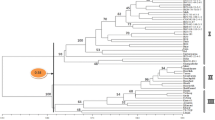
Abstract
Amplified fragment length polymorphism (AFLP) analysis is a rapid and efficient method for producing DNA fingerprints and molecular characterization. Our objectives were to: estimate genetic similarities (GS), marker indices, and polymorphic information contents (PICs) for AFLP markers in almond cultivars; assess the genetic diversity of almond cultivars and wild species, using GS estimated from AFLP fingerprints and molecular characterization; and facilitate the use of markers in inter-specific introgression and cultivar improvement. The genetic diversity of 45 almond cultivars from Iran, Europe, and America, were studied assaying 19 primer combinations. In addition, several agronomic traits were evaluated, including flowering and maturity times, self-incompatibility, and kernel and fruit properties. Out of the 813 polymerase chain reaction fragments that were scored, 781 (96.23%) were polymorphic. GS ranged from 0.5 to 0.96, marker indices ranged from 51.37 to 78.79, and PICs ranged from 0.56 to 0.86. Results allowed the unique molecular identification of all assayed genotypes. However, the correlation between genetic similarity clustering as based on AFLP and clustering for agronomic traits was low. Cluster analysis based on AFLP data clearly differentiated the genotypes and wild species according to their origin and pedigree, whereas, cluster analysis based on agronomic data differentiated according the pomological characterization. Our results showed the great genetic diversity of the almond cultivars and their interest for almond breeding.






Similar content being viewed by others
References
Aranzana M, Pineda A, Cosson P, Dirlewanger E, Ascasibar J, Cipriani G, Ryder C, Testolin R, Abbott A, King G, Arus P (2003) A set of simple sequence repeat (SSR) markers covering most of the Prunus genome. Theor Appl Genet 106:819–825
Arulsekar S, Parfitt DE, Kester DE, (1986) Comparison of Isozyme variability in peach and almond cultivars. J Hered 77:272–274
Bartolozzi F, Warburton ML, Arulsekar S, Gradziel TM (1998) Genetic characterization and relatedness among California almond cultivars and breeding line detected by randomly amplified polymorphic DNA (RAPD) analysis. J Am Sco Hort Sci 123:381–387
Bassam BJ, Caetano-Anolles G (1993) Silver staining of DNA in polyacrylamide gels. Appl Biochem Biotech 42:181–188
Berry ST, Allen RJ, Barnes SR, Caligari PDS (1994) Molecular marker analysis of Helianthus annus L. Restriction fragment length polymorphism between inbred lines of cultivated sunflower. Theor Appl Genet 89:435–441
Bortiri PE, Oh S, Jiang J, Baggett S, Granger A, Weeks C, Potter D, Parfitt DE (2001) Phylogeny and systematics of Prunus (Rosaceae) as determined by sequence analysis of ITS and the chloroplast trnL-trnF spacer DNA. Syetematic Bot 26:797–807
Browicz K, Zohary D (1996) The genus Amygdalus L. (Rosaceae): species relationships, distribution and evolution under domestication. Genet Resour Crop Evol 43:229–247
Brown-Guedira GL, Thompson JA, Nelson RL, Warburton ML (2000) Evaluation of genetic diversity of soybean introductions and North American ancestors using RAPD and SSR markers. Crop Sci 40:815–823
Caicedo AL, Gaitan E, Duque MC, Chica OT, Tohma J (1999) AFLP fingerprinting of Phaseolus lunatus L. and related wild species from South America. Crop Sci 39:1497–1507
Cerezo M, Socias I, Company R, Vargas F (1989) Identification of almond cultivars by pollen isoenzymes. J Am Soc Hort Sci 114:164–169
Channuntapipat C, Wirthensohn M, Ramesh SA, Batlle I, Arus P, Sedgley M, Collins G (2003) Identification of incompatibility genotypes in almond (Prunus dulcis Mill.) using specific primers based on the introns of the S-alleles. Plant Breed 122:164–168
Cipriani G, Lot G, Huang WG, Marrazzo MT, Peterlunger E, Testolin R (1999) AC/GT and AG/CT microsatellite repeats in peach (prunus Persica (L.) Batsch): isolation, characterization and cross-species amplification in prunus. Theor Appl Genet 100:713–722
De Nettancourt D (1997) Incompatibility in angiosperms. Monographs on theoretical and applied genetics, vol 3. Springer, Berlin
Denisov VP (1988) Almond genetics resources in the USSR and their use in production and breeding. Acta Hort 224:299–306
Dicenta F (1991) Mejora Genética del Almondro (Prunus dulcis Miller) por Cruzamientos intervarietales: Herencia de caracters y Selectión. Thesis. University Murcia, Spain, p 313
Doweny LD, Iezzoni AF (2000) Polymorphic DNA markers in black cherry are identified using sequences from sweet cherry, peach and sour cherry. J Am Soci Hort Sci 125:76–80
Evreinoff VA (1958) Contribution a letude de l’amandier. Fruits et Primeurs Afrique 28:99–104
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Gentzbittel L, Zhang Y-X, Vear F, Griveau B (1994) RFLP studies of genetics relationships among inbred lines of cultivated sunflower, Helianthus annus L. evidence for distinct restorer and maintainer germplasm pools. Theo Appl Genet 89:419–425
Gradziel TM, Kester DE (1998) Breeding for self-fertility in California almond cultivars. Acta Hort 470:109–117
Gradziel TM, Martínez-Gómez P, Dicenta F, Kester DE (2001) The utilization of Prunus species for almond variety improvement. J Am Pomol Soc 55:100–108
Grasselly C (1976) Les espéces sauvages ďamandier (in French). Options Méditerr 32:28–44
Hauagge R, Kester DE, Arulsekar S, Parfitt DE, Liu L (1987) Isozyme variation among California almond cultivars. II. Cultivar characterization and origins. J Am Sco Hort Sci 112:693–698
Hesse CO (1975) Peaches. In: Janick J, Moore JN (eds) Advances in fruit breeding. Purdue Univ. Press, West Lafayette, Ind, pp 285–326
Kester DE (1994) Almond cultivar and breeding programs in California. Acta Hort 373:13–28
Kester DE, Gradziel TM (1996) Almonds In: Janick J, Moore JN (eds) Fruit breeding, Almond, vol. 1. Wiley, New York, pp 1–97
Kester DE, Gradziel TM, Micke WC (1994) Identifying pollen incompatibility groups in California almond cultivars. J Am Sco Hort Sci 119:106–109
Kovaleff NV, Kostina KF (1935) A contribution of the study of the genus prunus Focke. Leningrad
Ladizinsky G (1999) On the origin of almond. Genet Res Crop Evol 46:143–147
Mantel N (1967) The detection of disease clustering and generalized regression approach. Cancer Res 27:209–220
Martínez de Toda E, Saccha JC (1997) Ampelographical characterization of red Vitis vinifera L. cultivars preserved in Rioja. Bultten de tow, Mars-Avril, 70:220–234
Martínez-Gómez P, Arulsekar S, Potter D, Gradziel TM (2003a) An extended inter-specific gene pool available to peach and almond breeding as characterized using simple sequence repeat (SSR) markers. Euphytica 131:313–322
Martínez-Gómez P, Arulsekar S, Potter D, Gradziel TM (2003b) Relationships among peach and almond and related species as detected by SSR markers. J Am Sco Hort Sci 128:667–671
Martínez-Gómez P, Sánchez-Pérez R, Dicenta F, Howad W, Arus P, Gradziel TM (2007) Almonds. In: Kole CR (ed) Genome mapping and molecular breeding, vol. 4: Fruits & Nuts. Springer. Heidelberg, Berlin, New York, Tokio, pp 229–242
Martins M, Farinha A, Ferreira E, Cordeiro V, Monterio A, Tenreiro R, Oliveira M (2001) Molecular analysis of the genetic variability of Portuguese almond collection. Act Hort 546:449–456
Martins M, Tenreiro R, Oliveira M (2003) Genetic relatedness of Portuguese almond collection assessed by RAPD and ISSR markers. Plant Cell Rep 22:71–78
Mnejja M, Garcia-Mas J, Howad W, Badenes ML, Arús P (2004) Simple-sequence repeat (SSR) markers of Japanese plum (Prunus salicina Lindl.) are highly polymorphic and transferable to peach and almond. Mol Ecol Notes 4:163–165
Mir Ali N, Nabulsi I (2003) Genetic diversity of almond (Prunus dulcis) using RAPD technique. Sci Hort 98:461–471
Murray HC, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res 8:4321–4325
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci (USA) 70:3321–3323
Powell W, Morgante M, Andre C, Hanafey Mm Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Reina A, Giorgio V, Godini A (1985) Autres types autocompatibles parmi la population ďamandiers des Pouilles (in Italian). Options Méditerr 85:25–29
Resta P, Corona MG, Fanizza G, Palasciano M, Godini A (1998) Random amplified DNA polymorphism in Amygdalus communis L. and A. webbii Spach. Acta Hort 470:82–90
Rohlf FJ (1998) NYSYS-pc. Numerical taxonomy and Multivariate Analysis System, Version 2.02 Exeter Software, Setauket, NY
Roland-Ruiz L, Vaneeuwijk FA, Gilliland TJ, Dubreil P, Dillman C, Lallemand J, De loost M, Barii CP (2001) A comparative study of molecular and morphological methods of describing relationships between perennial ryegrass (Lolium Perenne L.) varieties. Theor Appl Genet 103(8):1138–1150
Schneider S, Roessli D, Excoffier L (2001) Arlequin: a software for population genetics data analysis. Version 2.000. Genetics and Biometry Lab, Dep. of Athropology, University of Geneva, Geneva
Scorza R, Sherman WB (1996) Peaches, In: Janick J, Moore JN (eds) Fruit breeding, Wiely, New York, pp 285–326
Shiran B, Ameirbakhtiar N, Kiani S, Mohamadi Sh, Tabatabaei BES, Moradi H (2007) Molecular characterization and genetic relationships among almond cultivars assessed by RAPD and SSR markers, Scientia Horticulturae 111:280–292
Socias i Company R, Felipe AJ (1988) Self-incompatibility in almond: transmission and recent advances in breeding. Acta Hort 224:307–317
Van de Peer Y, De Wachter R (1994) TREECON for Windows: as software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci 10:569–570
Vavilov NI (1930) Wild progenitors of the fruit trees of Turkistan and Caucasus and the problem of the origin of fruit trees. In: Proceeding of the IX International Horticultural Congress report and proceeding. London, pp 271–286
Vezvaei A (2003) Isozyme diversity in Iranian almond. Acta Hort 622:451–456
Viruel MA, Messeguer R, de Vicente MC, Garcia-Mas J, Puigdomenaech P, Arus P (1995) A linkage map with RFLP and isozyme markers for almond. Thero Appl Genet 91:964–971
Vos P, Hogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Watkins R (1976) Cherry, Plum, Peach, apricot and almond. In: Simmonds NW (ed) Evolution of crop plants. Longman, London, pp 342–347
Weising K, Nybon H, Wolff K, Meyer W (1995) DNA fingerprinting in plants and fungi. CRC Press, Boca Raton, USA
Wishart D (1987) CLASTAN user manual, 3rd edn. Program Library Unit, Univ. of Edinburgh, Edinburgh
Wood MN (1925) Almond varieties in the United States. USDA Bul 1282:1–42
Woolley FM, Collins GG, Sedgly M (2000) Application of DNA fingerprinting for the classification of selected almond [prunus dulcis (Miller) D. A. Webb] cultivars. Aust J Exp Agr 40:995–1001
Xie H, Sui Y, Chang FQ, Xu Y, Ma RC (2006) SSR allelic variation in almond (Prunus dulcis Mill). Theor Appl Genet 112:366–372
Xu Y, Ma RC, Xie H, Cao MQ (2004) Development of SSR markers for the phylogenetic analysis of almond trees from China and the Mediterranean region. Genome 47:1091–1104
Zabeau M (1993) Selective restriction fragment amplification: a general method for DNA fingerprinting. European patent Application No.0–534-858-A1
Acknowledgement
The authors are grateful to Shahrekord University for financial assistance and Dr Ali Vezvaei for helpful suggestions. Thanks to the section of Horticulture, Agriculture and Natural Resources Research Center of Shahrekord for access to trees.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sorkheh, K., Shiran, B., Gradziel, T.M. et al. Amplified fragment length polymorphism as a tool for molecular characterization of almond germplasm: genetic diversity among cultivated genotypes and related wild species of almond, and its relationships with agronomic traits. Euphytica 156, 327–344 (2007). https://doi.org/10.1007/s10681-007-9382-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-007-9382-x