Abstract
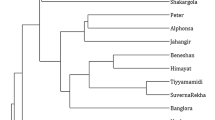
Garden asparagus (Asparagus officinalis L.) is known as a valuable genetic resource for both vegetables and medicinal purposes. However, little is known about the organization and diversity of repetitive DNA sequences, and the usefulness of such resources in analyzing genetic diversity and DNA fingerprinting in A. officinalis cultivars. In this study, a large-scale genome-wide identification of microsatellite molecular markers in A. officinalis genome was performed. Genetic diversity and DNA fingerprinting of 24 cultivars were carried out using the information obtained from polymorphic simple sequence repeats (SSRs) markers. The polymorphism information content were found in the range of 0.21–0.43 with an average of 0.35. The genetic distance of 24 cultivars were ranged from 0.26 to 1.07, with an average value of 0.75, indicating relatively high genetic diversity among these accessions. The dendrogram constructed by unweighted pair group method with arithmetic mean clustering method classified these cultivars into two discrete groups revealing the genetic relatedness and diversity among these cultivars. Furthermore, the 24 cultivars were fingerprinted using three SSR primers. Our results showed that the primer pair Asp-SSR-2-C6 enabled the identification of 12 cultivars, whereas Asp-SSR-3-C10 and Asp-SSR-6-C4 primers could differentiate 13 and 10 cultivars, respectively. The present study deciphered the reliability of SSR markers for DNA fingerprinting and genetic diversity analysis in asparagus cultivars, which will facilitate the conservation, breeding and future genetic studies.





Similar content being viewed by others
Data availability
All the data which is generated and analyzed during our study are included within the article and supplementary materials.
References
Bancroft I, Morgan C, Fraser F, Higgins J, Wells R, Clissold L, Baker D, Long Y, Meng J, Wang X (2011) Dissecting the genome of the polyploid crop oilseed rape by transcriptome sequencing. Nat Biotechnol 29(8):762–766
Boriah S, Chandola V, Kumar V Similarity measures for categorical data: a comparative evaluation. In: Proceedings of the 2008 SIAM international conference on data mining, 2008. SIAM, pp 243–254
Buschiazzo E, Gemmell NJ (2006) The rise, fall and renaissance of microsatellites in eukaryotic genomes. BioEssays 28(10):1040–1050
Cavagnaro PF, Senalik DA, Yang L, Simon PW, Harkins TT, Kodira CD, Huang S, Weng Y (2010) Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genom 11(1):1–18
Celik I, Gultekin V, Allmer J, Doganlar S, Frary A (2014) Development of genomic simple sequence repeat markers in opium poppy by next-generation sequencing. Mol Breed 34(2):323–334
Chase MW, Reveal JL, Fay MF (2009) A subfamilial classification for the expanded asparagalean families Amaryllidaceae, Asparagaceae and Xanthorrhoeaceae. Bot J Linn Soc 161(2):132–136
Chen H, Guo A, Wang J, Gao J, Zhang S, Zheng J, Huang X, Xi J, Yi K (2020) Evaluation of genetic diversity within asparagus germplasm based on morphological traits and ISSR markers. Physiol Mol Biol Plants 26(2):305–315
Duan Y, Liu J, Xu J, Bian C, Duan S, Pang W, Hu J, Li G, Jin L (2019) DNA fingerprinting and genetic diversity analysis with simple sequence repeat markers of 217 potato cultivars (Solanum tuberosum L.) in China. Am. J. Potato Res. 96(1):21–32
Gao Y, Zhu R, Liu C, Li W, Jiang H, Li C, Yao B, Hu G, Chen Q (2009) Establishment of molecular ID in soybean varieties in Heilongjiang, China. Acta Agron Sin 35:211–218
Garg G, Sharma V (2015) Assessment of fatty acid content and genetic diversity in Eruca sativa (L.) (Taramira) using ISSR markers. Biomass Bioenerg 76:118–129
Golestan Hashemi F, Rafii M, Razi Ismail M, Mohamed M, Rahim H, Latif M, Aslani F (2015) Opportunities of marker-assisted selection for rice fragrance through marker–trait association analysis of microsatellites and gene-based markers. Plant Biol 17(5):953–961
Gur-Arie R, Cohen CJ, Eitan Y, Shelef L, Hallerman EM, Kashi Y (2000) Simple sequence repeats in Escherichia coli: abundance, distribution, composition, and polymorphism. Genome Res 10(1):62–71
Han B, Wang C, Tang Z, Ren Y, Li Y, Zhang D, Dong Y, Zhao X (2015) Genome-wide analysis of microsatellite markers based on sequenced database in Chinese spring wheat (Triticum aestivum L.). PLoS One 10(11):e0141540
Harkess A, Zhou J, Xu C, Bowers JE, Van der Hulst R, Ayyampalayam S, Mercati F, Riccardi P, McKain MR, Kakrana A (2017) The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat Commun 8(1):1–10
Huo N, Lazo GR, Vogel JP, You FM, Ma Y, Hayden DM, Coleman-Derr D, Hill TA, Dvorak J, Anderson OD (2008) The nuclear genome of Brachypodium distachyon: analysis of BAC end sequences. Funct Integr Genom 8(2):135–147
Ii Y, Uno Y, Kanechi M, Inagaki N (2012) Screening of sex in asparagus at early growth stages. Horttechnology 22(1):77–82
Jena SN, Verma S, Nair KN, Srivastava AK, Misra S, Rana TS (2015) Genetic diversity and population structure of the mangrove lime (Merope angulata) in India revealed by AFLP and ISSR markers. Aquat Bot 120:260–267
Jiang C, Sink KC (1997) RAPD and SCAR markers linked to the sex expression locus M in asparagus. Euphytica 94(3):329–333
Kanno A, Yokoyama J (2011) Asparagus. Wild crop relatives: genomic and breeding resources. Springer, New York, pp 23–42
Kota R, Varshney R, Prasad M, Zhang H, Stein N, Graner A (2008) EST-derived single nucleotide polymorphism markers for assembling genetic and physical maps of the barley genome. Funct Integr Genom 8(3):223–233
Lee JW, Lee JH, Yu IH, Gorinstein S, Bae JH, Ku YG (2014) Bioactive compounds, antioxidant and binding activities and spear yield of Asparagus officinalis L. Plant Foods Hum Nutr 69(2):175–181
Li S-F, Gao W-J, Zhao X-P, Dong T-Y, Deng C-L, Lu L-D (2014) Analysis of transposable elements in the genome of Asparagus officinalis from high coverage sequence data. PLoS ONE 9(5):e97189
Li S, Zhang G, Li X, Wang L, Yuan J, Deng C, Gao W (2016) Genome-wide identification and validation of simple sequence repeats (SSRs) from Asparagus officinalis. Mol Cell Probes 30(3):153–160
Liu D, Guo X, Lin Z, Nie Y, Zhang X (2006) Genetic diversity of Asian cotton (Gossypium arboreum L.) in China evaluated by microsatellite analysis. Genet Resour Crop Evol 53(6):1145–1152
Lu C, Zou C, Zhang Y, Yu D, Cheng H, Jiang P, Yang W, Wang Q, Feng X, Prosper MA (2015) Development of chromosome-specific markers with high polymorphism for allotetraploid cotton based on genome-wide characterization of simple sequence repeats in diploid cottons (Gossypium arboreum L. and Gossypium raimondii Ulbrich). BMC Genom 16(1):1–12
Ma S, Han C, Zhou J, Hu R, Jiang X, Wu F, Tian K, Nie G, Zhang X (2020) Fingerprint identification of white clover cultivars based on SSR molecular markers. Mol Biol Rep 47(11):8513–8521
Mir RR, Zaman-Allah M, Sreenivasulu N, Trethowan R, Varshney RK (2012) Integrated genomics, physiology and breeding approaches for improving drought tolerance in crops. Theor Appl Genet 125(4):625–645
Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet 30(2):194–200
Musammilu K, Abdul-Muneer P, Gopalakrishnan A, Basheer V, Gupta H, Mohindra V, Lal KK, Ponniah A (2014) Identification and characterization of microsatellite markers for the population genetic structure in endemic red-tailed barb. Gonoproktopterus Curmuca Mol Biol Rep 41(5):3051–3062
Öztürk SC, Göktay M, Allmer J, Doğanlar S, Frary A (2018) Development of simple sequence repeat markers in hazelnut (Corylus avellana L.) by next-generation sequencing and discrimination of Turkish hazelnut cultivars. Plant Mol. Biol. Rep. 36(5):800–811
Panigrahi KK, Panigrahi P, Mohanty A, Mandal P, Satapathy B (2020) Development of new microsatellite markers for DNA fingerprinting pattern of black gram (Vigna mungo L. Hepper) and green gram (Vigna radiate L. Wilckzek). Genetika 52(3):1161–1179
Pickett BD, Karlinsey S, Penrod C, Cormier M, Ebbert MT, Shiozawa DK, Whipple C, Ridge PG (2016) SA-SSR: a suffix array-based algorithm for exhaustive and efficient SSR discovery in large genetic sequences. Bioinform Biol Insights 32(17):2707–2709
Polleys EJ, Freudenreich CH (2021) Homologous recombination within repetitive DNA. Curr Opin Genet Dev 71:143–153
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2(3):225–238
Qiu F, Guo L, Wen T-J, Liu F, Ashlock DA, Schnable PS (2003) DNA sequence-based „bar codes” for tracking the origins of expressed sequence tags from a maize cDNA library constructed using multiple mRNA sources. Plant Physiol 133(2):475–481
Ramakrishnan M, Ceasar SA, Duraipandiyan V, Al-Dhabi N, Ignacimuthu S (2016) Using molecular markers to assess the genetic diversity and population structure of finger millet (Eleusine coracana (L.) Gaertn.) from various geographical regions. Genet. Resour Crop Evol 63(2):361–376
Rana M, Arora K, Singh S, Singh AK (2013) Multi-locus DNA fingerprinting and genetic diversity in jute (Corchorus spp.) based on sequence-related amplified polymorphism. J Plant Biochem Biotechnol 22(1):1–8
Rohlf FJ (1998) NTSYSpc numerical taxonomy and multivariate analysis system version 2.0 user guide. Appl Biostat Inc, New York
Sim S-C, Durstewitz G, Plieske J, Wieseke R, Ganal MW, Van Deynze A, Hamilton JP, Buell CR, Causse M, Wijeratne S (2012) Development of a large SNP genotyping array and generation of high-density genetic maps in tomato. PLoS ONE 7(7):e40563
Singh A, Sinha B (2014) Asparagus racemosus and its phytoconstituents; an updated review. Asian J Biochem Pharm Res 4(4):230–240
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 109(6):1105–1114
Song GQ, Li MJ, Xiao H, Wang XJ, Tang RH, Xia H, Zhao CZ, Bi YP (2010) EST sequencing and SSR marker development from cultivated peanut (Arachis hypogaea L). Electron. J. Biotechnol. 13(3):7–8
Sorkheh K, Shiran B, Gradziel TM, Epperson B, Martínez-Gómez P, Asadi E (2007) Amplified fragment length polymorphism as a tool for molecular characterization of almond germplasm: genetic diversity among cultivated genotypes and related wild species of almond, and its relationships with agronomic traits. Euphytica 156(3):327–344
Taheri S, Lee Abdullah T, Yusop MR, Hanafi MM, Sahebi M, Azizi P, Shamshiri RRJM (2018) Mining and development of novel SSR markers using next generation sequencing (NGS) data in plants. Molecules 23(2):399
Umezawa T, Sakurai T, Totoki Y, Toyoda A, Seki M, Ishiwata A, Akiyama K, Kurotani A, Yoshida T, Mochida K (2008) Sequencing and analysis of approximately 40 000 soybean cDNA clones from a full-length-enriched cDNA library. DNA Res 15(6):333–346
Velvan S, Nagulendran K, Mahesh R, Begum V (2007) The chemistry, pharmacology and therapeutical applications of asparagus racemosus a review. Pharmacogn Rev 1(2):350–360
Wang J, Chen C, Na J-K, Yu Q, Hou S, Paull RE, Moore PH, Alam M, Ming R (2008) Genome-wide comparative analyses of microsatellites in papaya. Trop Plant Biol 1(3):278–292
Wei X, Wang L, Zhang Y, Qi X, Wang X, Ding X, Zhang J, Zhang X (2014) Development of simple sequence repeat (SSR) markers of sesame (Sesamum indicum) from a genome survey. Molecules 19(4):5150–5162
Wilkinson PA, Winfield MO, Barker GL, Allen AM, Burridge A, Coghill JA, Edwards KJ (2012) CerealsDB 2.0: an integrated resource for plant breeders and scientists. BMC Bioinform 13(1):1–6
Witkos TM, Krzyzosiak WJ, Fiszer A, Koscianska E (2018) A potential role of extended simple sequence repeats in competing endogenous RNA crosstalk. RNA Biol 15(11):1399–1409
Zhang L, Cai R, Yuan M, Tao A, Xu J, Lin L, Fang P, Qi J (2015) Genetic diversity and DNA fingerprinting in jute (Corchorus spp.) based on SSR markers. Crop J 3(5):416–422
Zhao C, Qiu J, Agarwal G, Wang J, Ren X, Xia H, Guo B, Ma C, Wan S, Bertioli DJ (2017) Genome-wide discovery of microsatellite markers from diploid progenitor species, Arachis duranensis and A. ipaensis, and their application in cultivated peanut (A. hypogaea). Front. Plant Sci. 8:1209
Zhu H, Senalik D, McCown B, Zeldin E, Speers J, Hyman J, Bassil N, Hummer K, Simon P, Zalapa J (2012) Mining and validation of pyrosequenced simple sequence repeats (SSRs) from American cranberry (Vaccinium macrocarpon Ait.). Theor. Appl. Genet. 124(1):87–96
Acknowledgements
The authors would like to thank the Shandong Ju Xin Yuan Agriculture Technology Co. Ltd and Shandong Yuncheng Jiuyuan Agriculture Technology Co. Ltd, Heze, China for providing plant materials for this study.
Funding
This work was supported by Shandong Agricultural Science and Technology Program (industrial upgrading of agricultural park) (2019YQ004), National Natural Science Foundation of China (31861143009), Taishan Scholar Project of Shandong Province (ts20190964), High-level Foreign Experts Introduction Program (GL20200123001), Agricultural scientific and technological innovation project of Shandong Academy of Agricultural Sciences (CXGC2018E13).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by N Ahmad, R Tian and J Lu. The study was conceptualized by S Zhao and X Wang. Formal analysis was conducted by G Li, J Sun, R Lin, and C Zhao. Plant materials were provided by C Zhou and H Chang. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ahmad, N., Tian, R., Lu, J. et al. DNA fingerprinting and genetic diversity analysis in Asparagus officinalis L. cultivars using microsatellite molecular markers. Genet Resour Crop Evol 70, 1163–1177 (2023). https://doi.org/10.1007/s10722-022-01493-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-022-01493-5