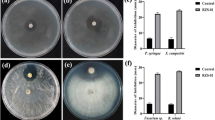
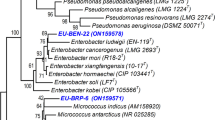
Abstract
Bacterial leaf streak (BLS) caused by Xanthomonas oryzae pv. oryzicola (Xoc), impacts the production of rice. However, several rice cultivars displayed resistance to Xoc in the field, but scarce information is available about the role of endophytic microbiota in disease resistance. In the present study, the endophytic bacterial communities of resistant and susceptible rice cultivars “CG2” and “IR24”, respectively, were analyzed using high throughput 16S rRNA gene amplified sequencing and culture dependent method was further used for bacterial isolation. A total of 452,716 high-quality sequences representing 132 distinct OTUs (Proteobacteria, Actinobacteria, Bacteroidetes, and Firmicutes) and 46 isolates of 16 genera were explored from rice leaves infected with Xoc. Community diversity of endophytic bacteria were higher in the leaves of the resistant cultivars compared to susceptible cultivars upon Xoc infection. Strikingly, this diversity might contribute to natural defense of the resistant cultivar against pathogen. Pantoea, which is pathogen antagonist, was frequently detected in two cultivars and higher abundance were recorded in resistant cultivars. Different abundance genus includes endophytic isolates with marked antagonistic activity to Xoc. The increased proportions of antagonistic bacteria, may contribute to resistance of rice cultivar against Xoc and the Pantoea genus was recruited by Xoc infection play a key role in suppressing the development of BLS disease in rice. Taken together, this work reveals the association between endophytic bacteria and BLS resistance in rice and identification of antagonism-Xoc bacterial communities in rice.






Similar content being viewed by others
References
Ahmed A, Munir S, He P, Li Y, He P, Yixin W, He Y (2020) Biocontrol arsenals of bacterial endophyte: an imminent triumph against clubroot disease. Microbiol Res 241:126565
Aurora S-G, Luisa AA, Espinosa-García J, González C, Kathleen T (2014) Diversity and communities of foliar endophytic fungi from different agroecosystems of Coffea arabica L. in two regions of Veracruz, Mexico. PLoS ONE 9:e98454
Bahroun A, Jousset A, Mhamdi R, Mrabet M, Mhadhbi H (2018) Anti-fungal activity of bacterial endophytes associated with legumes against Fusarium solani: assessment of fungi soil suppressiveness and plant protection induction. Appl Soil Ecol 124:131–140
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Cai Q, Zhou G, Ahmed W, Cao Y, Zhao M, Li Z, Zhao Z (2021) Study on the relationship between bacterial wilt and rhizospheric microbial diversity of flue-cured tobacco cultivars. Eur J Plant Pathol 160(2):265–276
Caporaso JG, Kuczynski J, Stombaugh J et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Dina B, Mamtaj S (2020) Endophytic microorganisms: colonization, plant-microbe interaction, diversity and their bioprospecting. Res J Biotechnol 15:151–179
Durán P, Thiergart T, Garrido-Oter R, Agler M, Kemen E, Schulze-Lefert P, Hacquard S (2018) Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 175:973–983
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Edwards J, Johnson C, Santos-Medellín C et al (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci 112:E911–E920
Enya J, Shinohara H, Yoshida S, Tsukiboshi T, Negishi H, Tsushima SS (2007) Culturable leaf-associated bacteria on tomato plants and their potential as biological control agents. Microb Ecol 53:524–536
Etesami H, Alikhani HA (2016) Evaluation of Gram-positive rhizosphere and endophytic bacteria for biological control of fungal rice (Oryzia sativa L.) pathogens. Eur J Plant Pathol 147:7–14
Felix MB, Fabrizia G, Alberto P, Edgloris M, Vittorio V (2018) Bacterial microbiota of rice roots: 16S-based taxonomic profiling of endophytic and rhizospheric diversity, endophytes isolation and simplified endophytic community. Microorganisms 6:14
Finkel OM, Salas-González I, Castrillo G, Law TF, Conway JM, Jones CD, Dangl JL (2019) Root development is maintained by specific bacteria–bacteria interactions within a complex microbiome. bioRxiv 645655
Fitzpatrick CR, Lu-Irving P, Copeland J et al (2018) Chloroplast sequence variation and the efficacy of peptide nucleic acids for blocking host amplification in plant microbiome studies. Microbiome 6:1–10
Free A (2018) Diversity-function relationships in natural, applied, and engineered microbial ecosystems. Adv Appl Microbiol 105:131–189
Fu L, Li H, Wei L et al (2018) Antifungal and biocontrol evaluation of four Lysobacter strains against clubroot disease. Indian J Microbiol 58:353–359
Ghoul M, Mitri S (2016) The ecology and evolution of microbial competition. Trends Microbiol 24:833–845
Gómez-Acata S, Esquivel-Ríos I, Pérez-Sandoval MV et al (2017) Bacterial community structure within an activated sludge reactor added with phenolic compounds. Appl Microbiol Biotechnol 101:3405–3414
Hamady M, Lozupone C, Knight R (2010) Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4:17–27
Hardoim PR, Andreote FD, Reinhold-Hurek B, Sessitsch A, van Overbeek LS, van Elsas JD (2011) Rice root-associated bacteria: insights into community structures across 10 cultivars. FEMS Microbiol Ecol 77:154–164
Harrison JG, Parchman TL, Cook D, Gardner DR, Forister ML (2018) A heritable symbiont and host-associated factors shape fungal endophyte communities across spatial scales. J Ecol 106:2274–2286
Hastuti RD, Lestari Y, Suwanto A, Saraswati R (2012) Endophytic Streptomyces spp. as biocontrol agents of rice bacterial leaf blight pathogen (Xanthomonas oryzae pv. oryzae). HAYATI J Biosci 19:155–162
Hossain MI, Sadekuzzaman M, Ha SD (2017) Probiotics as potential alternative biocontrol agents in the agriculture and food industries: a review. Food Res Int 100:63–73
Hu B, Wang W, Ou SJ et al (2015) Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat Genet 47:834–838
Huang X, Ren J, Li P, Feng S, Dong P, Ren M (2020) Potential of microbial endophytes to enhance the resistance to postharvest diseases of fruit and vegetables. J Sci Food Agric 101:1744–1757
Janssen S et al (2018) Phylogenetic placement of exact amplicon sequences improves associations with clinical information. Msystems 3:e00021-e118
Ji GH, Wei LF, He YQ, Wu YP, Bai XH (2008) Biological control of rice bacterial blight by Lysobacter antibioticus strain 13–1. Biol Control 45:288–296
Jiang YJ, Liang YT, Li CM et al (2016) Crop rotations alter bacterial and fungal diversity in paddy soils across East Asia. Soil Biol Biochem 95:250–261
Kanugala S, Kumar CG, Reddy RHK et al (2019) Chumacin-1 and Chumacin-2 from Pseudomonas aeruginosa strain CGK-KS-1 as novel quorum sensing signaling inhibitors for biocontrol of bacterial blight of rice. Microbiol Res 228:126301
Karmakar R, Bindiya S, Hariprasad P (2019) Convergent evolution in bacteria from multiple origins under antibiotic and heavy metal stress, and endophytic conditions of host plant. Sci Total Environ 650:858–867
Kim H, Lee Y-H (2020) The rice microbiome: a model platform for crop holobiome. Phytobiomes J 4:5–18
Kunda P, Dhal PK, Mukherjee A (2018) Endophytic bacterial community of rice (Oryza sativa L.) from coastal saline zone of West Bengal: 16S rRNA gene based metagenomics approach. Meta Gene 18:79–86
Li S-B, Fang M, Zhou R-C, Huang J (2012) Characterization and evaluation of the endophyte Bacillus B014 as a potential biocontrol agent for the control of Xanthomonas axonopodis pv. dieffenbachiae–induced blight of Anthurium. Biol Control 63:9–16
Li B, Liu BP, Shan CL et al (2013) Antibacterial activity of two chitosan solutions and their effect on rice bacterial leaf blight and leaf streak. Pest Manag Sci 69:312–320
Liu Y, Wang P, Pan G et al (2016) Functional and structural responses of bacterial and fungal communities from paddy fields following long-term rice cultivation. J Soils Sediments 16:1460–1471
Liu Y, Xu P, Yang F et al (2019) Composition and diversity of endophytic bacterial community in seeds of super hybrid rice ‘Shenliangyou 5814’(Oryza sativa L.) and its parental lines. Plant Growth Regul 87:257–266
Louca S, Jacques S, Pires A et al (2016) High taxonomic variability despite stable functional structure across microbial communities. Nat Ecol Evol 1:1–12
Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL (2013) Practical innovations for high-throughput amplicon sequencing. Nat Methods 10:999–1002
Lutz KA, Wang W, Zdepski A, Michael TP (2011) Isolation and analysis of high quality nuclear DNA with reduced organellar DNA for plant genome sequencing and resequencing. BMC Biotechnol 11:1–9
Ma Z, Liu C, Zhang Y, Qin G, Li Y, Tan J, Huang D (2015) Elevation of disease index and identification of resistance to rice bacterial leaf streak in field. Chin J Trop Agric 35:113–117
Majeed A, Muhammad Z, Ahmad H (2018) Plant growth promoting bacteria: role in soil improvement, abiotic and biotic stress management of crops. Plant Cell Rep 37:1599–1609
Marag PS, Suman A (2018) Growth stage and tissue specific colonization of endophytic bacteria having plant growth promoting traits in hybrid and composite maize (Zea mays L.). Microbiol Res 214:101–113
Matsumoto H, Fan X, Wang Y et al (2021) Bacterial seed endophyte shapes disease resistance in rice. Nat Plants 7:60–72
Mendes R, Garbeva P, Raaijmakers JM (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37:634–663
Mohammadi P, Tozlu E, Kotan R, Kotan MŞ (2017) Potential of some bacteria for biological control of postharvest citrus green mould caused by Penicillium digitatum. Plant Prot Sci 53:1–10
Munir S, Li Y, He P et al (2020) Core endophyte communities of different citrus varieties from citrus growing regions in China. Sci Rep 10:1–12
Munir S, Ahmed A, Li YM et al (2021) The hidden treasures of citrus: finding huanglongbing cure where it was lost. Crit Rev Biotechnol. https://doi.org/10.1080/07388551.2021.1942780
Niño-Liu D, Ronald PC, Bogdanove AJ (2006) Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol Plant Pathol 7:303–324
Pan X, Xu S, Wu J et al (2017) Screening and characterization of Xanthomonas oryzae pv. oryzae strains with resistance to pheazine-1-carboxylic acid. Pestic Biochem Physiol 145:8–14
Peng X, Xie J, Li W, Xie H, Cai Y, Ding X (2021) Comparison of wild rice (Oryza longistaminata) tissues identifies rhizome-specific bacterial and archaeal endophytic microbiomes communities and network structures. PLoS ONE 16:e0246687
Rana KL, Kour D, Kaur T et al (2020) Endophytic microbes from diverse wheat genotypes and their potential biotechnological applications in plant growth promotion and nutrient uptake. Proc Natl Acad Sci India Sect B Biol Sci 90:1–11
Reshmi U, Pious T (2015) Root-associated bacterial endophytes from Ralstonia solanacearum resistant and susceptible tomato cultivars and their pathogen antagonistic effects. Front Microbiol 6:255
Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK (2012) FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 30:460
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Sahu KP, Kumar A, Patel A, Kumar M, Gopalakrishnan S, Prakash G (2020) Rice blast lesions: an unexplored phyllosphere microhabitat for novel antagonistic bacterial species against Magnaporthe oryzae. Microb Ecol 81:1–15
Sakulkoo W, Osés-Ruiz M, Garcia EO et al (2018) A single fungal MAP kinase controls plant cell-to-cell invasion by the rice blast fungus. Science 359:1399–1403
Salazar C, Rand J (2020) Pesticide use, production risk and shocks. The case of rice producers in Vietnam. J Environ Manag 253:109705
Samain E, Van Tuinen D, Jeandet P, Aussenac T, Selim S (2017) Biological control of septoria leaf blotch and growth promotion in wheat by Paenibacillus sp. strain B2 and Curtobacterium plantarum strain EDS. Biol Control 114:87–96
Selim HM, Gomaa NM, Essa AM (2017) Application of endophytic bacteria for the biocontrol of Rhizoctonia solani (Cantharellales: ceratobasidiaceae) damping-off disease in cotton seedlings. Biocontrol Sci Tech 27:81–95
Shaver JM, Oldenburg DJ, Bendich AJ (2006) Changes in chloroplast DNA during development in tobacco, Medicago truncatula, pea, and maize. Planta 224:72–82
Shittu HO, Castroverde DC, Nazar RN, Robb J (2009) Plant-endophyte interplay protects tomato against a virulent Verticillium. Planta 229:415–426
Song L, Xie K (2020) Engineering CRISPR/Cas9 to mitigate abundant host contamination for 16S rRNA gene-based amplicon sequencing. Microbiome 8:1–15
Vannier N, Agler M, Hacquard S (2019) Microbiota-mediated disease resistance in plants. PLoS Pathog 15:e1007740
Walitang DI, Kim CG, Kim K, Kang Y, Kim YK, Sa T (2018) The influence of host genotype and salt stress on the seed endophytic community of salt-sensitive and salt-tolerant rice cultivars. BMC Plant Biol 18:1–16
Wan X, Yang J, Ahmed W, Liu Q, Wang Y, Wei L, Ji G (2021) Functional analysis of pde gene and its role in the pathogenesis of Xanthomonas oryzae pv. oryzicola. Infect Genet Evol 94:105008
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Wu N, Zhang L, Ren Y, Wang X (2020) Rice black-treaked dwarf virus: from multiparty interactions among plant–virus–vector to intermittent epidemics. Mol Plant Pathol 21:1007–1019
Yamamoto K, Shiwa Y, Ishige T et al (2018) Bacterial diversity associated with the rhizosphere and endosphere of two halophytes: Glaux maritima and Salicornia europaea. Front Microbiol 9:2878
Yan X, Wang Z, Mei Y et al (2018) Isolation, diversity, and growth-promoting activities of endophytic bacteria from tea cultivars of Zijuan and Yunkang-10. Front Microbiol 9:1848
Yang F, Zhang J, Zhang HY et al (2020a) Bacterial blight induced shifts in endophytic microbiome of rice leaves and the enrichment of specific bacterial strains with pathogen antagonism. Front Plant Sci 11:963
Yang J, Wang X, Wang Y, Liu Q, Wang Y, Wei L, Ji G (2020b) Pathotypes differentiation of Xanthomonas oryzae pv. oryzicola and identification of rice varieties resistant to bacterial leaf streak in Yunnan Province. Acta Phytopathol Sin 50:218–227
Zhang R, Liu YF, Luo CP et al (2012) Bacillus amyloliquefaciens LX-11, a potential biocontrol agent against rice bacterial leaf streak. J Plant Pathol 94:609–619
Zhang J, Zhang C, Yang J et al (2019) Insights into endophytic bacterial community structures of seeds among various Oryza sativa L. rice genotypes. J Plant Growth Regul 38:93–102
Zhang J, Wei L, Yang J, Ahmed W, Wang Y, Fu L, Ji G (2020) Probiotic consortia: reshaping the rhizospheric microbiome and its role in suppressing root-rot disease of Panax notoginseng. Front Microbiol 11:701
Acknowledgements
We thank the Agricultural Institute of Dehong State for providing rice seeds and the Dr. Fenghuan Yang of Chinese Academy of Agricultural Sciences for technical support.
Funding
This research was funded by the National Natural Science Foundation of China (32060601) and the National Key R&D Program of China (2019YFD1002000) and the Yunnan Ten Thousand Talents Plan leading talents of industrial technology project (YNWR-CYJS-2019-046). PhD research startup foundation of Chuxiong Normal University, China (BSQD2105).
Author information
Authors and Affiliations
Contributions
GHJ and LFW conceived and designed the experiments. JY, XYW, and XW performed the experiments. JY, and ZLD collect and analyzed the data. JY, and SM wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declared that they have no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, J., Dai, Z., Wan, X. et al. Insights into the relevance between bacterial endophytic communities and resistance of rice cultivars infected by Xanthomonas oryzae pv. oryzicola. 3 Biotech 11, 434 (2021). https://doi.org/10.1007/s13205-021-02979-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-021-02979-2