Abstract
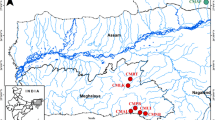
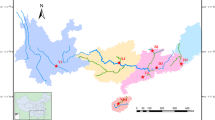
Gonoproktopterus curmuca is an endangered red tailed barb found in Southern part of Western Ghat, India. As a part of stock-specific, propagation assisted rehabilitation and management program, polymorphic microsatellites markers were used to study the genetic diversity and population structure of this species from the three River systems of Southern Western Ghats, such as Periyar River, the Chalakkudy River, and the Chaliyar River. From selected eight polymorphic microsatellite markers, the number of alleles per locus ranged from 2 to 8, and the average number of alleles among 3 populations ranged from 5.0 to 5.75. The mean observed (Hob) and expected (Hex) heterozygosity ranged from 0.5148 to 0.5360 and from 0.5996 to 0.6067, respectively. Significant deviations from Hardy–Weinberg Equilibrium expectation were found at majority of the loci (except Gcur MFW72 and Gcur MFW19) and in all three populations in which heterozygote deficits were apparent. The analysis of molecular variance indicates that the percent of variance among populations and within populations were 6.73 and 93.27, respectively. The pairwise FST values between populations indicate that there were significant deviations in genetic differentiations for the red-tailed barb populations from these three Rivers of the Western Ghats, India. The microsatellites methods reported a low degree of gene diversity and lack of genetic heterogeneity in the population of G. curmuca, which strongly emphasize the need of fishery management, conservation and rehabilitation of G. curmuca.




Similar content being viewed by others
Abbreviations
- AMOVA:
-
Analysis of molecular variance
- CAMP:
-
Conservation assessment management plan
- CHD:
-
Chalakkudy River
- CLR:
-
Chaliyar River
- DNA:
-
Deoxyribo nucleic acid
- FIS :
-
Coefficient of inbreeding or heterozygosity deficient
- FST :
-
Coefficient of genetic differentiation
- Hex :
-
Expected heterozygosity
- Hob :
-
Observed heterozygosity
- HWE:
-
Hardy–Weinberg equilibrium
- IUCN:
-
International Union for Conservation of Nature and Natural Resources
- Na:
-
Number of observed alleles
- NBFGR:
-
National Bureau of Fish Genetic Resources
- NCBI:
-
National Centre for Biotechnology Information
- Ne:
-
Number of expected alleles
- OD:
-
Optical density
- PCR:
-
Polymerase chain reaction
- PER:
-
Periyar River
- UPGMA:
-
Unweighted pair-group method with arithmetic mean
References
Jayaram KC (1999) The freshwater fishes of the Indian Region. Narendra Publishing House, Delhi
Talwar PK, Jhingran AG (1991) Inland fishes of India and adjacent countries. AA Balkema, Rotterdam
Shaji CP, Easa PS, Gopalakrishnan A (2000) Freshwater fish diversity of the Western Ghats. In: Ponniah AG, Gopalakrishnan A (eds) Endemic fish diversity of the Western Ghats. NBFGR-NATP Publication-1, National Bureau of Fish Genetic Resources, Lucknow, pp 33–55
Menon AGK, Remadevi K (1995) Hypselobarbus kurali (Pisces: Cyprinidae) a new large barb from the south western rivers of Peninsular India. J Bom Nat Hist Soc 92(3):389–393
Knight JDM, Rai A, D'Souza RKP (2013) On the identities of Barbus mussullah Sykes and Cyprinus curmuca Hamilton with notes on the status of Gobio canarensis Jerdon (Teleostei: Cyprinidae). Zootaxa 3750(3):201–215
Chhapgar BF, Manakadan R (2000) Ecology of Hill Streams of Western Ghats with special reference to fish community. US Fish and Wildlife Service & Bombay Natural History Society, Bombay
Daniels RJR (2002) Freshwater Fishes of Peninsular India. Universities Press (India) Private Limited, Hyderabad
Musammilu KK (2006) Molecular genetic characterization of endemic endemic red: tailed barb, Gonoproktopterus curmuca (Hamilton: Buchanan, 1807). PhD Thesis, Cochin University of Science and Technology, Cochin, p 268
CAMP (1998) Conservation Assessment and Management Plan (CAMP) for freshwater fishes of India 1997. Zoo Outreach Organization (ZOO) and National Bureau of Fish Genetic Resources (NBFGR), Lucknow, India. Zoo Outreach Organization, Coimbatore, India, p 156
Tautz D (1989) Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res 17:6463–6471
Litt M, Luty JA (1989) A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Hum Genet 4:397–401
Knapik EW, Goodman A, Ekker M, Chevrette M, Delgado J, Neuhauss S, Shimoda N, Driever W, Fishman MC, Jacob HJ (1998) A microsatellite genetic linkage map for zebrafish (Danio rerio). Nat Genet 18:338–343
Gupta A, Lal KK, Punia P, Singh RK, Mohindra V, Sah RS, Kumar R, Luhariya RK, Dwivedi AK, Masih P, Mishra RM, Jena JK (2013) Characterization of polymorphic microsatellite markers and genetic diversity in wild bronze featherback, Notopterus notopterus (Pallas, 1769). Mol Biol Rep 40(12):6625–6631
Kumla S, Doolgindachbaporn S, Sudmoon R, Sattayasai N (2012) Genetic variation, population structure and identification of yellow catfish, Mystus nemurus (C&V) in Thailand using RAPD, ISSR and SCAR marker. Mol Biol Rep 39(5):5201–5210
Matallanas B, Ochando MD, Alonso F, Callejas C (2013) Phylogeography of the white-clawed crayfish (Austropotamobius italicus) in Spain: inferences from microsatellite markers. Mol Biol Rep 40(9):5327–5338
An HS, Lee JW, Park JY, Jung HT (2013) Genetic structure of the Korean black scraper Thamnaconus modestus inferred from microsatellite marker analysis. Mol Biol Rep 40(5):3445–3456
Abdul-Muneer PM, Gopalakrishnan A, Musammilu KK, Mohindra V, Basheer VS, Lakra WS (2009) Genetic variation and population structure of endemic yellow catfish, Horabagrus brachysoma (Bagridae) among three populations of Western Ghat region using RAPD and microsatellite markers. Mol Biol Rep 36(7):1779–1791
Abdul-Muneer PM, Gopalakrishnan A, Sivanandan R, Basheer VS, Ponniah AG (2011) Genetic variation and phylogenetic relationships between two species of yellow catfish, Horabagrus brachysoma and H. nigricollaris using RAPD and microsatellite markers. Mol Biol Rep 38(4):2225–2232
Abdul-Muneer PM, Sivanandan R, Gopalakrishnan A, Basheer VS, Musammilu KK, Ponniah AG (2011) Identification and characterization of RAPD and microsatellite markers for genetic variation analysis in critically endangered yellow catfish, Horabagrus nigricollaris. Biochem Genet 49(1):83–95
Abdul-Muneer PM, Gopalakrishnan A, Musammilu KK, Basheer VS, Mohindra V, Lal KK, Padmakumar KG, Ponniah AG (2012) Comparative assessment of genetic variability in the populations of endemic and endangered yellow catfish, Horabagrus brachysoma (Hoarabagridae) based on allozymes, RAPD and microsatellite markers. Biochem Genet 50(3–4):192–212
Mohindra V, Singh A, Barman AS, Tripathi R, Sood N, Lal KK (2012) Development of EST derived SSRs and SNPs as a genomic resource in Indian catfish, Clarias batrachus. Mol Biol Rep 39(5):5921–5931
Gopalakrishnan A, Musammilu KK, Basheer VS, John L, Padmakumar KG, Lal KK, Mohindra V, Punia P, Dinesh K, Manjebrayakath H, Ponniah AG, Lakra WS (2009) Low genetic differentiation in the populations of the Malabar Carp Labeo dussumieri as revealed by allozymes, microsatellites and RAPD. Asian Fisher Sci 22(2):359–391
Taggart JB, Hynes RA, Prodohl PA, Ferguson A (1992) A simplified protocol for routine total DNA isolation from salmonid fishes. J Fish Biol 40:963–965
Naish KA, Skibinski DOF (1998) Tetranucleotide microsatellite loci for Indian major carp. J Fish Biol 53:886–889
Crooijmans RPMA, Bierbooms VAF, Komen J, Van der Poel JJ, Groenen MAM (1997) Microsatellite markers in common carp (Cyprinus carpio L.). Anim Genet 28:129–134
Yue GH, Ho MY, Orban L, Komen J (2004) Microsatellites within genes and ESTs of common carp and their applicability in silver crucian carp. Aquaculture 234:85–98
Chenuil A, Galtier N, Berrebi P (1999) A test of the hypothesis of an autopolyploid vs. allopolyploid origin for a tetraploid lineage: application to the Genus Barbus (Cyprinida). Heredity 82:373–380
Dimsoski P, Gregory PT, Mark JB (2000) Microsatellite characterization in central stoneroller Campostoma anomalum (Pisces: Cyprinidae). Mol Ecol 9:2187–2189
Das P, Barat A, Meher PK, Ray PP, Majumdar D (2005) Isolation and characterization of polymorphic microsatellites in Labeo rohita and their cross-species amplification in related species. Mol Ecol Notes 5:231–233
Bessert ML, Orti G (2003) Microsatellite loci for paternity analysis in the fathead minnow, Pimephales promelas (Teleostei: Cyprinidae). Mol Ecol Notes 3(4):532–534
Raymond M, Rousset F (1998) GENEPOP (ver. 3.1): a population genetics software for exact test and ecumenicism. J Hered 86:248–249
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (1996–2004) GENETIX 4.05, logicielsous Windows TM pour la génétique des populations. France. http://www.genetix.univ-montp2.fr/genetix/genetix.htm
Yeh FC, Yang RC, Boyle T (1999) POPGENE 32: version 1.31. Population genetics software. Hyperlink http://www.ualberta.ca/~fyeh/fyeh/
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Schneider S, Roessli D, Excoffier L (2000) Arlequin: a software for population genetics data analysis, version 2000. Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva, Geneva
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotying errors in microsatellite data. Mol Ecol Notes 4:535–538
Van Oosterhout C, Weetman D, Hutchinson WF (2006) Estimation and adjustment of microsatellite null alleles in non equilibrium populations. Mol Ecol Notes 6:255–256
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Frankham R (2003) Genetics and conservation biology. C R Biol 326:S22–S29
Nazia AK, Siti Azizah MN (2014) Isolation of microsatellites in the bighead catfish, Clarias macrocephalus and cross-amplification in selected Clarias species. Mol Biol Rep. doi:10.1007/s11033-013-2965-9
Huang W, Liang X, Qu C, Cao J, Zhao C, Cao L (2013) Development and characterization of novel polymorphic microsatellite loci in Siniperca scherzeri Steindachner and Siniperca chuatsi (Basilewsky). Mol Biol Rep 40(2):751–756
Chaturvedi A, Mohindra V, Singh RK, Lal KK, Punia P, Bhaskar R, Mandal A, Narain L, Lakra WS (2011) Population genetic structure and phylogeography of cyprinid fish, Labeo dero (Hamilton, 1822) inferred from allozyme and microsatellite DNA marker analysis. Mol Biol Rep 38(5):3513–3529
Chauhan T, Lal KK, Mohindra V, Singh RK, Punja P, Gopalakrishnan A, Shanna PC, Lakra WS (2007) Evaluating genetic differentiation in wild populations of the Indian major carp, Cirrhimis rrmgala (Hamilton–Buchanan, 1882): evidence from allozyme and microsatellite markers. Aquaculture 269(1/4):135–149
Na-Nakorn U, Taniguchi N, Nugroho E, Seki S, Kamonrat W (1999) Isolation and characterization of microsatellite loci of Clarias macrocephalus and their application to genetic diversity study. Fish Sci 65(4):520–526
Yang G, Xiao M, Yu Y, Xu S (2012) Genetic variation at mtDNA and microsatellite loci in Chinese longsnout catfish (Leiocassis longirostris). Mol Biol Rep 39(4):4605–4617
Neff BD, Gross MR (2001) Microsatellite evolution in vertebrates: inference from AC dinucleotide repeats. Evolution Int J Org Evolution 55(9):1717–1733
Watanabe K, Watanabe T, Nishida M (2001) Isolation and characterization of microsatellite loci from the endangered bagrid catfish, Pseudobagrus ichikawai. Mol Ecol Notes 1:61–63
Usmani S, Tan SG, Siraj SS, Yusoff K (2001) Isolation and characterisation of microsatellites in the Southeast Asian River catfish Mystus nemurus. Mol Ecol Notes 1:264–266
David L, Rajasekaran P, Fang J, Hillel J, Lavi U (2001) Polymorphism in ornamental and common carp strains (Cyprinus carpio L.) as revealed by AFLP analysis and a new set of microsatellite markers. Mol Genet Genomics 266:353–362
Abdul-Muneer PM (2006) Molecular genetic characterization of endemic yellow catfish, Horabagrus brachysoma (Gunther). PhD thesis, Cochin University of Science and Technology, Cochin, p 225
Carvalho GR, Hauser I (1994) Molecular genetics and the stock concept in fisheries. Rev Fish Biol Fish 4:326–350
Liu ZJ, Cordes JF (2004) DNA marker technologies and their applications in aquaculture genetics. Aquaculture 238:1–37
Zhou J, Wu Q, Wang Z, Ye Y (2004) Genetic variation analysis within and among six varieties of common carp (Cyprinus carpio L.) in China using microsatellite markers. Russ J Genet (Genet) 40:1144–1148
Castric V, Bernatchez L, Belkhir K, Bonhomme F (2002) Heterozygote deficiencies in small lacustrine populations of brook charr Salvelinus fontinalis Mitchill (Pisces, Salmonidae): a test of alternative hypotheses. Heredity 89:27–35
Gopalakrishnan A, Abdul Muneer PM, Lal KK, Mohindra V, Kapoor D, Ponniah AG (2006) Primers from the orders Siluriform and Osteoglossiform detect polymorphic microsatellite loci in sun-catfish, Horabagrus brachysoma. J Appl Ichthyol 22:456–458
Gopalakrishnan A, Abdul Muneer PM, Thomas PC, Lal KK, Mohindra V, Kapoor D, Ponniah AG (2006) Identification of allozyme markers for population structure analysis in yellow catfish, Horabagrus brachysoma (Gunther, 1864). Indian J fish 53(3):253–261
Abdul-Muneer PM, Gopalakrishnan A, Lal KK, Mohindra V (2007) Population genetic structure of endemic and endangered yellow catfish, Horabagrus brachysoma using allozyme markers. Biochem Genet 45(9–10):637–645
Abdul-Muneer PM, Gopalakrishnan A, Basheer VS, Lakra WS (2008) Identification of RAPD markers in endemic yellow catfish, Horabagrus brachysoma (Gunther, 1864). Asian Fish Sci 21:293–304
Nagarajan M, Haniffa MA, Gopalakrishnan A, Basheer VS, Abdul-Muneer PM (2006) Genetic variability of Channa punctatus populations using randomly amplified polymorphic DNA. Aquac Res 37:1151–1155
Acknowledgments
This study was financially supported by NATP-ICAR (MM-III-18) to AG. The authors are thankful to Dr. WS Lakra, Dr. MJ Modayil and Dr. SP Singh for encouragement, support and guidance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Musammilu, K.K., Abdul-Muneer, P.M., Gopalakrishnan, A. et al. Identification and characterization of microsatellite markers for the population genetic structure in endemic red-tailed barb, Gonoproktopterus curmuca . Mol Biol Rep 41, 3051–3062 (2014). https://doi.org/10.1007/s11033-014-3164-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3164-z