241–253
Lu YZ, Boonmee S, Liu JK, Hyde KD, McKenzie EHC, Eungwanichayapant PD, Kang JC (2017b) Multi-gene phylogenetic analyses reveals Neohelicosporium gen. nov. and five new species of helicosporous hyphomycetes from aquatic habitats. Mycol Prog 17:631–646
Lu YZ, Liu JK, Hyde KD, Jeewon R, Kang JC, Fan C, Boonmee S, Bhat DJ, Luo ZL, Lin CG (2018) A taxonomic reassessment of Tubeufiales based on multi-locus phylogeny and morphology. Fungal Divers 92:131–344
Lücking R, Hodkinson BP, Leavitt SD (2017) The 2016 classification of lichenized fungi in the Ascomycota and Basidiomycota-Approaching one thousand genera. Bryologist 119:361–417
Lumbsch HT, Huhndorf SM (2007) Outline of ascomycota—2007. Myconet 13:1–58
Lumbsch HT, Huhndorf SM (2010) Myconet Volume 14. Part One. Outline of Ascomycota—2009. Part Two. Notes on Ascomycete Systematics. Nos. 4751–5113. Fieldiana Life Earth Sci 1:1–64
Luo J, Zhuang WY (2007) A new species and two new Chinese records of Bionectria (Bionectriaceae, Hypocreales). Mycotaxon 101:315–323
Luo ZL, Bhat DJ, Jeewon R, Boonmee S, Bao DF, Zhao YC, Chai HM, Su HY, Su XJ, Hyde KD (2017) Molecular phylogeny and morphological characterization of asexual fungi (Tubeufiaceae) from freshwater habitats in Yunnan, China. Cryptogr Mycol 38:27–53
Luo ZL, Hyde KD, Liu JK, Maharachchikumbura SSN, Jeewon R, Bao DF, Bhat DJ, Lin CG, Li WL, Yang J, Liu NG, Lu YZ, Jayawardena RS, Li JF, Su HY (2019) Freshwater Sordariomycetes. Fungal Divers 99:451–660
Luttrell ES (1951) Taxonomy of Pyrenomycetes. Univ Mo Stud 24(3):1–120
Machouart M, Samerpitak K, De Hoog GS, Gueidan C (2014) A multigene phylogeny reveals that Ochroconis belongs to the family Sympoventuriaceae (Venturiales, Dothideomycetes). Fungal Divers 65:77–88
Maharachchikumbura SSN, Hyde KD, Groenewald JZ, Xu JC, Crous PW (2014) Pestalotiopsis revisited. Stud Mycol 79:121–186
Maharachchikumbura SSN, Hyde KD, Jones EBG, McKenzie EHC, Huang SK, Abdel-Wahab MA, Daranagama DA, Dayarathne M, D’souza MJ, Goonasekara ID, Hongsanan S, Jayawardena RS, Kirk PM, Konta S, Liu JK, Liu ZY, Norphanphoun C, Pang KL, Perera RH, Senanayake IC, Shang QJ, Shenoy BD, Xiao YP, Bahkali AH, Kang JC, Somrothipol S, Suetrong S, Wen TC, Xu JC (2015) Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers 72:199–301
Maharachchikumbura SSN, Hyde KD, Jones EBG, McKenzie EHC, Bhat JD, Dayarathne MC, Huang SK, Norphanphoun C, Senanayake IC, Perera RH, Shang QJ, Xiao YP, D’souza MJ, Hongsanan S, Jayawardena RS, Daranagama DA, Konta S, Goonasekara ID, Zhuang WY, Jeewon R, Phillips AJL, Abdel-Wahab MA, Al-Sadi AM, Bahkali AH, Boonmee S, Boonyuen N, Cheewangkoon R, Dissanayake AJ, Kang J, Li QR, Liu JK, Liu XZ, Liu ZY, Luangsa-ard JJ, Pang KL, Phookamsak R, Promputtha I, Suetrong S, Stadler M, Wen TC, Wijayawardene NN (2016) Families of Sordariomycetes. Fungal Divers 79:1–317
Maharachchikumbura SSN, Ariyawansa HA, Wanasinghe DN, Day-arathne MC, Al-Saady NA, Al-Sadi AM (2019) Phylogenetic classification and generic delineation of Hydeomyces desertipleosporoides gen. et sp. nov. (Phaeosphaeriaceae) from Jebel Akhdar Mountain in Oman. Phytotaxa 391:28–38
Malloch D, Cain RF (1971) New cleistothecial Sordariaceae and a new family, Coniochaetaceae. Can J Bot 49:869–880
Malysheva V, Spirin V (2017) Taxonomy and phylogeny of the Auriculariales (Agaricomycetes, Basidiomycota) with stereoid basidiocarps. Fungal Biol 121:689–715
Malysheva V, Spirin V, Miettinen O, Motato-Vásquez V, Hernawati Seelan JSS, Karl-Henrik Larsson KH (2018) Revision of Protohydnum (Auriculariales, Basidiomycota). Mycol Prog 17:805–814
Manawasinghe IS, Phillips AJL, Hyde KD, Chethana KWT, Zhang W, Zhao WS, Yan JY, Li XH (2016) Mycosphere Essays 14: Assessing the aggressiveness of plant pathogenic Botryosphaeriaceae. Mycosphere 7:883–892
Manawasinghe IS, Dissanayake A, Liu M, Wanasinghe D, Xu J, Zhao W, Wei Z, Zhou Y, Hyde KD, Brooks S, Yan J (2019) High genetic diversity and species complexity of Diaporthe associated with Grapevine Dieback in China. Front Microbiol 10:1936
Mapook A, Boonmee S, Ariyawansa HA, Tibpromma S, Campesori E, Jones EG, Bahkali AH, Hyde KD (2016a) Taxonomic and phylogenetic placement of Nodulosphaeria. Mycol Prog 15:34
Mapook A, Hyde KD, Dai DQ, Li J, Jones EB, Bahkali AH, Boonmee S (2016b) Muyocopronales, ord. nov., (Dothideomycetes, Ascomycota) and a reappraisal of Muyocopron species from northern Thailand. Phytotaxa 265:225–237
Marincowitz S, Crous PW, Groenewald JZ, Wingfield MJ (2008) Microfungi occurring on the Proteaceae in the fynbos: 1–166
Marin-Felix Y, Groenewald JZ, Cai L, Chen Q, Marincowitz S, Barnes I, Bensch K, Braun U, Camporesi E, Damm U, De Beer ZW (2017) Genera of phytopathogenic fungi: GOPHY 1. Stud Mycol 86:99–216
Marin-Felix Y, Hernández-Restrepo M, Iturrieta-González I, García D, Gené J, Groenewald JZ, Cai L, Chen Q, Quaedvlieg W, Schumacher RK, Taylor PWJ, Ambers C, Bonthond G, Edwards J, Krueger-Hadfield SA, Luangsa-ard JJ, Morton L, Moslemi A, Sandoval-Denis M, Tan YP, Thangavel R, Vaghefi N, Cheewangkoon R, Crous PW (2019a) Genera of phytopathogenic fungi: GOPHY 3. Stud Mycol 94:1–124
Marin-Felix Y, Hernández-Restrepo M, Wingfield MJ, Akulov A, Carnegie AJ, Cheewangkoon R, Gramaje D, Groenewald JZ, Guarnaccia V, Halleen F, Lombard L, Luangsa-ard J, Marincowitz S, Moslemi A, Mostert L, Quaedvlieg W, Schumacher RK, Spies CFJ, Thangavel R, Taylor PWJ, Wilson AM, Wingfield BD, Wood AR, Crous PW (2019b) Genera of phytopathogenic fungi: GOPHY 2. Stud Mycol 92:47–133
Martin GW (1952) Revision of the North Central Tremellales. Univ Iowa Stud Nat Hist 19:1–122
Marvanová L, Bärlocher F (2001) Hyphomycetes from Canadian streams. VI. Rare species in pure cultures. Czech Mycol 53:1–28
Matsushima T (1971) Some interesting fungi imperfecti. Nat Sci Mus Bull Tokyo 14:460–480
Matsushima T (1983) Matsushima mycological memoirs No. 3. Published by the author, Kobe, Japan
Matsushima T (1993) Matsushima mycological memoirs No. 7. Matsushima Fungus Collection, Kobe, Japan
McDonald GK, Peck D (2009) Effects of crop rotation, residue retention and sowing time on the incidence and survival of ascochyta blight and its effect on grain yield of field peas (Pisum sativum L.). Field Crops Res 111:11–21
Mehrabi M, Hemmati R, Vasilyeva LN, Trouillas FP (2016) Diatrypella macrospora sp. nov. and new records of diatrypaceous fungi from Iran. Phytotaxa 252:43–55
Mercado SA, Heredia G, Mena PJ (1995) New species of dematiaceous hyphomycetes from Veracruz, Mexico. Mycotaxon 55:491–499
Meyer SLF, Luttrell LS (1986) Ascoma morphology of Pseudopeziza trifolii forma specialis medicaginis-sativae (Dermateaceae) on alfalfa. Mycologia 78:529–542
Meyer SLF, Luttrell LS (1987) Host-parasitic relationship between Pseudopeziza trifolii f. sp. medicaginis-sativae. Phytopathology 77:309–319
Mitteilung VI (1939) Beschreibung einiger neuer Pilzarten aus dem Centraalbureau voor Schimmelcultures, Baarn (Nederland). Antonie Van Leeuwenhoek 6:263–290
Moberg R (1983) Studies on Physciaceae (Lichen) II. The genus Pyxine in Europe. Lichenologist 15:161–167
Mongkolsuk P, Meesim S, Poengsungnoen V, Kalb K (2012) The lichen family Physciaceae in Thailand: I. The genus Pyxine. Phytotaxa 59:32–54
Monkai J, Liu JK, Boonmee S, Chomnunti P, Chukeatirote E, Jones EG, Wang Y, Hyde KD (2013) Planistromellaceae (Botryosphaeriales). Cryptogr Mycol 34:45–78
Monkai J, McKenzie EHC, Phillips AJL, Hongsanan S, Pem D, Liu JK, Chethana KWT, Tian Q, Ekanayaka AH, Lestari AS, Zeng M, Zhao Q, Norphanphoun C, Abeywikrama PD, Maharachchikumbura SSN, Jayawardena RS, Chen YJ, Zhao R-L, He M-Q, Raspé O, Kirk PM, Gentekaki E, Hyde KD (2019) https://fungalgenera.org/: a comprehensive database providing webbased information for all fungal genera. Asian J Mycol 2: 298–305
Moral J, Lichtemberg PS, Papagelis A, Sherman J, Michailides TJ (2018) Didymella glomerata causing leaf blight on pistachio. Eur J Plant Pathol 151:1095–1099
Mukhtar I, Li H, Quan X, Chou T, Jiang S, Chen B, Wen Z, Xie B (2019) First Report of Pestalotiopsis theae Causing Leaf Spot of Ixora chinensis in China. Plant Dis 103:370
Nagy LG, Ohm RA, Kovács GM, Floudas D, Riley R, Gácser A, Sipiczki M, Davis JM, Doty SL, de Hoog GS, Lang BF, Spatafora JW, Martin FM, Grigoriev IV, Hibbett DS (2014) Latent homology and convergent regulatory evolution underlies the repeated emergence of yeasts. Nat Commun 15:4471
Nakabonge G, Gryzenhout M, Roux J, Wingfield BD, Wingfield MJ (2006) Celoporthe dispersa gen. et sp. nov. from native Myrtales in South Africa. Stud Mycol 55:255–267
Ňancucheo I, Johnson DB (2012) Acidophilic algae isolated from mine-impacted environments and their roles in sustaining heterotrophic acidophiles. Front Microbiol 3:325
Nannfeldt JA (1932) Studien über die Morphologie und Systematik der nicht-lichenisierten inoperculaten Discomyceten. Nova Acta Regiae Soc Sci Upsal Ser IV 8:1–368
Nawawi A, Kuthubutheen AJ (1989) Canalisporium, a new genus of lignicolous hyphomycetes from Malaysia. Mycotaxon 34:475–487
Niessl G (1872) Beiträge zur Kenntniss der Pilze. Beschreibung neuerund wenig bekannter Pilze. Verh Naturforschenden Vereines Brünn 10:153–217
Nilsson RH, Hyde KD, Pawlowska J, Ryberg M, Tedersoo L, Aas AB, Alias SA, Alves A, Anderson CL, Antonelli A, Arnold AE, Bahnmann B, Bahram M, Bengtsson-Palme J, Berlin A, Branco S, Chomnunti P, Dissanayake A, Drenkhan R, Friberg H, Frøslev TG, Halwachs B, Hartmann M, Henricot B, Jayawardena R, Jumpponen A, Kauserud H, Koskela S, Kulik T, Liimatainen K, Lindahl BD, Lindner D, Liu J-K, Maharachchikumbura S, Manamgoda D, Martinsson S, Neves MA, Niskanen T, Nylinder S, Pereira OL, Pinho DB, Porter TM, Queloz V, Riit T, Sánchez-García M, de Sousa F, Stefańczyk S, Tadych M, Takamatsu S, Tian Q, Udayanga D, Unterseher M, Wang Z, Wikee S, Yan J, Larsson E, Larsson K-H, Kõljalg U, Abarenkov K (2014) Improving ITS sequence data for identification of plant pathogenic fungi. Fungal Divers 67:11–19
Niskanen T, Kytövuori (2012) Key F: Subgen. Telamonia sects Bovini Liimat., Niskanen & Kytöv. in press, Illumini Liimat., Niskanen & Kytöv. in press, Saturnini Möenne-Locc. & Reumaux, Sciophyl-li (Moënne-Locc. & Reumaux) Liimat. & Niskanen in press, Subbalaustini Liimat., Niskanen & Kytöv. in press. and Sordescentes Melot. 2nd ed. In: Knudsen H, Vesterholt J., eds. Funga Nordica. Agaricoid, boletoid, clavarioid, cyphelloid and gastroid genera. Copenhagen, Denmark: Nordsvamp
Norphanphoun C, Doilom M, Daranagama DA, Phookamsak R, Wen TC, Bulgakov TS, Hyde KD (2017) Revisiting the genus Cytospora and allied species. Mycosphere 8:51–97
Norphanphoun C, Raspé O, Jeewon R, Wen TC, Hyde KD (2018) Morphological and phylogenetic characterisation of novel Cytospora species associated with mangroves. MycoKeys 38:93–120
Norphanphoun C, Jayawardena RS, Chen Y, Wen TC, Meepol W, Hyde KD (2019) Morphological and phylogenetic characterization of novel pestalotioid species associated with mangroves in Thailand. Mycosphere 10:531–578
Peever TL, Barve MP, Stone LJ, Kaiser WJ (2007) Evolutionary relationships among Ascochyta species infecting wild and cultivated hosts in the legume tribes Cicereae and Vicieae. Mycologia 99:59–77
Peintner U, Bougher NL, Castellano MA, Moncalvo JM, Moser MM, Trappe JM, Vilgalys R (2001) Multiple origins of sequestrate fungi related to Cortinarius (Cortinariaceae). Am J Bot 88:2168–2179
Peintner U, Horak E, Moser M, Vilgalys R (2002) Phylogeny of Rozites, Cuphocybe and Rapacea inferred from ITS and LSU rDNA sequences. Mycologia 94:620–629
Peintner U, Knapp M, Fleischer V, Walch G, Dresch P (2016) Myrmecridium hiemale sp. nov. from snow-covered alpine soil is the first eurypsychrophile in this genus of anamorphic fungi. Int J Syst Evol Microbiol 66:2592–2598
Pem D, Hongsanan S, Doilom M, Tibpromma S, Wanasinghe DN, Dong W, Ningguo L, Phookamsak R, Phillips AJL, Jeewon R, Hyde KD (2019a) https://www.dothideomycetes.org: an online taxonomic resource for the classification, identification, and nomenclature of Dothideomycetes. Asian J Mycol 2(1):287–297
Pem D, Jeewon R, Bhat DJ, Doilom M, Boonmee S, Hongsanan S, Promputtha I, Xu JC, Hyde KD (2019b) Mycosphere Notes 275–324: a morpho-taxonomic revision and typification of obscure Dothideomycetes genera (incertae sedis). Mycosphere 10:1115–1246
Pem D, Jeewon R, Gafforov Y, Hongsnanan S, Phukhamsakda C, Promputtha I, Doilom M, Hyde KD (2019c) Melanocamarosporioides ugamica gen. et sp. nov., a novel member of the family Melanommataceae from Uzbekistan. Mycol Prog 18:471–481
Perera RH, Hyde KD, Dissanayake AJ, Jones EBG, Liu JK, Wei D, Liu ZY (2018a) Diaporthe collariana sp. nov., with prominent collarettes associated with Magnolia champaca fruits in Thailand. Studies in Fungi 3:141–151
Perera RH, Maharachchikumbura SS, Hyde KD, Bhat DJ, Camporesi E, Jones EBG, Senanayake IC, Al-Sadi AM, Saichana N, Liu JK, Liu ZY (2018b) An appendage-bearing coelomycete Pseudotruncatella arezzoensis gen. and sp. nov. (Amphisphaeriales genera incertae sedis) from Italy, with notes on Monochaetinula. Phytotaxa 338:177–188
Persoon CH (1795) Observationes mycologicae. Annalen der Botanik (Usteri). 15:1–39
Pfister DH (2015) Chapter 2. Pezizomycotina: Pezizomycetes, Orbiliomycetes The Mycota VII (B). In: Mclaughlin DJ, Spatafora JW (eds) Systematics and evolution. Springer, Berlin, Heidelberg, pp 35–55
Pfister DH, Kimbrough JW (2001) Discomycetes. In: McLaughlin EG, Lemke PA, McLaughlin DJ (eds) The mycota VII part A. Systematics and evolution. Springer, Berlin
Phillips AJL, Alves A, Abdollahzadeh J, Slippers B, Wingfield MJ, Groenewald JZ, Crous PW (2013) The Botryosphaeriaceae: genera and species known from culture. Stud Mycol 76:51–167
Phillips AJL, Hyde KD, Alves A, Liu JK (2019) Families in Botryosphaeriales: a phylogenetic, morphological and evolutionary perspective. Fungal Divers 94:1–22
Phookamsak R, Liu JK, McKenzie EHC, Manamgoda DS, Ariyawansa HA, Thambugala KM, Dai DQ, Camporesi E, Chukeatirote E, Wijayawardene NN, Bahkali AH, Mortimer PE, Xu JC, Hyde KD (2014) Revision of Phaeosphaeriaceae. Fungal Divers 68:159–238
Phookamsak R, Manamgoda DS, Li W-J et al (2015) Poaceascoma helicoides gen et sp. nov., a new genus with scolecospores in Lentitheciaceae. Cryptogr Mycol 36:225–236
Phookamsak R, Wanasinghe DN, Hongsanan S, Phukhamsakda C, Huang S-K, Tennakoon DS, Norphanphoun C, Camporesi E, Bulgakov TS, Promputtha I, Mortimer PE, Xu JC, Hyde KD (2017) Towards a natural classification of Ophiobolus and ophiobolus-like taxa; introducing three novel genera Ophiobolopsis, Paraophiobolus and Pseudoophiobolus in Phaeosphaeriaceae (Pleosporales). Fungal Divers 87:299–339
Phookamsak R, Hyde KD, Jeewon R, Bhat DJ, Jones EBG, Maharachchikumbura SSN, Raspé O, Karunarathna SC, Wanasinghe DN, Hongsanan S, Doilom M, Tennakoon DS, Machado AR, Firmino AL, Ghosh A, Karunarathna A, Mešić A, Dutta AK, Thongbai B, Devadatha B, Norphanphoun C, Senwanna C, Wei DP, Pem D, Ackah FK, Wang GN, Jiang HB, Madrid H, Lee HB, Goonasekara ID, Manawasinghe IS, Kušan I, Cano J, Gené J, Li JF, Das K, Acharya K, Raj KNA, Latha KPD, Chethana KWT, He MQ, Dueñas M, Jadan M, Martín MP, Samarakoon MC, Dayarathne MC, Raza M, Park MS, Telleria MT, Chaiwan N, Matočec N, de Silva NI, Pereira OL, Singh PN, Manimohan P, Uniyal P, Shang QJ, Bhatt RP, Perera RH, Alvarenga RLM, Nogal-Prata S, Singh SK, Vadthanarat S, Oh SY, Huang SK, Rana S, Konta S, Paloi S, Jayasiri SC, Jeon SJ, Mehmood T, Gibertoni TB, Nguyen TTT, Singh U, Thiyagaraja V, Sarma VV, Dong W, Yu XD, Lu YZ, Lim YW, Chen Y, Tkalčec Z, Zhang ZF, Luo ZL, Daranagama DA, Thambugala KM, Tibpromma S, Camporesi E, Bulgakov TS, Dissanayake AJ, Senanayake IC, Dai DQ, Tang LZ, Khan S, Zhang H, Promputtha I, Cai L, Chomnunti P, Zhao RL, Lumyong S, Boonmee S, Wen TC, Mortimer PE, Xu JC (2019) Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers 95:1–273
Phoulivong S, Cai L, Chen H, McKenzie EHC, Abdelsalam K, Chukeatirote E, Hyde KD (2010) Colletotrichum gloeosporioides is not a common pathogen on tropical fruits. Fungal Divers 44:33–43
Phukhamsakda C, Ariyawansa HA, Phookamsak R, Chomnunti P, Bulgakov TS, Yang JB, Bhat DJ, Bahkali AH, Hyde KD (2015) Muriphaeosphaeria galatellae gen. et sp. nov. in Phaeosphaeriaceae (Pleosporales). Phytotaxa 227:55–65
Phukhamsakda C, Hongsanan S, Ryberg M, Ariyawansa H, Chomnunti P, Bahkali A, Hyde K (2016) The evolution of Massarineae with Longipedicellataceae fam. nov. Mycosphere 7:1713–1731
Phukhamsakda C, Jeewon R, McKenzie EHC, Xu JC (2019) Morphology and phylogeny of Phaeoseptum mali sp. nov. (Phaeoseptaceae, Pleosporales) on bark of Malus halliana. AJOM 2:118–128
Põldmaa K (2011) Tropical species of Cladobotryum and Hypomyces producing red pigments. Stud Mycol 68:1–34
Pratibha J, Prabhugaonkar A (2017) Torula goaensis, a new asexual ascomycetous fungus in Torulaceae. Webbia 72:171–175
Preuss CGT (1851) Übersicht untersuchter Pilze, besonders aus der Umgegend von Hoyerswerda. Linnaea 24:99–153
Prieto M, Wedin M (2017) Phylogeny, taxonomy and diversification events in the Caliciaceae. Fungal Divers 82:221–238
Punithalingam E (1979) Sphaeropsidales in culture from humans. Nova Hedwigia 31:119–158
Quaedvlieg W, Verkley GJM, Shin HD, Barreto RW, Alfenas AC, Swart WJ, Groenewald JZ, Crous PW (2013) Sizing up Septoria. Stud Mycol 75:307–390
Ranojevic N (1914) Dritter beitrag zur pilzflora serbiens. Ann Mycol 12:393–421
Rao RA (1964) A new species of Diatrypella from India. Mycopathologia 23:263–265
Rao PR, Rao D (1964) Some species of Camposporium Harkn. from India. Antonie Van Leeuwenhoek 30:60–64
Rappaz F (1992) Phomatospora berkeleyi, P. arenaria and their Sporothrix anamorphs. Mycotaxon 45:323–330
Réblová M (2009) Teleomorph of Rhodoveronaea (Sordariomycetidae) discovered and re-evolution of Pleurophragmium. Fungal Divers 36:129–139
Réblová M, Seifert KA (2004) Conioscyphascus, a new ascomycetous genus for holomorphs with Conioscypha anamorphs. Stud Mycol 50:95–108
Réblová M, Gams W, Seifert KA (2011) Monilochaetes and allied genera of the Glomerellales, and a reconsideration of families in the Microascales. Stud Mycol 68:163–191
Réblová M, Fournier J, Štěpánek V (2016a) Two new lineages of aquatic ascomycetes: Atractospora gen. nov. and Rubellisphaeria gen. et sp. nov., and a sexual morph of Myrmecridium montsegurinum sp. nov. Mycol Prog 15:21
Réblová M, Miller AN, Rossman AY, Seifert KA et al (2016b) Recommendations for competing sexual—asexually typified generic names in Sordariomycetes (except Diaporthales, Hypocreales, and Magnaporthales). IMA Fungus 7:131–153
Réblová M, Seifert KA, Fournier J, Štěpánek V (2016c) Newly recognized lineages of perithecial ascomycetes: the new orders Conioscyphales and Pleurotheciales. Persoonia 37:57–81
Rehm H (1888) Rabenhorst’s Kryptogamen-Flora, Pilze - Ascomyceten. 1:65–208
Riley R, Haridas S, Wolfe KH, Lopes MR, Hittinger CT, Göker M, Salamov AA, Wisecaver JH, Long TM, Calvey CH, Aerts AL, Barry KW, Choi C, Clum A, Coughlan AY, Deshpande S, Douglass AP, Hanson SJ, Klenk HP, LaButti KM, Lapidus A, Lindquist EA, Lipzen AM, Meier-Kolthoff JP, Ohm RA, Otillar RP, Pangilinan JL, Peng Y, Rokas A, Rosa CA, Scheuner C, Sibirny AA, Slot JC, Stielow JB, Sun H, Kurtzman CP, Blackwell M, Grigoriev IV, Jeffries TW (2016) Comparative genomics of biotechnologically important yeasts. Proc Nat Acad Sci USA 113:9883–9887
Rogers JD (2000) Thoughts and musings on tropical Xylariaceae. Mycol Res 104:1412–1420
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rossman AY, Samuels GJ, Rogerson CT, Lowen R (1999) Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes). Stud Mycol 42:1–248
Rossman AY, Farr DF, Castlebury LA (2007) A review of the phylogeny and biology of the Diaporthales. Mycoscience 48:135–144
Rossman AY, Seifert KA, Samuels GJ, Minnis AM, Schroers HJ, Lombard L, Crous PW, Põldmaa K, Cannon PF, Summerbell RC, Geiser DM, Zhuang WY, Hirooka Y, Herrera C, Salgado-Salazar C, Chaverri P (2013) Genera in Bionectriaceae, Hypocreaceae, and Nectriaceae (Hypocreales) proposed for acceptance or rejection. IMA Fungus 4:41–51
Saccardo PA (1883) Sylloge Pyrenomycetum, Vol. II. Sylloge Fungorum. 2: 1–813
Saccardo PA (1912) Notae mycologicae. Series XIV. 2. Fungi ex Italia. Ann Mycol 10:316–322
Saccardo PA (1913) Notae mycologicae. Series XVII. Ann Mycol 11:546–568
Sadowski EM, Beimforde C, Gube M, Rikkinen J, Singh H, Seyfullah LJ, Heinrichs J, Nascimbene PC, Reitner J, Schmidt AR (2012) The anamorphic genus Monotosporella (Ascomycota) from Eocene amber and from modern Agathis resin. Fungal Biol 116:1099–1110
Saikia UN, Sarbhoy AK (1981) Hyphomycetes of north-eastern India—IV. The genus Melanographium. Indian Phytopathol 34:356–367
Samarakoon MC, Gafforov Y, Liu NG, Maharachchikumbura SSN, Bhat JD, Liu JK, Promputtha I, Hyde KD (2018) Combined multi-gene backbone tree for the genus Coniochaeta with two new species from Uzbekistan. Phytotaxa 336:43–58
Samerpitak K, Van der Linde E, Choi HJ, van den Ende AG, Machouart M, Gueidan C, de Hoog GS (2014) Taxonomy of Ochroconis, genus including opportunistic pathogens on humans and animals. Fungal Divers 65:89–126
Samson RA (1974) Paecilomyces and some allied hyphomycetes. Stud Mycol 6:1–119
Samuels GJ, Blackwell M (2001) Pyrenomycetes—fungi with perithecia. In: McLaughlin DJ, McLaughlin EG, Lemke PA (eds) Systematics and Evolution. Springer, Berlin, Heidelberg, pp 221–255
Samuels GJ, Müller E (1979) Life-history studies of Brazilian Ascomycetes. 7. Rhytidhysteron rufulum and the genus Eutryblidiella. Sydowia 32:277–292
Sanchez RM, Miller AN, Bianchinotti MV (2012) A new species of Acanthostigma (Tubeufiaceae, Dothideomycetes) from the southern hemisphere. Mycologia 104:223–231
Schmidt JC, Kunze G (1817) Mykologische Hefte. 1:1–109
Schmull M, Miadlikowska J, Pelzer M, Stocker-Wörgötter E, Hofstetter V, Fraker E, Hodkinson BP, Reeb V, Kukwa M, Lumbsch HT, Kauff F, Lutzoni F (2011) Phylogenetic affiliations of members of the heterogeneous lichen-forming fungi of the genus Lecidea Sensu Zahlbruckner (Lecanoromycetes, Ascomycota). Mycologia 103:983–1003
Schoch CL, Shoemaker RA, Seifert KA, Hambleton S, Spatafora JW, Crous PW (2006) A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98:1041–1052
Schoch CL, Crous PW, Groenewald JZ, Boehm EWA, Burgess TI, de Gruyter J, de Hoog GS, Dixon LJ, Grube M, Gueidan C, Harada Y, Hatakeyama S, Hirayama K, Hosoya T, Huhndorf SM, Hyde KD, Jones EBG, Kohlmeyer J, Kruys Å, Li YM, Lücking R, Lumbsch HT, Marvanová L, Mbatchou JS, McVay AH, Miller AN, Mugambi GK, Muggia L, Nelsen MP, Nelson P, Owensby CA, Phillips AJL, Phongpaichit S, Pointing SB, Pujade-Renaud V, Raja HA, Rivas Plata E, Robbertse B, Ruibal C, Sakayaroj J, Sano T, Selbmann L, Shearer CA, Shirouzu T, Slippers B, Suetrong S, Tanaka K, Volkmann-Kohlmeyer B, Wingfield MJ, Wood AR, Woudenberg JHC, Yonezawa H, Zhang Y, Spatafora JW (2009) A class-wide phylogenetic assessment of Dothideomycetes. Stud Mycol 64:1–1
Schroers HJ (2001) A monograph of Bionectria (Ascomycota, Hypocreales, Bionectriaceae) and its Clonostachys anamorphs. Stud Mycol 46:1–214
Seifert K, Morgan-Jones G, Gams W, Kendrick B (2011) The Genera of Hyphomycetes. CBS Biodiversity Series, Utrecht, p 997
Senanayake IC, Al-Sadi AM, Bhat JD, Camporesi E, Dissanayake AJ, Lumyong S, Maharachchikumbura SSN, Hyde KD (2016) Phomatosporales ord. nov. and Phomatosporaceae fam. nov., to accommodate Lanspora, Phomatospora and Tenuimurus, gen. nov. Mycosphere 7:628–641
Senanayake IC, Crous PW, Groenewald JZ, Maharachchikumbura SSN, Jeewon R, Phillips AJL, Bhat JD, Perera RH, Li QR, Li WJ, Tangthirasunun N, Norphanphoun C, Karunarathna SC, Camporesi E, Manawasighe IS, Al-Sadi AM, Hyde KD (2017) Families of Diaporthales based on morphological and Phylogenetic evidence. Stud Mycol 86:217–296
Senanayake IC, Jeewon R, Camporesi E, Hyde KD, Zeng YJ, Tian SL, Xie N (2018a) Sulcispora supratumida sp. nov. (Phaeosphaeriaceae, Pleosporales) on Anthoxanthum odoratum from Italy. Mycokeys 38:35–46
Senanayake IC, Jeewon R, Chomnunti P, Wanasinghe DN, Norphanphoun C, Karunarathna A, Pem D, Perera RH, Camporesi E, McKenzie EHC, Hyde KD, Karunarathna SC (2018b) Taxonomic circumscription of Diaporthales based on multi-gene phylogeny and morphology. Fungal Divers 93:241–443
Senwanna C, Phookamsak R, Doilom M, Hyde KD, Cheewangkoon R (2017) Novel taxa of Diatrypaceae from Para rubber (Hevea brasiliensis) in northern Thailand; introducing a novel genus Allocryptovalsa. Mycosphere 8:1835–1855
Seyedmousavi S, Guillot J, de Hoog GS (2013) Phaeohyphomycoses, emerging opportunistic diseases in animals. Clin Microbiol Rev 26:19–35
Shang QJ, Hyde KD, Phookamsak R, Doilom M, Bhat DJ, Maharachchikumbura SS, Promputtha I (2017) Diatrypella tectonae and Peroneutypa mackenziei sp. nov. (Diatrypaceae) from northern Thailand. Mycol Prog 16:463–476
Shang QJ, Hyde KD, Jeewon R, Khan S, Promputtha I, Phookamsak R (2018) Morpho-molecular characterization of Peroneutypa (Diatrypaceae, Xylariales) with two novel species from Thailand. Phytotaxa 356:1–8
Shearer CA (1973) Fungi of the Chesapeake Bay and its Tributaries II. The Genus Conioscypha. Mycologia 65:128–136
Shearer CA (1974) Fungi of the Chesapeake Bay and its tributaries IV. Three new species from the Patuxent River. Mycologia 66:16–24
Shearer CA, Motta JJ (1973) Ultrastructure and conidiogenesis in Conioscypha (Hyphomycetes). Can J Bot 51:1747–1751
Shen XX, Opulente DA, Kominek J, Zhou X et al (2018) Tempo and mode of genome evolution in the budding yeast subphylum. Cell 175(6):1533.e20–1545.e20
Shenoy BD, Jeewon R, Wu WP, Bhat DJ, Hyde KD (2006) Ribosomal and RPB2 DNA sequence analyses suggest that Sporidesmium and morphologically similar genera are polyphyletic. Mycol Res 110:916–928
Shenoy BD, Jeewon R, Hyde KD (2007) Impact of DNA sequence-data on the taxonomy of anamorphic fungi. Fungal Divers 26:1–54
Shenoy BD, Jeewon R, Wang H, Amandeep K, Ho WH, Bhat DJ, Crous PW, Hyde KD (2010) Sequence data reveals phylogenetic affinities of fungal anamorphs Bahusutrabeeja, Diplococcium, Natarajania, Paliphora, Polyschema, Rattania and Spadicoides. Fungal Divers 44:161–169
Shi CH, Bau T, Li Y (2016) Newly recorded genus and species of Pezizales in China. Mycosystema 35:1348–1356
Shoemaker RA, Babcock CE (1989) Phaeosphaeria. Can J Bot 67:1500–1599
Shoemaker RA, Hambleton S, Lacroix M, Tesolin M, Coulombe J (2002) Fungi Canadenses No. 344: Rhexocercosporidium carotae. Can J Plant Pathol 24:59–362
Simmons EG (1948) Studies in the stromatic Sphaeriales of Indiana—II. Proc Indiana Acad Sci 58:82–91
Singtripop C, Camporesi E, Ariyawansa HA, Wanasinghe DN, Bahkall AH, Chomnunti P, Boonmee S, Hyde KD (2015) Keissleriella dactylidis, sp. nov., from Dactylis glomerata and its phylogenetic placement. Sci Asia 41:295–304
Sipiczki M, Horváth E, Pfliegler WP (2018) Birth-and-death evolution and reticulation of ITS segments of Metschnikowia andauensis and Metschnikowia fructicola rDNA repeats. Front Microbiol 9:1193
Sivanesan A (1984) The bitunicate ascomycetes and their anamorphs. J. Cramer, Berlin-Stuttgart
Smerlis E (1966) Notes on the nomenclature of Nothophacidium phyllophilum comb. nov., its pathogenicity and relationship to Pezizella minuta. Can J Bot 44:563–565
Smith JDG, Liew ECY, Hyde KD (2001) The Xylariales: a monophyletic order containing 7 families. Fungal Divers 13:175–208
Somrithipol S, Jones EBG (2005) An addition to the hyphomycete genus Melanographium from Thailand. Fungal Divers 19:137–144
Song J, Liang J-F, Mehrabi-Koushki M, Krisai-Greilhuber I, Ali B, Bhatt VK, CernaMendoza A, Chen B, Chen Z-X, Chu H-L, Corazon-Guivin MA, da Silva GA, De Kesel A, Dima B, Dovana F, Farokhinejad R, Ferisin G, Guerrero-Abad JC, Guo T, Han L-H, Ilyas S, Iqbal J, Justo A, Khalid AN, Khodadadi-Pourarpanahi S, Li T-H, Liu C, Lorenzini M, Lu J-K, Mumtaz AS, Oehl F, Pan X-Y, Papp V, Qian W, Razaq A, Semwal KC, Tang L-Z, Tian X-L, Uddin S, Vallejos-Tapullima A, van der Merwe NA, Wang S-K, Wang C-Q, Yang R-H, Yu F, Zapparoli G, Zhang M, Antonín V, Aptroot A, Aslan A, Banerjee A, Chatterjee S, Dirks AC, Ebrahimi L, Fotouhifar K, Ghosta Y, Kalinina LB, Liu J, Karahan D, Maiti M, Mookherjee A, Nath PS, Panja B, Saha J, Ševčíková H, Voglmayr H, Yazıcı K, Haelewaters D (2019) Fungal systematics and evolution. 5. Sydowia 71:141–245
Species Fungorum (2020) http://www.speciesfungorum.org/Index.htm
Spegazzini C (1881) Fungi argentini additis nonnullis brasiliensibus montevideensibusque. Pugillus quartus (Continuacion). An Soc Cient Argent 12:174–189
Spegazzini C (1911) Mycetes Argentinenses (Series V). An Mus Nac Hist Nat B Aires 20:329–467
Spirin V, Malysheva V, Roberts P, Trichies G, Savchenko A, Larsson KH (2019) A convolute diversity of the Auriculariales (Agaricomycetes. Nord J Bot, Basidiomycota) with sphaeropedunculate basidia
Sprague R (1950) Diseases of cereals and grasses in North America. Diseases of cereals and grasses in North America
Sri-Indrasutdhi V, Boonyuen N, Suetrong S, Chuaseeharonnachai C, Sivichai S, Jones EBG (2010) Wood-inhabiting freshwater fungi from Thailand: Ascothailandia grenadoidia gen. et sp. nov., Canalisporium grenadoidia sp. nov. with a key to Canalisporium species (Sordariomycetes, Ascomycota). Mycoscience 51:411–420
Srivastava N, Srivastava AK, Kamal Rai AN (1995) New hyphopodiate hyphomycetes from North-Eastern Uttar Pradesh, India. Mycol Res 99:395–396
Stadler M (2011) Importance of secondary metabolites in the Xylariaceae as parameters for assessment of their taxonomy, phylogeny, and functional biodiversity. Curr Res Environ Appl Mycol 1:75–133
Stadler M, Hellwig V (2005) Chemotaxonomy of the Xylariaceae and remarkable bioactive compounds from Xylariales and their associated asexual stages. Phytochemistry 9:41–93
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-Analysis of large Phylogenies. Bioinformatics 30:1312–1313
Su H, Hyde KD, Maharachchikumbura SSN, Ariyawansa HA, Luo Z, Promputtha I, Tian Q, Lin C, Shang Q, Zhao Y, Chai H, Liu X, Bahkali AH, Bhat JD, McKenzie EHC, Zhou D (2016) The families Distoseptisporaceae fam. nov., Kirschsteiniotheliaceae, Sporormiaceae and Torulaceae, with new species from freshwater in Yunnan Province, China. Fungal Divers 80:375–409
Su XJ, Luo ZL, Jeewon R, Bhat DJ, Bao DF, Li WL, Hao YE, Su HY, Hyde KD (2018) Morphology and multigene phylogeny reveal new genus and species of Torulaceae from freshwater habitats in northwestern Yunnan, China. Mycol Prog 17:531–545
Subramanian CV (1957) Two new species of Petrakia. Beihefte zur Sydowia 1:14–15
Subramanian CV (1994) Hyphomycetes from South East Asia: novelties from Singapore and Malaysia. Kavaka 22(23):52–76
Suetrong S, Schoch CL, Spatafora JW, Kohlmeyer J, Volkmann-Kohlmeyer B, Sakayaroj J, Phongpaichit S, Tanaka K, Hirayama K, Jones EBG (2009) Molecular systematics of the marine Dothideomycetes. Stud Mycol 64:155–173
Suetrong S, Hyde KD, Ahang Y, Bahkali AB, Jones EBG (2011) Trematosphaeriaceae fam. nov. (Dothideomycetes, Ascomycota). Cryptogr Mycol 32:343–358
Suh SO, Blackwell M (2004) Three new beetle-associated yeast species in the Pichia guilliermondii clade. FEMS Yeast Res 5:87–95
Suh SO, Nguyen NH, Blackwell M (2005) Nine new Candida species near C. membranifaciens isolated from insects. Mycol Res 109:1045–1056
Suh SO, Blackwell M, Kurtzman CP, Lachance MA (2006a) Phylogenetics of Saccharomycetales, the ascomycete yeasts. Mycologia 98:1006–1017
Suh SO, Nguyen NH, Blackwell M (2006b) A yeast clade near Candida kruisii uncovered: nine novel Candida species associated with basidioma-feeding beetles. Mycol Res 110:1379–1394
Summerbell RC (2003) Ascomycetes: Aspergillus, Fusarium, Sporothrix, Piedraia, and their relatives. In: Howard DH (ed) Pathogenic fungi in humans and animals, 2nd edn. Marcel Dekker, New York, pp 237–498
Summerbell RC, Gueidan C, Schroers HJ, De Hoog GS, Starink M, Rosete YA, Guarro J, Scott JA (2011) Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium, and Trichothecium. Stud Mycol 68:139–162
Sun JZ, Liu XZ, Hyde KD, Zhao Q, Maharachchikumbura SSN, Camporesi E, Bhat J, Nilthong S, Lumyong S (2017) Calcarisporium xylariicola sp. nov. and introduction of Calcarisporiaceae fam. nov. in Hypocreales. Mycol Prog 16:433–445
Sun JZ, Liu XZ, McKenzie EHC, Jeewon R, Liu JK, Zhang XL, Zhao Q, Hyde KD (2019) Fungicolous fungi: terminology, diversity, distribution, evolution, and species checklist. Fungal Divers 95:337–430
Takamatsu S, Ito H, Shiroya Y, Kiss L, Heluta V (2015) First comprehensive phylogenetic analysis of the genus Erysiphe (Erysiphales, Erysiphaceae) I. The Microsphaera lineage. Mycologia 107:475–489
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tanaka K, Hirayama K, Yonezawa H, Hatakeyama S, Harada Y, Sano T, Shirouzu T, Hosoya T (2009) Molecular taxonomy of bambusicolous fungi: Tetraplosphaeriaceae, a new pleosporalean family with tetraploa-like anamorphs. Stud Mycol 64:175–209
Tanaka K, Hirayama K, Yonezawa H, Sato G, Toriyabe A, Kudo H, Hashimoto A, Matsumura M, Harada Y, Kurihara Y, Shirouzu T (2015) Revision of the Massarineae (Pleosporales, Dothideomycetes). Stud Mycol 82:75–136
Tanney JB, Douglas B, Seifert KA (2016) Sexual and asexual states of some endophytic Phialocephala species of Picea. Mycologia 108(2):255–280
Tennakoon DS, Hyde KD, Phookamsak R, Wanasinghe DN, Cam-poresi E E, Promputtha I (2016) Taxonomy and phylogeny of Juncaceicola gen. nov. Phaeosphaeriaceae, Pleosporinae, Pleosporales. Cryptogam, Mycol 37:135–156
Tennakoon DS, Phookamsak R, Wanasinghe DN, Yang JB, Lumyong S, Hyde KD (2017) Morphological and phylogenetic insights resolve Plenodomus sinensis (Leptosphaeriaceae) as a new species. Phytotaxa 324:73–82
Tennakoon DS, Jeewon R, Kuo CH, Hyde KD (2018a) Phylogenetic and morphological characterization of Byssosphaeria macarangae sp. nov., and B. taiwanense sp. nov. from Macaranga tanarius. Phytotaxa 364:211–226
Tennakoon DS, Kuo CH, Jeewon R, Thambugala KM, Hyde KD (2018b) Saprobic Lophiostomataceae (Dothideomycetes): Pseudolophiostoma mangiferae sp. nov. and Neovaginatispora fuckelii, a new record from Mangifera indica. Phytotaxa 364:157–171
Thakur RS, Rai AN, Dudey MK (2014) A novel addition of fungal taxa from Indian subcontinent. IJPAES 4:117–121
Thambugala KM, Camporesi E, Ariyawansa HA, Phookamsak R, Liu ZY, Hyde KD (2014) Phylogeny and morphology of Phaeosphaeriopsis triseptata sp. nov., and Phaeosphaeriopsis glaucopunctata. Phytotaxa 176:238–250
Thambugala KM, Hyde KD, Tanaka K, Tian Q, Wanasinghe DN, Ariyawansa HA, Jayasiri SC, Boonmee S, Camporesi E, Hashimoto A, Hirayama K (2015) Towards a natural classification and backbone tree for Lophiostomataceae, Floricolaceae, and Amorosiaceae fam. nov. Fungal Divers 74:199–266
Thambugala KM, Hyde KD, Eungwanichayapant PD, Romero AI, Liu ZY (2016) Additions to the genus Rhytidhysteron in Hysteriaceae. Cryptogr Mycol 37:99–116
Thambugala KM, Wanasinghe DN, Phillips AJL, Camporesi E, Bulgakov TS, Phukhamsakda C, Ariyawansa HA, Goonasekara ID, Phookamsak R, Dissanayake A, Tennakoon DS, Tibpromma S, Chen YY, Liu ZY, Hyde KD (2017) Mycosphere notes 1–50: Grass (Poaceae) inhabiting Dothideomycetes. Mycosphere 8:697–796
Thambugala KM, Hyde KD, Zhang JF, Liu ZY (2018) Didymella eriobotryae sp. nov. (Didymellaceae) and Arthrinium arundinis (Apiosporaceae) from fruit of Eriobotrya japonica (loquat) in China. Phytotaxa 382:136–147
Than PP, Prihastuti H, Phoulivong S, Taylor PWJ, Hyde KD (2008) Chilli anthracnose disease caused by Colletotrichum species. J Zhejiang Univ Sci B 9:764–788
Thongbai B, Rapior S, Hyde KD, Wittstein K, Stadler M (2015) Hericium erinaceus, an amazing medicinal mushroom. Mycol Prog 14:91
Tian Q, Liu JK, Hyde KD, Wanasinghe DN, Boonmee S, Jayasiri SC, Luo ZL, Taylor JE, Phillips AJL, Bhat DJ, Li WJ, Ariyawansa H, Thambugala KM, Jones EBG, Chomnunti P, Bahkali AH, Xu JC, Camporesi E (2015) Phylogenetic relationships and morphological reappraisal of Melanommataceae (Pleosporales). Fungal Divers 74:267–324
Tibpromma S, Hyde KD, Jeewon R, Maharachchikumbura SSN, Liu JK, Bhat DJ, Jones EBG, McKenzie EHC, Camporesi E, Bulgakov TS, Doilom M, de Azevedo Santiago ALCM, Das K, Manimohan P, Gibertoni TB, Lim YW, Ekanayaka AH, Thongbai B, Lee HB, Yang J-B, Kirk PM, Sysouphanthong P, Singh SK, Boonmee S, Dong W, Raj KNA, Latha KPD, Phookamsak R, Phukhamsakda C, Konta S, Jayasiri SC, Norphanphoun C, Tennakoon DS, Li J, Dayarathne MC, Perera RH, Xiao Y, Wanasinghe DN, Senanayake IC, Goonasekara ID, de Silva NI, Mapook A, Jayawardena RS, Dissanayake AJ, Manawasinghe IS, Chethana KWT, Luo Z-L, Hapuarachchi KK, Baghela A, Soares AM, Vizzini A, Meiras-Ottoni A, Mešić A, Dutta AK, de Souza CAF, Richter C, Lin C-G, Chakrabarty D, Daranagama DA, Lima DX, Chakraborty D, Ercole E, Wu F, Simonini G, Vasquez G, da Silva GA, Plautz HL Jr, Ariyawansa HA, Lee H, Kušan I, Song J, Sun J, Karmakar J, Hu K, Semwal KC, Thambugala KM, Voigt K, Acharya K, Rajeshkumar KC, Ryvarden L, Jadan M, Hosen MI, Mikšík M, Samarakoon MC, Wijayawardene NN, Kim NK, Matočec N, Singh PN, Tian Q, Bhatt RP, de Oliveira RJV, Tulloss RE, Aamir S, Kaewchai S, Marathe SD, Khan S, Hongsanan S, Adhikari S, Mehmood T, Bandyopadhyay TK, Svetasheva TY, Nguyen TTT, Antonín V, Li W-J, Wang Y, Indoliya Y, Tkalčec Z, Elgorban AM, Bahkali AH, Tang AMC, Su HY, Zhang H, Promputtha I, Luangsa-ard J, Xu J, Yan J, Ji-Chuan K, Stadler M, Mortimer PE, Chomnunti P, Zhao Q, Phillips AJL, Nontachaiyapoom S, Wen TC, Karunarathna SC (2017) Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 83:1–261
Tibpromma S, Hyde KD, McKenzie EH, Bhat DJ, Phillips AJ, Wanasinghe DN, Samarakoon MC, Jayawardena RS, Dissanayake AJ, Tennakoon DS, Doilom M, Phookamsak R, Tang AMC, Xu J, Mortimer PE, Promputtha I, Maharachchikumbura SSN, Khan S, Karunarathna SC (2018) Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal Divers 93:1–60
Tode HJ (1791) Fungi Mecklenburgenses selecti. Fasc. II, Generum novorum appendicem, Lüneburg
Travadon R, Lawrence DP, Rooney-Latham S, Gubler WD, Wilcox WF, Rolshausen PE, Baumgartner K (2015) Cadophora species associated with wood-decay of grapevine in North America. Fungal Biol 119:53–66
Tsui CKM, Berbee ML (2006) Phylogenetic relationships and convergence of helicosporous fungi inferred from ribosomal DNA sequences. Mol Phylogenet Evol 39:587–597
Tsui K, Goh T, Hyde K (1997) A news species of Dactylaria from Hong Kong. Sydowia 49:182–186
Tsui CKM, Sivichai S, Berbee ML (2006) Molecular systematics of Helicoma, Helicomyces and Helicosporium and their teleomorphs inferred from rDNA sequences. Mycologia 98:94–104
Tulasne LR, Tulasne C (1865) Selecta Fungorum Carpologia: Nectriei-Phacidiei-Pezizei 3:3
Udayanga D, Liu X, McKenzie EHC, Chukeatirote E, Bahkali AH, Hyde KD (2011) The genus Phomopsis: biology, applications, species concepts and names of common phytopathogens. Fungal Divers 50:189
Udayanga D, Liu X, Crous PW, McKenzie EH, Chukeatirote E, Hyde KD (2012) A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis). Fungal Divers 56:157–171
Valenzuela-Lopez N, Cano-Lira JK, Guarro J, Sutton DA, Wiederhold N, Crous PW, Stchigel AM (2018) Coelomycetous Dothideomycetes with emphasis on the families Cucurbitariaceae and Didymellaceae. Stud Mycol 90:1–69
Vasilyeva LN, Stephenson SL (2005) Pyrenomycetes of the Great Smoky Mountains National Park. II. Cryptovalsa Ces. et De Not. and Diatrypella (Ces. et De Not.) Nitschke (Diatrypaceae). Fungal Divers 19:189–200
Vera DI, Murray TD (2016) Occurrence and Survival of Apothecia of the Eyespot Pathogens Oculimacula acuformis and O. yallundae on Wheat Stubble in the U.S. Pacific Northwest. Plant Disease 100:991–995
Videira SI, Groenewald JZ, Nakashima C, Braun U, Barreto RW, de Wit PJ, Crous PW (2017) Mycosphaerellaceae-Chaos or clarity? Stud Mycol 87:257–421
Voglmayr H, Jaklitsch WM (2019) Stilbocrea walteri sp. nov., an unusual species of Bionectriaceae. Mycol Prog 18:91–105
Voglmayr H, Friebes G, Gardiennet A, Jaklitsch WM (2018) Barrmaelia and Entosordaria in Barrmaeliaceae (fam. nov., Xylariales) and critical notes on Anthostomella-like genera based on multigene phylogenies. Mycol Prog 17:155–177
Von Arx JA (1951) On the genus Laestadia and Gnomoniaceae. Antonie Van Leeuwenhoek 17:259
Von Höhnel F (1904) Mycologische Fragmente. Ann Mycol 2:38–60
Von Höhnel F (1919) Fragmente zur Mykologie. XXIII Mitteilung, Nr. 1154 bis 1188. Sitzungsberichte der Kaiserlichen Akademie der Wissenschaften Math.-naturw. Klasse Abt I 128:535–625
Von Matheis W (1977) Dennisiodiscus hippocastani (Richon) combo nov., der Violette Roßkastanien-Wollbecherling. Z Pilzkunde 43:213–216
Vujanovic V, St-Arnaud M, Barabé D, Thibeault G (2000) Phialocephala victorinii sp. nov., endophyte of Cypripedium parviflorum. Mycologia 92:571–576
Walsh E, Duan W, Mehdi M, Naphri K (2018) Cadophora meredithiae and C. interclivum, new species from roots of sedge and spruce in a western Canada subalpine forest. Mycologia 110:201–214
Wanasinghe DN, Jones EBG, Camporesi E, Boonmee S, Karunarathna SC, Thines M, Mortimer PE, Xu JC, Hyde KD (2014) Dematiopleospora mariae gen. sp. nov., from Ononis spinosa in Italy. Cryptogr Mycol 35:105–117
Wanasinghe DN, Hyde KD, Jeewon R, Crous PW, Wijayawardene NN, Jones EBG, Bhat DJ, Phillips AJL, Groenewald JZ, Dayarathne MC, Phukhamsakda C, Thambugala KM, Bulgakov TS, Camporesi E, Gafforov Y, Mortimer PE, Karunarathna SC (2017a) Phylogenetic revision of Camarosporium (Pleosporinae, Dothideomycetes) and allied genera. Stud Mycol 87:207–256
Wanasinghe DN, Hyde KD, Konta S, To-Anun C, Jones EG (2017b) Saprobic Dothideomycetes in Thailand: Neoaquastroma gen. nov. (Parabambusicolaceae) introduced based on morphological and molecular data. Phytotaxa 302:133–144
Wanasinghe DN, Phukhamsakda C, Hyde KD, Jeewon R, Lee HB, Gareth Jones EB, Tibpromma S, Tennakoon DS, Dissanayake AJ, Jayasiri SC, Gafforov Y, Camporesi E, Bulgakov TS, Ekanayake AH, Perera RH, Samarakoon MC, Goonasekara ID, Mapook A, Li WJ, Senanayake IC, Li J, Norphanphoun C, Doilom M, Bahkali AH, Xu J, Mortimer PE, Tibell L, Tibell S, Karunarathna SC (2018) Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers 89:1–236
Wang CL, Lin CC (2004) Five new records of Ascomycetes in Taiwan. Fungal Sci 19:21–29
Wang Z, Binder M, Schoch CL, Johnston PR, Spatafora JW, Hibbett DS (2006a) Evolution of helotialean fungi (Leotiomycetes, Pezizomycotina): a nuclear rDNA phylogeny. Mol Phylogenet Evol 41:295–312
Wang Z, Johnston PR, Takamatsu S, Spatafora JW, Hibbett DS (2006b) Toward a phylogenetic classification of the Leotiomycetes based on rDNA data. Mycologia 98:1065–1075
Wedin M, Grube M (2002) Parsimony analyses of mtSSU and nIT rDNA sequences reveal the natural relationships of the lichen families Physciaceae and Caliciaceae. Taxon 51:655–660
Wei XL, Hur JS (2007) Foliose genera of Physciaceae (lichenized Ascomycota) of South Korea. Mycotaxon 102:127–137
Weir BS, Johnston PR, Damm U (2012) The Colletotrichum gloeosporioides species complex. Stud Mycol 73:115–180
Wendt L, Sir EB, Kuhnert E, Heitkämper S, Lambert C, Hladki AI, Romero AI, Luangsa-ard JJ, Srikitikulchai P, Peršoh D, Stadler M (2018) Resurrection and emendation of the Hypoxylaceae, recognised from a multigene phylogeny of the Xylariales. Mycol Prog 17:115–154
Whalley AJ, Edwards RL (1995) Secondary metabolites and systematic arrangement within the Xylariaceae. Can J Bot 73(S1):802–810
Whitton SR, McKenzie EHC, Hyde KD (2002) Microfungi on the Pandanaceae: two new species of Camposporium and key to the genus. Fungal Divers 11:177–187
Wijayawardene NN, Hyde KD, Al-Ani LKT, Tedersoo L, Rajeshkumar KC, Zhao RL, Aptroot A, Leontyev DV, Saxena RK, Tokarev YS, Dai DQ, Letcher PM, Stephenson SL, Ertz D, Lumbsch HT, Haelewaters D, Bensch K, Madrid H, Kukwa M, Issi IV, Lateef AA, Phillips AJL, Selbmann L, Pfliegler WP, Horváth E, Pereira OL, Kirk PM, Kolaříková Z, Raja HA, Radek R, Papp V, Dima B, Ma J, Malosso E, Takamatsu S, Rambold G, Gannibal PB, Triebel D, Gautam AK, Avasthi S, Suetrong S, Timdal E, Fryar SC, Delgado G, Réblová M, Doilom M, Dolatabadi S, Pawłowska J, Humber RA, Kodsueb R, Sánchez-Castro I, Goto BT, Silva DKA, de Souza FA, Oehl F, da Silva GA, Silva IR, Błaszkowski J, Jobim K, Maia LC, Barbosa FR, Fiuza PO, Divakar PK, Shenoy BD, Castañeda-Ruiz RF, Somrithipo S, Karunarathna SC, Tibpromma S, Mortimer PE, Wanasinghe DN, Phookamsak R, Xu JC, Wang Y, Fenghua T, Alvarado P, Li DW, Kušan I, Matočec N, Maharachchikumbura SSN, Papizadeh M, Heredia G, Wartchow F, Bakhshi M, Boehm E, Youssef N, Hustad VP, Lawrey JD, Santiago ALCMA, Bezerra JDP, Souza-Motta CM, Firmino AL, Tian Q, Houbraken J, Hongsanan S, Tanaka K, Dissanayake AJ, Monteiro JS, Grossart HP, Suija A, Weerakoon G, Etayo J, Tsurykau A, Kuhnert E, Vázquez V, Mungai P, Damm U, Li QR, Zhang H, Boonmee S, Lu YZ, Becerra AG, Kendrick B, Brearley FQ, Motiejūnaitė J, Sharma B, Khare R, Gaikwad S, Wijesundara DSA, Tang LZ, He MQ, Flakus A, Rodriguez-Flakus P, Zhurbenko MP, McKenzie EHC, Stadler M, Bhat DJ, Liu JK (Jack), Raza M, Jeewon R, Nassonova ES, Prieto M, Jayalal RGU, Yurkov A, Schnittler M, Shchepin ON, Novozhilov YK, Liu P,. Cavender JC, Kang YQ, Mohammad S, Zhang LF, Xu RF, Li YM, Dayarathne MC, Ekanayaka AH, Wen TC, Deng CY, Navathe S (2020) Outline of Fungi (including fossil fungi) and fungi-like taxa. Mycosphere (in press)
Wijayawardene NN, Hyde KD, Bhat DJ, Camporesi E, Schumacher RK, Chethana KWT, Wikee S, Bahkali AH, Wang Y (2014) Camarosporium-like species are polyphyletic in Pleosporales; introducing Paracamarosporium and Pseudocamarosporium gen. nov. in Montagnulaceae. Cryptogr Mycol 35:177–198
Wijayawardene NN, Hyde KD, Wanasinghe DN, Papizadeh M, Goonasekara ID, Camporesi E, Bhat DJ, McKenzie EHC, Phillips AJL, Diederich P, Tanaka K, Li WJ, Tangthirasunun N, Phookamsak R, Dai DQ, Dissanayake AJ, Weerakoon G, Maharachchikumbura SSN, Hashimoto A, Matsumura M, Bahkali AH, Wang Y (2016a) Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Divers 77:1–316
Wijayawardene NN, Tibpromma S, Hyde KD, An YL, Camporesi E, Wang Y (2016b) Additions to Pseudocamarosporium; two new species from Italy. Stud Fungi 1:1–10
Wijayawardene NN, Hyde KD, Rajeshkumar KC, Hawksworth DL, Madrid H, Kirk PM, Braun U, Singh RV, Crous PW, Kukwa M, Lücking R, Kurtzman CP, Yurkov A, Haelewaters D, Aptroot A, Lumbsch HT, Timdal E, Ertz D, Etayo J, Phillips AJL, Groenewald JZ, Papizadeh M, Selbmann L, Dayarathne MC, Weerakoon G, Jones EBG, Suetrong S, Tian Q, Castañeda-Ruiz RF, Bahkali AH, Pang K-L, Tanaka K, Dai DQ, Sakayaroj J, Hujslová M, Lombard L, Shenoy BD, Suija A, Maharachchikumbura SSN, Thambugala KM, Wanasinghe DN, Sharma BO, Gaikwad S, Pandit G, Zucconi L, Onofri S, Egidi E, Raja HA, Kodsueb R, Cáceres MES, Pérez-Ortega S, Fiuza PO, Monteiro JS, Vasilyeva LN, Shivas RG, Prieto M, Wedin M, Olariaga I, Lateef AA, Agrawal Y, Fazeli SAS, Amoozegar MA, Zhao GZ, Pfliegler WP, Sharma G, Oset M, Abdel-Wahab MA, Takamatsu S, Bensch K, de Silva NI, De Kese A, Karunarathna A, Boonmee S, Pfister DH, Lu YZ, Luo ZL, Boonyuen N, Daranagama DA, Senanayake IC, Jayasiri SC, Samarakoon MC, Zeng XY, Doilom M, Quijada L, Rampadarath S, Heredia G, Dissanayake AJ, Jayawardana RS, Perera RH, Tang LZ, Phukhamsakda C, Hernández-Restrepo M, Ma X, Tibpromma S, Gusmao LFP, Weerahewa D, Karunarathna SC (2017a) Notes for genera Ascomycota. Fungal Divers 86:1–594
Wijayawardene NN, Hyde KD, Tibpromma S, Wanasinghe DN, Thambugala KM, Tian Q, Wang Y (2017b) Towards incorporating asexual fungi in a natural classification: checklist and notes 2012–2016. Mycosphere 8:1457–1554
Wijayawardene NN, Hyde KD, Lumbsch HT, Liu JK, Maharachchikumbura SS, Ekanayaka AH, Tian Q, Phookamsak R (2018a) Outline of Ascomycota: 2017. Fungal Divers 88:167–263
Wijayawardene NN, Pawłowska J, Letcher PM, Kirk PM, Humber RA, Schüßler A, Wrzosek M, Muszewska A, Okrasińska A, Istel Ł, Gęsiorska A, Mungai P, Lateef AA, Rajeshkumar KC, Singh RV, Radek R, Walther G, Wagner L, Walker C, Wijesundara DSA, Papizadeh M, Dolatabadi S, Shenoy BD, Tokarev YS, Lumyong S, Hyde KD (2018b) Notes for genera: basal clades of Fungi (including Aphelidiomycota, Basidiobolomycota, Blastocladiomycota, Calcarisporiellomycota, Caulochytriomycota, Chytridiomycota, Entomophthoromycota, Glomeromycota, Kickxellomycota, Monoblepharomycota, Mortierellomycota, Mucoromycota, Neocallimastigomycota, Olpidiomycota, Rozellomycota and Zoopagomycota). Fungal Divers 92:43–129
Wikee S, Lombard L, Nakashima C, Motohashi K, Chukeatirote E, Cheewangkoon R, McKenzie EHC, Hyde KD, Crous PW (2013) A phylogenetic re-evaluation of Phyllosticta (Botryosphaeriales). Stud Mycol 76:1–29
Winter G (1885) Pilze-Ascomyceten. In GL Rabenhorst’s Kryptogamen-Flora von Deutschland, Oesterreich und der Schweiz 1:65–528
Wisitrassameewong K, Karunarathna SC, Thongklang N, Zhao R, Callac P, Moukha S, Férandon C, Chukeatirote E, Hyde KD (2012) Agaricus subrufescens: a review. Saudi J Biol Sci 19:131–146
Woudenberg JHC, Aveskamp MM, De Gruyter J, Spiers AG, Crous PW (2009) Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia 22:56–62
Xavier KV, KC AN, Crous PW, Groenewald JZ, Vallad GE (2019) Dwiroopa punicae sp. nov. (Dwiroopaceae fam. nov., Diaporthales), associated with leaf spot and fruit rot of pomegranate (Punica granatum). Fungal Systematics and Evolution 4: 33–41
Yang CS, Korf RP (1985) A monograph of the genus Tricharina and of a new, segregate genus, Wilcoxina (Pezizales). Mycotaxon 24:467–531
Yang Q, Fan XL, Crous PW, Liang YM, Tian CM (2015) Cytospora from Ulmus pumila in northern china. Mycol Prog 14:74
Yang J, Liu JK, Hyde KD, Jones EBG, Liu ZY (2017) Two new species in Fuscosporellaceae from freshwater habitats in Thailand. Mycosphere 8:1893–1903
Yang J, Liu JK, Hyde KD, Jones EBG, Liu ZY (2018a) New species in Dictyosporium, new combinations in Dictyocheirospora and an updated backbone tree for Dictyosporiaceae. MycoKeys 36:83–105
Yang J, Liu NG, Liu JK, Hyde KD, Jones EBG, Liu ZY (2018b) Phylogenetic placement of Cryptophiale, Cryptophialoidea, Nawawia,Neonawawia gen. nov. and Phialosporostilbe. Mycosphere 9:1132–1150
Yang CL, Xu XL, Wanasinghe DN, Jeewon R, Phookamsak R, Liu YG, Liu LJ, Hyde KD (2019a) Neostagonosporella sichuanensis gen. et sp. nov. (Phaeosphaeriaceae, Pleosporales) on Phyllostachys heteroclada (Poaceae) from Sichuan Province, China. MycoKeys 46:119–150
Yang MX, Wang XY, Liu D, Zhang YY, Li LJ, Yin AC, Scheidegger C, Wang LS (2019b) New species and records of Pyxine (Caliciaceae) in China. MycoKeys 45:93–109
Zelski SE, Raja HA, Miller AN, Shearer CA (2015) Conioscypha peruviana sp. nov., its phylogenetic placement based on 28S rRNA gene, and a report of Conioscypha gracilis from Peru. Mycoscience 56:319–325
Zhang Y, Schoch CL, Fournier J, Crous PW, de Gruyter J, Woudenberg JHC, Hirayama K, Tanaka K, Pointing SB, Spatafora JW, Hyde KD (2009) Multi-locus phylogeny of the Pleosporales: a taxonomic, ecological and evolutionary re-evaluation. Stud Mycol 64:85–102
Zhang Y, Crous PW, Schoch CL, Bahkali AH, Guo LD, Hyde KD (2011) A molecular, morphological and ecological re-appraisal of Venturiales—a new order of Dothideomycetes. Fungal Divers 51:249–277
Zhang Y, Crous PW, Schoch CL, Hyde KD (2012) Pleosporales. Fungal Divers 53:1–221
Zhang Y, Fournier J, Phookamsak R, Bakhali AH, Hyde KD (2013) Halotthiaceae fam. nov. (Pleosporales) accommodates the new genus Phaeoseptum and several other aquatic genera. Mycologia 40:107–110
Zhang Y, Zhang J, Wang Z, Fournier J, Crous PW, Zhang X, Li W, Ariyawansa HA, Hyde KD (2014) Neotypification and phylogeny of Kalmusia. Phytotaxa 176:64–173
Zhang T, Deng X, Yu Y, Zhang M, Zhang Y (2016) Pseudochaetosphaeronema ginkgonis sp. nov., an endophyte isolated from Ginkgo biloba. Int J Syst Evol Microbiol 66:4377–4381
Zhang JF, Liu JK, Hyde KD, Yang W, Liu ZY (2017) Fungi from Asian Karst formations II. Two new species of Occultibambusa (Occultibambusaceae, Dothideomycetes) from Karst landforms of China. Mycosphere 8:550–559
Zhang KK, Hongsanan S, Tennakoon DS, Tian SL, Xie N (2019a) Phaeosphaeria chinensis sp. nov. (Phaeosphaeriaceae) with an asexual/sexual morph connection from GuangDong Province, China. Phytotaxa 419:28–38
Zhang SN, Abdel-Wahab MA, Jones EBG, Hyde KD, Liu JK (2019b) Additions to the genus Savoryella (Savoryellaceae), with the asexual morphs Savoryella nypae comb. nov. and S. sarushimana sp. nov. Phytotaxa 408:195–207
Zhao GZ, Liu X, Wu W (2007) Helicosporous hyphomycetes from China. Fungal Divers 26:313–524
Zhao Q, Zhang XL, Li SH, Chai HM, Bahkali AH, Hyde KD (2016) New species and records of saddle fungi (Helvella, Helvellaceae) from Jiuzhaigou Natural Reserve, China. Mycoscience 57:422–430
Zhuang WY, Zeng ZQ (2017) Cocoonihabitus sinensis gen. et sp. nov. on remaining leaf veins of cocoons in a new family (Cocoonihabitaceae fam. nov.) of Hypocreales. Mycosystema 36:1591–1598
Acknowledgements
The authors would like to thank Yunnan Provincial Key Programs of Yunnan Eco-friendly Food International Cooperation Research Center Project under Grant 2019ZG00908 and Key Research Program of Frontier Sciences “Response of Asian mountain ecosystems to global change”, CAS, Grant No. QYZDY-SSW-SMC014”. We also thank to the director Jun-Bo Yang and Plant Germplasm and Genomics Center in Germplasm Bank of Wild Species, Kunming Institute of Botany for the molecular laboratory support. Kevin D. Hyde thanks the 2019 high-end foreign expert introduction plan to Kunming Institute of Botany (Granted by the Ministry of Science and Technology of the People’s Republic of China, Grant Number G20190139006), Thailand Research Grants entitled Biodiversity, phylogeny and role of fungal endophytes on above parts of Rhizophora apiculata and Nypa fruticans (Grant No. RSA5980068), the future of specialist fungi in a changing climate: baseline data for generalist and specialist fungi associated with ants, Rhododendron species and Dracaena species (Grant No. DBG6080013), Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion (Grant No. RDG6130001). Kevin D. Hyde also thanks Chiang Mai University for the award of visiting Professor. The authors extend their appreciation to the International Scientific Partnership Program ISPP at King Saud University for funding this research work through ISPP#0089. Rungtiwa Phookamsak thanks CAS President’s International Fellowship Initiative (PIFI) for young staff (Grant No. Y9215811Q1), the Yunnan Provincial Department of Human Resources and Social Security (Grant No. Y836181261), and National Science Foundation of China (NSFC) Project Code 31850410489 (Grant No. Y81I982211) for financial support. Dr. Shaun Pennycook and Prof Eric H.C. McKenzie are thanked for his essential nomenclatural review. Rajesh Jeewon thanks Mae Fah Luang University for the award of a Visiting Scholar and University of Mauritius for research support. Jian-Kui Liu thanks the National Natural Science Foundation of China (NSFC 31600032). Chaynard Phukhamsakda would like to thank the Royal Golden Jubilee PhD Program under Thailand Research Fund (RGJ) for a personal grant to C. Phukhamsakda (The Scholarship No. PHD/0020/2557 to study towards a Ph.D.). This research work was partially supported by Chiang Mai University. Ausana Mapook thanks to Research and Researchers for Industries (RRI) under Thailand Research Fund for a personal Grant (PHD57I0012), with the German Academic Exchange Service (DAAD) for a joint TRF-DAAD (PPP 2017–2018) academic exchange grant to K.D. Hyde and M. Stadler. Witoon Purahong and Tesfaye Wubet are thanked for funding support of Molecular work and also thanks to Katalee Jariyavidyanont, Maitree Malaithong and Benjawan Tanunchai for their valuable help. Saowaluck Tibpromma would like to thank the International Postdoctoral Exchange Fellowship Program (Number Y9180822S1), CAS President’s International Fellowship Initiative (PIFI) (Number 2020PC0009), China Postdoctoral Science Foundation and Yunnan Human Resources and Social Security Department Foundation for funding her postdoctoral research. V.V. Sarma would like to thank SERB, Department of Science and Technology, Government of India, for funding a project (SERB/SB/SO/PS/18/2014 dt.19.5.2015) and Ministry of Earth Sciences (MOES), Govt. of India for funding a project (Sanction order: MOES/36/OO1S/Extra/40/2014/PC-IV dt. 14.01.2015); the Department of Biotechnology, Pondicherry University for facilities; forest departments of Andaman and Nicobar Islands and Tamil Nadu, India are thanked for providing permission to collect samples. M. Niranjan thanks SERB, Govt. of India for a fellowship and B. Devadatha thanks MOES, Govt. of India for a fellowship. Napalai Chaiwan would like to thank the Thailand Research Fund (PHD60K0147). Danushka S. Tennakoon would like to thank Lakmali Dissanayake and Binu Samarakoon for their support. Dhanushka N. Wanasinghe would like to thank CAS President’s International Fellowship Initiative (PIFI) for funding his postdoctoral research (Number 2019PC0008). Peter E. Mortimer and Dhanushka N. Wanasinghe thank the National Science Foundation of China and the Chinese Academy of Sciences for financial support under the following Grants: 41761144055, 41771063 and Y4ZK111B01. Mingkwan Doilom would like to thank the 5th batch of Postdoctoral Orientation Training Personnel in Yunnan Province (grant no.: Y934283261) and the 64th batch of China Postdoctoral Science Foundation (grant no.: Y913082271). Amanda Lucia Alves acknowledges scholarships from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Ana Carla da Silva Santos acknowledges scholarships from the Conselho Nacional de Pesquisa (CNPq) and Patricia Vieira Tiago acknowledges financial support from the Pró-Reitoria de Pesquisa e Pós-Graduação (Propesq). Dan-Feng Bao thanks Dr. Zong-Long Luo and Prof. Dr. Hong-Yan Su for their available suggestions on fungal taxonomy as well as providing partial financial research support. Shi-Ke Huang thanks Prof. Dr. Ting-Chi Wen for partially support on research study. Danny Haelewaters was funded for fieldwork in Panama by the David Rockefeller Center for Latin American Studies (2017 Summer Research Travel Grant), Smithsonian Tropical Research Institute (2017 Short-Term Research Fellowship), Mycological Society of America (2016 Graduate Research Fellowship, 2017 Robert W. Lichtwardt Award), and through the Harvard University Herbaria (Fernald Fund). D. Haelewaters thanks W. Owen McMillan (Smithsonian Tropical Research Institute, Panama) and Edilma Gomez (Molecular Multi-User’s Lab, Panama) for providing lab space at STRI. Walter P. Pfliegler and Enikő Horváth are deeply indebted to Matthias Sipiczki (University of Debrecen, Hungary) for his support for generations of yeasts researchers, as well as to Ida Miklós (University of Debrecen, Hungary) for a continuous support for yeast studies and to Anita Csabainé Oláh (University of Debrecen, Hungary) for excellent technical support. Alexandra Imre was supported by the ÚNKP-19-3-I-234 New National Excellence Program of the Ministry of Human Capacities of Hungary. Walter P. Pfliegler, Enikő Horváth, and Alexandra Imre are deeply thankful to Gábor Péter for his comments on yeast taxonomy. Walter P. Pfliegler was supported by the Albert Szent-Györgyi Young Investigator Award. Kunhiraman C. Rajeshkumar thanks SERB, Department of Science and Technology, Government of India for providing financial support under the Project YSS/2015/001590 and Dr. Prashant K. Dhakephalkar, Director, Agharkar Research Institute for providing the facility. Sanjay K. Singh and Shiv Mohan Singh thank Dr. Prashant K. Dhakephalkar, Director, Agharkar Research Institute and Head, Department of Botany, Banaras Hindu University (BHU), Varanasi (UP) for providing necessary facilities. Shiwali Rana thanks SP Pune University and UGC New Delhi for Fellowship (JRF). Kunthida Phutthacharoen would like to thank the Royal Golden Jubilee PhD Program under Thailand Research Fund (RGJ) No. PHD/0002/2560. Saranyaphat Boonmee would like to thank the Thailand Research Fund (No. TRG6180001) and Plant Genetic Conservation Project under the Royal Initiation of Her Royal Highness Princess Maha Chakri Sirindhorn-Mae Fah Luang University. Qi Zhao and Ming Zeng are supported by the open research project of “Cross-Cooperative Team” of the Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Science, and The Biodiversity Survey and Assessment Project of the Ministry of Ecology and Environment, China (2019HJ2096001006). Subodini N. Wijesinghe offers her profound gratitude to Dr. Samantha C. Karunarathne for financial support on molecular work under the National Science Foundation of China (NSFC) Project Code: 31750110478 as well as Prof. Dr. Yong Wang, Dr. Udeni Jayalal and Achala R. Rathnayaka for their valuable suggestions. Renato Lúcio Mendes Alvarenga and Tatiana Baptista Gibertoni acknowledge Ailton Matheus for the specimen, Pós-Graduação em Biologia de Fungos (UFPE, Brazil) for support, CNPq (PQ 307601/2015-3) for financing this research and CAPES and CNPq for the PhD scholarship of RLM Alvarenga. Wei Dong would like to thank Huang Zhang for supporting this work under the National Natural Science Foundation of China (Project ID: NSF 31500017). Jing Yang would like to thank Prof. Zuoyi Liu for his support and great help on the lab work.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hyde, K.D., Dong, Y., Phookamsak, R. et al. Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100, 5–277 (2020). https://doi.org/10.1007/s13225-020-00439-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13225-020-00439-5