Abstract
Etiological studies have shown genetic disorders to be a major cause of sensorineural hearing loss, but there are a limited number of comprehensive etiological reports based on genetic analysis. In the present study, the same platform using a diagnostic DNA panel carrying 63 deafness genes and the same filtering algorithm were applied to 10,047 samples obtained from social health insurance-based genetic testing of hearing loss. The most remarkable result obtained in this comprehensive study was that the data first clarified the genetic epidemiology from congenital/early-onset deafness to late-onset hearing loss. The overall diagnostic rate was 38.8%, with the rate differing for each age group; 48.6% for the congenital/early-onset group (~5y.o.), 33.5% for the juvenile/young adult-onset group, and 18.0% for the 40+ y.o. group. Interestingly, each group showed a different kind of causative gene. With regard to the mutational spectra, there are certain recurrent variants that may be due to founder effects or hot spots. A series of haplotype studies have shown many recurrent variants are due to founder effects, which is compatible with human migration. It should be noted that, regardless of differences in the mutational spectrum, the clinical characteristics caused by particular genes can be considered universal. This comprehensive review clarified the detailed clinical characteristics (onset age, severity, progressiveness, etc.) of hearing loss caused by each gene, and will provide useful information for future clinical application, including genetic counseling and selection of appropriate interventions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hearing loss is an extremely heterogenous disorder, and more than 120 genes are currently considered to be implicated in non-syndromic hearing loss, making the screening strategy difficult. Targeted genome resequencing using massively parallel DNA sequencing (MPS) has become a powerful strategy for the identification of causative genes from among the large numbers of genes in rare Mendelian disorders such as deafness. This sequencing technology followed by an appropriate filtering algorithm will be able to identify rare responsible genes for individual hearing loss patients. Although genetic disorders are thought to be a major cause of sensorineural hearing loss, there are a limited number of comprehensive etiological reports based on genetic analysis. In addition, it is difficult to draw conclusions without using the same analysis platform, filtering method, and pathogenicity assessment for samples collected by the same criteria. Through collaborative study with 102 collaborative centers in Japan, over 10,000 samples obtained from social health insurance-based testing and detailed clinical data have been collected. In this paper, we not only reviewed our series of studies based on analysis by MPS conducted over the last decade, but also performed comprehensive verification using the same analytic method and criteria.
Genetic epidemiology based on genetic testing
Etiological studies have shown that genetic causes are the most common etiology of deafness, and approximately two-thirds of congenital/early-onset sensorineural hearing loss in developed countries is estimated to be due to genetic causes (Morton and Nance 2006). Recent studies have indicated that a significant portion of late-onset hearing loss is also due to genetic causes (Kitano et al. 2017; Kobayashi et al. 2018; Shinagawa et al. 2020a; Yasukawa et al. 2019; Oka et al. 2020; Miyajima et al. 2020). A series of etiological studies has demonstrated genetic disorders to be a common cause of all types of sensorineural hearing loss, but there has been no detailed genetic epidemiological data covering a wide range of ages.
Hereditary hearing impairment is an extremely heterogenous disorder that involves more than 120 distinct genes, thereby making the precise diagnosis and appropriate intervention difficult. Recent advances in targeted genome resequencing using massively parallel DNA sequencing (MPS) has provided a powerful new strategy and revolutionized the elucidation of genetic defects causing monogenic disorders. We have shown that this approach is appropriate for identifying causative genes/variants and actually demonstrated that various genes/gene variants are involved in hearing loss in Japanese patients (Miyagawa et al. 2013; Nishio et al. 2015). Meanwhile, both the number of DNA samples and detailed clinical data are increasing, and genetic and clinical data from over 10,000 patients has been collected from 102 collaborative centers. Based on this large-cohort data, we have published a series of studies demonstrating the mutational spectrum and clinical features caused by the representative deafness genes, including GJB2 (Tsukada et al. 2010), CDH23 (Miyagawa et al. 2012), KCNQ4 (Naito et al. 2013), OTOF (Iwasa et al. 2013, 2019), mitochondrial 1555A > G and 3243A > G (Yano et al. 2014), SLC26A4 (Miyagawa et al. 2014), LRTOMT (Ichinose et al. 2015), GRXCR1 (Mori et al. 2015a), PTPRQ (Sakuma et al. 2015), COCH (Tsukada et al. 2015a), TMPRSS3 (Miyagawa et al. 2015d), STRC (Moteki et al. 2016; Yokota et al. 2019), LOXHD1 (Mori et al. 2015b; Maekawa et al. 2019), ACTG1 (Miyagawa et al. 2015b; Miyajima et al. 2020), MYO15A (Miyagawa et al. 2015a), POU4F3 (Kitano et al. 2017), WFS1 (Kobayashi et al. 2018), CLDN14 (Kitano et al. 2019), EYA4 (Shinagawa et al. 2020a, b), TECTA (Yasukawa et al. 2019), OTOA (Sugiyama et al. 2019), and MYO6 (Miyagawa et al. 2015c; Oka et al., 2020). These studies were performed between 2010 and 2020. It seems that the reported clinical characteristics of hearing loss caused by each gene do not significantly differ from the contents of our original papers, but the present review seeks to summarize the findings as more accurate and comprehensive results can now be obtained for the following reasons; (1) the samples and clinical data from hearing loss patients are increasing (approximately 700 samples per year), (2) the public database for normal controls is being updated, (3) disease-specific databases can be referenced, and (4) the results for the same hearing loss population using the same filtering algorithm can be compared.
Unbiased samples obtained from social health insurance-based testing
In Japan, genetic testing for deafness, which has been reimbursed by the social health insurance system since 2012, has become a standard diagnostic tool for deafness. Collecting DNA samples for research purposes inevitably results in biased samples, but the biggest advantage of the social health insurance-based testing is that it is accessible to everyone and, therefore, more unbiased samples can be collected. This is important when discussing etiology.
Currently, DNA samples as well as clinical data from 10,047 patients have been collected from 102 collaborative centers participating in the deafness consortium, and the relationships between causative gene variants and clinical features have become clear. This review summarizes the findings, including mutational spectra and genotype/phenotype correlations, obtained from the large-cohort data.
The samples used in this review were as follows. With regard to inheritance mode, 2243 subjects were from autosomal dominant or mitochondrial inherited families, 6163 subjects from autosomal recessive families or sporadic cases, and 1641 showed unknown inheritance mode. Patients for whom the onset age (the age of awareness) was available numbered 3877 prelingual hearing loss (below age 6), 2698 post-lingual hearing loss (aged between 6 and 39), and 1057 late-onset hearing loss (after the age of 40) cases. Hearing levels were classified based on the better hearing ear as normal, < 20 dB; mild hearing loss, 21–40 dB (n = 1162); moderate hearing loss, 41–70 dB (n = 2746); severe hearing loss, 71–95 dB (n = 1622); and profound hearing loss, > 95 dB (n = 1660). All subjects had presumed non-syndromic sensorineural hearing loss (SNHL). The samples were obtained primarily from bilateral SNHL, but also contained 220 samples from unilateral SNHL patients. Written informed consent was obtained from all patients (or from their next of kin, caretaker or guardian in case of minors or children), and the study was approved by the Shinshu University Ethical Committee as well as the relevant bodies of the other participating institutions of the Deafness Gene Study Consortium. Clinical information and peripheral blood samples were obtained from each subject and from all their consenting relatives. This study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of Shinshu University School of Medicine (No. 387 ~ 4 September 2012, No. 576 ~ 2 May 2017).
Sequencing strategy and filtering algorithm
Sixty-three genes (shown in Supplementary Table S1; Ion AmpliSeq™ Hearing Loss Research Panel v1, Thermo Fisher Scientific, Waltham, MA USA), reported to be causative of non-syndromic hearing loss (Hereditary Hearing loss Homepage; http://hereditaryhearingloss.org/), were analyzed in this study. The detailed protocols for targeted genome enrichment and massively parallel DNA sequencing have been described elsewhere (Nishio et al. 2015). In addition to MPS, copy number variation (CNV) analysis was performed using the same platform as for MPS in accordance with our previous report (Nishio et al. 2018).
For the filtering algorithm used in the present MPS analysis, (1) allele frequency in a public database, (2) a disease-specific database, (3) variant type, and (4) in silico prediction score were used. For clinical setting and practical genetic diagnosis, a great deal of the above-mentioned information needs to be integrated and evaluated. To integrate (1)–(4), we have developed an integrated database of clinical information and genetic information from patients with genetic deafness (Nishio and Usami 2017). It should be emphasized that this database is particularly powerful for selecting candidate gene variants, and we have already collected detailed clinical information and genetic analysis data from more than 10,000 hearing loss patients. In the case of rare diseases such as hereditary hearing loss, it is often difficult to determine whether the variant found in a specific patient is pathogenic. However, the pathogenicity can be determined efficiently using a database containing information obtained from large number of hearing loss patients.
The pathogenicity of the identified variants was evaluated in accordance with the American College of Medical Genetics (ACMG) standards and guidelines (Richards et al. 2015) with the ClinGen hearing loss clinical domain working group expert specification (Oza et al. 2018). Variants were defined as likely causative variants if the following criteria was fulfilled; (1) for the variants previously reported as “pathogenic” or “likely pathogenic” and did not show any contradictory evidence, we applied the same pathogenicity classification as the previous report (HGMD professional), with reference to the pathogenicity classification in the Deafness variation database ver. 9 (Azaiez et al. 2018) and ClinVar database ver. 20210501 (Landrum et al. 2018), (2) novel variants classified as “pathogenic” or “likely pathogenic” were considered as strong candidates for each case, (3) variants of “uncertain significance” (VUS) identified as the only candidate after the filtering procedure with no other candidate variants in the other 62 genes were also included, (4) two variants found in recessive inheritance cases, (5) there was no contradiction with the results of family analysis, and (6) in cases with conflicting findings between our pathogenicity classification and the HGMD professional, Deafness variation database and ClinVar database, we manually curated the data based on the ACMG guidelines with reference to the patient’s clinical information.
Responsible genes in the Japanese hearing loss population
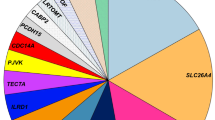
Using simultaneous screening for 63 deafness genes, we identified 51 genes that are definitely involved in the Japanese hearing loss population (Table S2). Further, using the criteria mentioned above, the overall diagnostic rate was 38.8% (3896/10,047); i.e., one causative variant in autosomal dominant or mitochondrial inheritance cases, and biallelic causative variants in recessive inheritance or sporadic cases (Fig. 1A).
To determine which genes among those identified have the greatest impact on the etiology of deafness, the number of variants and variant types (missense, nonsense, frameshift) were calculated. Results showed that the number of mutations in GJB2 was exceptionally high, followed by mutations in CDH23, SLC26A4, STRC, KCNQ4, mitochondrial m. 3243A > G, MYO7A, mitochondrial m.1555A > G, MYO6, TECTA, WFS1, MYO15A, OTOF, POU4F3 and USH2A. (Fig. 1A, B). GJB2 is the most prevalent causative gene, with several major (commonly found) gene mutations causing deafness in 30–35% of cases, while the remaining cases of hearing loss are the result of various rare genes/mutations that have been efficiently diagnosed by the present sequencing and CNV detection approach (Fig. 1A, B). The high prevalence of GJB2 mutations in the hearing loss population is evident regardless of ethnic differences. It is hypothesized that having variants in GJB2 was beneficial for survival. GJB2 is also known to be expressed on the skin, and the skin of subjects with GJB2 mutations is thicker (the skin is a barrier against pathogen invasion, trauma, and insect bites), which was beneficial to survival associated with human evolution (Meyer et al. 2002). This hypothesis could be supported from the prevalence of LOF (loss of function) mutations observed in the Genome Aggregation Database (gnomAD, https://gnomad.broadinstitute.org). Based on the gnomAD, in contrast to the other deafness genes where the number of “observed” LOFs (loss of function variants) is equal to that of “expected” LOFs, the number of “observed” LOFs (n = 17) in GJB2 is exceptionally high compared to that of “expected” LOFs (n = 6.5) (see GJB2 on the gnomAD home page), indicating that there was a certain benefit in having GJB2 mutations in ancient times.
Table 1 shows a summary of the previous papers describing the results of genetic screening using MPS. It is a consensus fact that variants in GJB2 are predominant across all ethnic groups, but there are also some exceptions as shown in Table 1. For example, MYO15A and TMC1 are predominant in consanguineous Egyptian families and Sephardi Jews, respectively (Brownstein et al. 2020; Budde et al. 2020). This specific causative gene rate is probably the result of consanguineous marriage, which is common in these regions.
Several causative genes, such as SLC26A4, MYO15A, MYO7A, and CDH23, are frequently found across different populations, although there are some ethnic differences in the second and third most frequent genes. These differences are most likely due to founder mutations occurring during human migration, as these ethnic differences are evident even in analyses using the same screening platform (Sloan-Heggen et al. 2016). However, it is important to keep in mind that such differences may also be caused by other factors, including sampling bias, method used, differences in filtering procedure, and differences in the definitions of pathogenicity. In connection with sampling bias and the methodology used, when CNV analysis is performed or when mild-to-moderate deafness is included, a certain number of patients with hearing loss caused by CNV (copy loss) of the STRC gene could be identified (Sloan-Heggen et al. 2016; this study).
Regardless of the number of genes in the diagnostic panel (number ranges from 12 to 327 or the whole exome), the number of mutated genes is somewhat limited (maximum: 57). These results converge to around 50–60 genes, even when many genes are analyzed. There probably are, however, many extremely rare genes in the various hearing loss populations.
The present comprehensive study revealed certain tendencies in terms of variant type. In general, gene mutations in AR deafness tend to have more LOF mutations; GJB2 (frequency of LOF: 76.7%), STRC (97.2%), MYO15A (44.8%), LOXHD1 (71.4%), USH2A (64.9%), and PCDH15 (75.0%). However, even in AR genes, if the founder mutation is a missense mutation, the number of LOF mutations is slightly lower; for example, SLC26A4 (p. His723Arg, a known founder mutation, 28.1%) and OTOF (p.Arg1939Gln a known founder mutation, 18.5%). Also, CDH23 has a very low LOF percentage (9.5%), probably as mutations lead to Usher syndrome. In the case of the gene mutations in AD deafness, there are two patterns regarding variant type; (1) genes that have many missense and a low frequency of LOF mutations, such as TECTA (7.1%), WFS1 (0%), ACTG1 (0%), COCH (6.1%), and MYH9 (10.5%), and (2) genes that have a high frequency of LOF and few missense mutations, such as KCNQ4 (62.3%), MYO6 (75.9%), POU4F3 (51.3%), and EYA4 (86.3%). The mechanisms by which the mutations cause deafness are thought to differ; a dominant negative mechanism for the former and haploinsufficiency for the latter group.
Samples collected in a more unbiased manner from social health insurance-based testing cover a wide range of age groups and reveal the comprehensive genetic epidemiology in Japanese. With regard to age at onset, we divided the samples into 3 groups by age; congenital/early-onset (~ 5 y.o.), juvenile/young adult-onset (6–39 y.o.), and middle aged/aged-onset (40 y.o ~). The diagnostic rate was higher in the congenital/early-onset group (~ 5 y.o.) (48.6%; 1885/3877) (Fig. 2A), with variants in GJB2, SLC26A4, CDH23, STRC, and MYO15A frequently observed. The diagnostic rate in the juvenile/young adult-onset group was lower than that in the congenital/early-onset group, but the responsible genes were detected in a significant portion of the patients (Fig. 2A) (33.5%; 904/2698). It is well known that many different genes are involved in congenital/early-onset hearing loss, and the present study revealed that many genes are also involved in late-onset hearing loss. Interestingly, the responsible genes differed from those found in the congenital/early-onset group. The present analysis revealed that genes such as KCNQ4, mitochondrial mutations (m.1555A > G and m.3243A > G), GJB2, CDH23, MYO6 and MYO7A were frequently found in the juvenile/young adult-onset group. The causative gene was found in only a few patients in the over 40 y.o. group (18.0%; 190/1057) (Figs. 2A, 3) and genetic involvement was relatively low in the subjects over 40 years, suggesting that other factors such as environmental factors (exposure to noise, aging, etc.), or other unknown genes (either Mendelian or multifactorial inheritance) may be involved. However, it is still interesting to note that the characteristic genes in this group were CDH23, mitochondrial m.3243A > G, MYO6, MYO7A, POU4F3 and COCH. GJB2 and CDH23 were found in all groups, but it should be noted that the identified mutations differed; i.e., p.Val37Ile in GJB2, and p.Arg1588Trp and p.Arg2029Trp in CDH23 were predominantly found in the oldest group. As p.Val37Ile, the causative variant of the milder phenotype as shown in our previous study (Tsukada et al. 2010), was found in older patients, it probably reflects the age at diagnosis by school and workplace annual health examinations, rather than the age at onset. Actually, in this study, the proportion of p.Val37Ile patients among those with GJB2 mutations was 10.2% (86/846) in the congenital group, compared to 52.9% (45/85) in the juvenile and young adult-onset (6–39 y.o.) group, supporting our previous results.
A Responsible genes in each age group (congenital/6–39 y.o./over 40 y.o.). B Responsible genes found in autosomal dominant (AD) and autosomal recessive (AR) patients. Among 89 GJB2-associated hearing loss cases identified from autosomal dominant families, 23 carried autosomal dominant inheritance variants and 66 carried biallelic autosomal recessive variants (pseudo-dominant cases). Similarly, among 75 MYO7A-associated hearing loss cases identified from autosomal dominant families, 71 carried autosomal dominant inheritance variants and only 4 carried biallelic autosomal recessive variants (pseudo-dominant cases). C Responsible genes found in groups classified by severity of HL. It was revealed that the types of genes differed according to severity
GJB2, SLC26A4, CDH23, STRC, OTOF and MYO15A were commonly found to be the responsible genes in autosomal recessive/sporadic inheritance cases, and KCNQ4, mitochondrial m.3243A > G, MYO6, MYO7A, WFS1, POU4F3 and TECTA were frequently identified in autosomal dominant inheritance cases (Figs. 2B, 3).
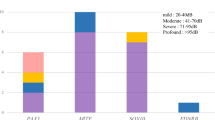
It was also revealed that the types of genes differed in the groups according to severity; i.e., GJB2, SLC26A4, and CDH23 were more commonly found in the severe-to-profound hearing loss group, while GJB2, STRC, KCNQ4, WFS1, MYO7A, and MYO6 were found to be more common in the mild-to-moderate hearing loss group (Fig. 2C).
The fact that the causative gene was only found in one case (mitochondrial m.1555A > G) out of the 220 unilateral hearing loss cases suggested that factors other than genetic factors were involved in unilateral hearing loss.
Recurrent variants: founder effect or mutational hot spot?
The identified variants are shown in Supplemental Table S2, which shows that our series of studies revealed certain recurrent mutations, which is crucial for molecular diagnosis to allow decision-making with regard to the appropriate intervention (Ohtsuka et al. 2003; Miyagawa et al. 2014). Based on such recurrent mutations in the Japanese population, we have developed an Invader screening assay that has been proven to give satisfactory results (Abe et al. 2007; Usami et al, 2012b). It is generally accepted that recurrent genetic mutations occur via two mechanisms: one is a founder effect and the other is a mutational hot spot, and haplotype analysis was performed to reveal whether these mutations occur by a founder effect or mutational hot spot.
The most readily understandable example is GJB2, the most common deafness gene. Numerous pathogenic variants have been demonstrated over the past two decades and the reported mutational spectra are known to be ethnicity specific. There are great variations in the allele frequency of patients with GJB2 mutations in each population, suggesting that the allele frequency in the population, which reflects a founder effect, strongly affects the status of the GJB2 gene in the hearing loss population (Tsukada et al. 2015b). As certain mutations are clearly found in only limited ethnic populations, it is possible to predict that those mutations are due to a founder effect (Fig. 4A).
A The spectrum of GJB2 mutations. A larger circle indicates a larger number of mutated alleles (from Tsukada et al. 2015b). B Suspected origin of the GJB2 variants (modified from Tsukada et al. 2015b). Suspected origin of GJB2 variants are marked on the human Y-chromosomal haplogroup tree (Karafet et al. 2008), which is applicable to the investigation of human migration. Lineages associated with haplogroup IJ in the Y-haplotype tree based on Karafet et al. (2008). From geographical distribution and haplotype analysis, it is speculated that p. Arg143Trp may have occurred in the Y-chromosomal haplogroup B, p.Val37Ile in D and E, and c.235delC, p.[G45E; Y136X], and c.176_191del, c.299_300delAT in N and O, respectively. C GJB2 founder variants from a human migration perspective based on Y chromosome haplotypes: c.235 del C is considered to have occurred in haplotype N/O from the viewpoint of its geographic distribution. On the other hand, p.Val37Ile is consistent with the area where the haplotype C/D/E was found, so it is probable that it occurred within this haplotype
We have reviewed the reported GJB2 variants in different populations and considered them from the perspective of human migration (Tsukada et al. 2015b). Founder effects have received special emphasis from this perspective. The c.35delG variant, which is predominant throughout Europe, the Middle East, North Africa, North and South America, and Australia, has been proven by haplotype analysis to be due to a founder effect occurring 11,000 years ago (van Laer et al. 2001). With regard to six frequent GJB2 variants observed in Japanese hearing loss patients (i.e., c.235delC, p.Val37Ile, p.[Gly45Glu; Tyr136Ter], p.Arg143Trp, c.176_191del, and c.299_300delAT), we concluded that the six variants were derived from founder effects as they were observed in a specific haplotype. The c.235delC variant, most frequently found in Japanese hearing loss population, was observed in countries in East and Central Asia, such as Japan, Korean, China, Mongolia, and Thailand, whereas it is rarely observed in other populations. Such an uneven distribution of this variant suggests that it was caused by a founder effect. The c.235delC variant is estimated to have first occurred at around 6500 years ago (Shinagawa et al. 2020b). Our haplotype analysis, together with their distribution patterns, indicated that the p.Arg143Trp and p.Val37Ile variants may have occurred as multiple events, suggesting that both a founder effect and hot spot may be involved in these variants. With regard to the founder age of frequent GJB2 variants, each variant may have occurred at a different time, with the oldest, p.Val37Ile, considered to have occurred around 14,500 years ago, and the most recent, c.176_191del, considered to have occurred around 4000 years ago (Shinagawa et al. 2020b).
A comparative analysis of the results of clustering analysis and the phylogenetic tree based on the results of Y-chromosomal haplogroup analysis indicates that many variant distributions are well explained by founder effects in ancient human lineages (Fig. 4B, C). The p.Arg143Trp and p.Val37Ile variants are spread widely across the globe and are speculated to have occurred at a very early stage in human migration. As these two mutations are frequently found in Japanese, as stated above, it is considered that the two mutations occurred multiple times. Our haplotype analysis indicated that p. Arg143Trp and p.Val37Ile are thought to have occurred in the Y-chromosomal haplogroup N as well as O (in the same haplotype as c.235delC, p. [Gly45Glu; Tyr136Ter], c.176_191del, and c.299_300delAT).
To date, various other genes have been extensively studied in a number of ethnic groups and certain recurrent mutations have been confirmed to be due to founder effects, such as CDH23 (Kim et al. 2015), MYO15A (Palombo et al. 2017), and TMC1 (Ramzan et al. 2020). Several founder mutations have also been reported in Japanese patients with hearing loss. A series of haplotype analyses indicated that p. His723Arg in SLC26A4 (Park et al. 2003), p.Arg1939Gln in OTOF (Matsunaga et al. 2012), c.211delC in KCNQ4 (Naito et al. 2013), and c.4212 + 1G > A in LOXHD1 (Maekawa et al. 2019) are suspected to be founder variants. In contrast, c.5597C > T in TECTA (Yasukawa et al. 2019) and p.Ala716Thr, p.Lys836Thr, and p.Glu864Lys in WFS1 (Kobayashi et al. 2018) occurred in mutational hot spots. As mentioned above, p.Val37Ile and p.Arg143Trp in GJB2 are due to both a founder effect and a hot spot.
Clinical characteristics
Regarding hereditary hearing loss, it is known that age at onset, severity, audiogram configuration, progressiveness, and presence of associated symptoms differ depending on the type of causative gene and variant, and site on the variant. We have collected data from a large number of Japanese hearing loss patients and clarified the clinical characteristics and genotype/phenotype correlation for each causative gene.
As to the onset age, Fig. 5 clearly shows that hearing loss related to GJB2, CDH23, SLC26A4, STRC, TECTA, MYO15A, OTOF, USH2A, and LOXHD1 is congenital/early-onset. In contrast, a significant proportion of cases related to KCNQ4, mitochondrial m.3243A > G, MYO6, POU4F3, ACTG1, EYA4 and COCH are adult-onset.
Age at onset (awareness) of each gene. Hearing loss related to GJB2, CDH23, SLC26A4, STRC, TECTA, MYO15A, OTOF, USH2A, and LOXHD1 is shown to be congenital/early-onset. In contrast, a significant portion of cases with mutations in KCNQ4, mitochondrial m.3243A > G, MYO6, POU4F3, ACTG1, EYA4, and COCH showed adult-onset hearing loss
Regarding severity and audiogram configuration, it has been clarified that there is a genotype/phenotype correlation for GJB2, which is the gene most frequently observed in congenital hearing loss (Snoeckx et al. 2005; Tsukada et al. 2010). Also, particular genes such as TECTA and WFS1 are associated with characteristic audiogram configurations that show mid-frequency- and low-frequency-involved hearing loss, respectively (Taylor et al. 2013). Such characteristic audiogram configurations have also been found to be related to the mutated domain; i.e., if the mutation is located in the ZP domain of the TECTA gene, a dish-shaped audiogram configuration is observed (Yasukawa et al. 2019). With regard to the causative gene of mild-to-moderate hearing loss, as mentioned above, the GJB2 gene has been known to have genotype/phenotype correlations, and p.V37I variant-associated hearing loss is mild-to-moderate (Tsukada et al. 2015b). Recent CNV analyses have shed light on the etiology of this group. CNV analysis revealed the CNV of the STRC gene is the second most common etiology in cases of mild-to-moderate hearing loss after the GJB2 gene (Yokota et al. 2019). We have developed an efficient way to identify CNV using the same MPS platform used for the social health insurance-based genetic testing (Nishio et al. 2018). In addition, the OTOA gene, which encodes otoancorin and has a role in anchoring the extracellular matrix of the tectorial membrane to the edge of the spiral plate, has also been clarified as the causative gene of mild-to-moderate deafness (Sugiyama et al. 2019).
With regard to progression, in the case of the GJB2 and STRC, hearing is rather stable and progression of hearing loss is rarely observed (Tsukada et al. 2010; Yokota et al. 2019). On the other hand, hearing loss due to SLC26A4 (Suzuki et al. 2007; Miyagawa et al. 2014), CDH23 (Miyagawa et al. 2012), TMPRSS3 (Miyagawa et al. 2015c), LOXHD1 (Maekawa et al. 2019), KCNQ4 (Naito et al. 2013), ACTG1 (Miyajima et al. 2020), POU4F3 (Kitano et al. 2017), EYA4 (Shinagawa et al. 2020a), MYO6 (Oka et al. 2020), and mitochondrial m.1555A > G (Usami et al. 1997) requires a good deal of attention as it is progressive. Figure 6 demonstrates the progressive or non-progressive nature of each gene, and supports our previous data regarding progressiveness. From a clinical perspective, the most important information revealed by the analysis using the large-cohort data is whether the deafness is progressive and, if so, the progression rate. As deafness is a rare disease, it is difficult to analyze the progression of hearing loss from a small amount of data, with it only being possible from such large-cohort data as used in this study. This information is crucial for choosing an appropriate therapeutic intervention strategy, such as a hearing aid, electric acoustic stimulation (EAS) or conventional cochlear implantation (CI).
The average audiogram configuration and progressiveness. Hearing in each age group was plotted on the audiogram for each gene. In the case of the GJB2 and STRC genes, hearing is rather stable. On the other hand, hearing loss due to SLC26A4, CDH23, TMPRSS3, LOXHD1, KCNQ4, ACTG1, POU4F3, EYA4, MYO6, and mitochondrial m.1555A > G showed progression with age
With regard to intervention, cochlear implantation is currently the standard therapeutic option for severe-to-profound hearing loss patients. The indications for CI are being expanded to patients with residual hearing and EAS, which uses both electric and acoustic stimulation at the same time, is indicated for such patients. Although CI provides a good outcome in the majority of cases, the outcomes still vary among patients. It is presumed that a number of factors are involved in such variability. Among them, genetic factors, which represent the most common etiology in severe-to-profound hearing loss, might be one of the key determinants of outcomes for CI and EAS (Usami et al. 2012a, 2020; Miyagawa et al. 2016). Causative mutations were successfully identified in 60% of patients with prelingual-onset hearing loss and in 36% of those with post-lingual hearing loss (Miyagawa et al. 2016). Based on our series of studies, good performance can be expected if the intra-cochlear etiology can be proven by gene identification (Miyagawa et al. 2016; Usami et al. 2020). Thus, determination of the involved regions inside/outside of the cochlea by identification of the responsible gene is crucial for selecting the appropriate intervention strategy.
In conclusion, although there are some ethnic differences in the mutational spectra, the genetic epidemiology and clinical features revealed by the present review study are believed to contain a general rule that transcends such ethnic differences.
References
Abe S, Yamaguchi T, Usami SI (2007) Application of deafness diagnostic screening panel based on deafness mutation/gene database using invader assay. Genet Test 11:333–340
Abu Rayyan A, Kamal L, Casadei S et al (2020) Genomic analysis of inherited hearing loss in the Palestinian population. Proc Natl Acad Sci USA 117:20070–20076
Azaiez H, Booth KT, Ephraim SS et al (2018) Genomic landscape and mutational signatures of deafness-associated genes. Am J Hum Genet 103:484–497
Bademci G, Foster J, Mahdieh N et al (2016) Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort. Genet Med 18:364–371
Baux D, Vaché C, Blanchet C et al (2017) Combined genetic approaches yield a 48% diagnostic rate in a large cohort of French hearing-impaired patients. Sci Rep 7:16783
Brownstein Z, Gulsuner S, Walsh T et al (2020) Spectrum of genes for inherited hearing loss in the Israeli Jewish population, including the novel human deafness gene ATOH1. Clin Genet 98:353–364
Budde BS, Aly MA, Mohamed MR et al (2020) Comprehensive molecular analysis of 61 Egyptian families with hereditary nonsyndromic hearing loss. Clin Genet 98:32–42
Cabanillas R, Diñeiro M, Cifuentes GA et al (2018) Comprehensive genomic diagnosis of non-syndromic and syndromic hereditary hearing loss in Spanish patients. BMC Med Genom 11:58
García-García G, Berzal-Serrano A, García-Díaz P et al (2020) Improving the Management of Patients with Hearing Loss by the Implementation of an NGS Panel in Clinical Practice. Genes 11:1467
Ichinose A, Moteki H, Hattori M et al (2015) Novel mutations in LRTOMT associated with moderate progressive hearing loss in autosomal recessive inheritance. Ann Otol Rhinol Laryngol 124:142S-147S
Iwasa YI, Nishio SY, Yoshimura H et al (2013) OTOF mutation screening in Japanese severe to profound recessive hearing loss patients. BMC Med Genet 14:95
Iwasa YI, Nishio SY, Sugaya A et al (2019) OTOF mutation analysis with massively parallel DNA sequencing in 2,265 Japanese sensorineural hearing loss patients. PLoS ONE 14:e0215932
Karafet TM, Mendez FL, Meilerman MB et al (2008) New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res 18:830–838
Kim SY, Kim AR, Kim NK et al (2015) Strong founder effect of p. P240L in CDH23 in Koreans and its significant contribution to severe-to-profound nonsyndromic hearing loss in a Korean pediatric population. J Trans Med 13:263
Kitano T, Miyagawa M, Nishio SY et al (2017) POU4F3 mutation screening in Japanese hearing loss patients: Massively parallel DNA sequencing-based analysis identified novel variants associated with autosomal dominant hearing loss. PLoS ONE 12:e0177636
Kitano T, Kitajiri SI, Nishio SY et al (2019) Detailed clinical features of deafness caused by a claudin-14 variant. Int J Mol Sci 20:4579
Kobayashi M, Miyagawa M, Nishio SY et al (2018) WFS1 mutation screening in a large series of Japanese hearing loss patients: Massively parallel DNA sequencing-based analysis. PLoS ONE 13:e0193359
Landrum MJ, Lee JM, Benson M et al (2018) ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 46:D1062–D1067
Maekawa K, Nishio SY, Abe S et al (2019) Mutational spectrum and clinical features of patients with LOXHD1 variants identified in an 8074 hearing loss patient cohort. Genes 10:735
Matsunaga T, Mutai H, Kunishima S et al (2012) A prevalent founder mutation and genotype–phenotype correlations of OTOF in Japanese patients with auditory neuropathy. Clin Genet 82:425–432
Meyer CG, Amedofu GK, Brandner JM et al (2002) Selection for deafness? Nat Med 8:1332–1333
Miyagawa M, Nishio SY, Usami SI (2012) Prevalence and clinical features of hearing loss patients with CDH23 mutations: a large cohort study. PLoS ONE 7:e40366
Miyagawa M, Naito T, Nishio SY et al (2013) Targeted exon sequencing successfully discovers rare causative genes and clarifies the molecular epidemiology of Japanese deafness. PLoS ONE 8:e71381
Miyagawa M, Nishio SY, Usami SI (2014) Mutation spectrum and genotype–phenotype correlation of hearing loss patients caused by SLC26A4 mutations in the Japanese: a large cohort study. J Hum Genet 59:262–268
Miyagawa M, Nishio SY, Hattori M et al (2015a) Mutations in the MYO15A Gene are a significant cause of nonsyndromic hearing loss: massively parallel DNA sequencing–based analysis. Ann Otol Rhinol Laryngol 124:158S-168S
Miyagawa M, Nishio SY, Ichinose A et al (2015b) Mutational spectrum and clinical features of patients with ACTG1 mutations identified by massively parallel DNA sequencing. Ann Otol Rhinol Laryngol 124:84S-93S
Miyagawa M, Nishio SY, Kumakawa K et al (2015c) Massively parallel DNA sequencing successfully identified seven families with deafness-associated MYO6 mutations: the mutational spectrum and clinical characteristics. Ann Otol Rhinol Laryngol 124:148S-157S
Miyagawa M, Nishio SY, Sakurai Y et al (2015d) The patients associated with TMPRSS3 mutations are good candidates for electric acoustic stimulation. Ann Otol Rhinol Laryngol 124:193S-204S
Miyagawa M, Nishio SY, Usami SI (2016) A comprehensive study on the etiology of patients receiving cochlear implantation with special emphasis on genetic epidemiology. Otol Neurotol 37:e126–e134
Miyajima H, Moteki H, Day T et al (2020) Novel ACTG1 mutations in patients identified by massively parallel DNA sequencing cause progressive hearing loss. Sci Rep 10:7056
Morgan A, Lenarduzzi S, Cappellani S et al (2018) Genomic studies in a large cohort of hearing impaired Italian patients revealed several new alleles, a rare case of uniparental disomy (UPD) and the importance to search for copy number variations. Front Genet 9:681
Morgan A, Lenarduzzi S, Spedicati B et al (2020) Lights and shadows in the genetics of syndromic and non-syndromic hearing loss in the Italian population. Genes 11:1237
Mori K, Miyanohara I, Moteki H et al (2015a) Novel Mutations in GRXCR1 at DFNB25 lead to progressive hearing loss and dizziness. Ann Otol Rhinol Laryngol 124:129S-134S
Mori K, Moteki H, Kobayashi Y et al (2015b) Mutations in LOXHD1 gene cause various types and severities of hearing loss. Ann Otol Rhinol Laryngol 124:135S-141S
Morton CC, Nance WE (2006) Newborn hearing screening—a silent revolution. N Engl J Med 354:2151–2164
Moteki H, Azaiez H, Sloan-Heggen CM et al (2016) Detection and confirmation of deafness-causing copy number variations in the STRC gene by massively parallel sequencing and comparative genomic hybridization. Ann Otol Rhinol Laryngol 125:918–923
Naito T, Nishio SY, Iwasa YI et al (2013) Comprehensive genetic screening of KCNQ4 in a large autosomal dominant nonsyndromic hearing loss cohort: genotype-phenotype correlations and a founder mutation. PLoS ONE 8:e63231
Nishio SY, Usami SI (2015) Deafness gene variations in a 1120 nonsyndromic hearing loss cohort: molecular epidemiology and deafness mutation spectrum of patients in Japan. Ann Otol Rhinol Laryngol 124:49S-60S
Nishio SY, Usami SI (2017) The clinical next-generation sequencing database: a tool for the unified management of clinical information and genetic variants to accelerate variant pathogenicity classification. Hum Mutat 38:252–259
Nishio SY, Hayashi Y, Watanabe M et al (2015) Clinical application of a custom AmpliSeq library and ion torrent PGM sequencing to comprehensive mutation screening for deafness genes. Genet Test Mol Biomark 19:209–217
Nishio SY, Moteki H, Usami SI (2018) Simple and efficient germline copy number variant visualization method for the Ion AmpliSeqTM custom panel. Mol Genet Genomic Med 6:678–686
Ohtsuka A, Yuge I, Kimura S et al (2003) GJB2 deafness gene shows a specific spectrum of mutations in Japan, including a frequent founder mutation. Hum Genet 112:329–333
Oka SI, Day TF, Nishio SY et al (2020) Clinical characteristics and in vitro analysis of MYO6 variants causing late-onset progressive hearing loss. Genes 11:273
Oza AM, DiStefano MT, Hemphill SE et al (2018) Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum Mutat 39:1593–1613
Palombo F, Al-Wardy N, Ruscone GAG et al (2017) A novel founder MYO15A frameshift duplication is the major cause of genetic hearing loss in Oman. J Hum Genet 62:259–264
Park HJ, Shaukat S, Liu XZ et al (2003) Origins and frequencies of SLC26A4 (PDS) mutations in east and south Asians: global implications for the epidemiology of deafness. J Med Genet 40:242–248
Ramzan K, Al-Owain M, Al-Numair NS et al (2020) Identification of TMC1 as a relatively common cause for nonsyndromic hearing loss in the Saudi population. Am J Med Genet B Neuropsychiatr Genet 183:172–180
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–424
Safka Brozkova D, Poisson Marková S, Mészárosová AU et al (2020) Spectrum and frequencies of non GJB2 gene mutations in Czech patients with early non-syndromic hearing loss detected by gene panel NGS and whole-exome sequencing. Clin Genet 98:548–554
Sakuma N, Moteki H, Azaiez H et al (2015) Novel PTPRQ mutations identified in three congenital hearing loss patients with various types of hearing loss. Ann Otol Rhinol Laryngol 124:184S-192S
Seco CZ, Wesdorp M, Feenstra I et al (2017) The diagnostic yield of whole-exome sequencing targeting a gene panel for hearing impairment in The Netherlands. Eur J Hum Genet 25:308–314
Shinagawa J, Moteki H, Nishio SY et al (2020a) Prevalence and clinical features of hearing loss caused by EYA4 variants. Sci Rep 10:3552
Shinagawa J, Moteki H, Nishio SY et al (2020b) Haplotype analysis of GJB2 mutations: founder effect or mutational hot spot? Genes 11:250
Sloan-Heggen CM, Bierer AO, Shearer AE et al (2016) Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet 135:441–450
Snoeckx RL, Huygen PL, Feldmann D et al (2005) GJB2 mutations and degree of hearing loss: a multicenter study. Am J Hum Genet 77:945–957
Sommen M, Schrauwen I, Vandeweyer G et al (2016) DNA diagnostics of hereditary hearing loss: a targeted resequencing approach combined with a mutation classification system. Hum Mutat 37:812–819
Sugiyama K, Moteki H, Kitajiri SI et al (2019) Mid-frequency hearing loss is characteristic clinical feature of OTOA-associated hearing loss. Genes 10:715
Sun Y, Yuan J, Wu L et al (2019) Panel-based NGS reveals disease-causing mutations in hearing loss patients using BGISEQ-500 platform. Medicine 98:e14860
Suzuki H, Oshima A, Tsukamoto K et al (2007) Clinical characteristics and genotype–phenotype correlation of hearing loss patients with SLC26A4 mutations. Acta Oto-Laryngol 127:1292–1297
Taylor KR, Deluca AP, Shearer AE et al (2013) AudioGene: predicting hearing loss genotypes from phenotypes to guide genetic screening. Hum Mutat 34:539–545
Tsukada K, Nishio S, Usami S et al (2010) A large cohort study of GJB2 mutations in Japanese hearing loss patients. Clin Genet 78:464–470
Tsukada K, Ichinose A, Miyagawa M et al (2015a) Detailed hearing and vestibular profiles in the patients with COCH mutations. Ann Otol Rhinol Laryngol 124:100S-110S
Tsukada K, Nishio SY, Hattori M et al (2015b) Ethnic-specific spectrum of GJB2 and SLC26A4 mutations: their origin and a literature review. Ann Otol Rhinol Laryngol 124:61S-76S
Usami S, Abe S, Kasai M et al (1997) Genetic and clinical features of sensorineural hearing loss associated with the 1555 mitochondrial mutation. Laryngoscope 107:483–490
Usami SI, Miyagawa M, Nishio SY et al (2012a) Patients with CDH23 mutations and the 1555A>G mitochondrial mutation are good candidates for electric acoustic stimulation (EAS). Acta Otolaryngol 132:377–384
Usami S, Nishio S, Nagano M et al (2012b) Simultaneous screening of multiple mutations by invader assay improves molecular diagnosis of hereditary hearing loss: a multicenter study. PLoS One 7:e31276
Usami SI, Nishio SY, Moteki H et al (2020) Cochlear implantation from the perspective of genetic background. Anat Rec 303:563–593
Van Laer L, Coucke P, Mueller RF et al (2001) A common founder for the 35delG GJB2 gene mutation in connexin 26 hearing impairment. Med Genet 38:515–518
Wu CC, Tsai CY, Lin YH et al (2019) Genetic epidemiology and clinical features of hereditary hearing impairment in the Taiwanese population. Genes 10:772
Yan D, Tekin D, Bademci G et al (2016) Spectrum of DNA variants for non-syndromic deafness in a large cohort from multiple continents. Hum Genet 135:953–961
Yang T, Wei X, Chai Y et al (2013) Genetic etiology study of the non-syndromic deafness in Chinese Hans by targeted next-generation sequencing. Orphanet J Rare Dis 8:85
Yano T, Nishio SY, Usami SI (2014) Frequency of mitochondrial mutations in non-syndromic hearing loss as well as possibly responsible variants found by whole mitochondrial genome screening. J Hum Genet 59:100–106
Yasukawa R, Moteki H, Nishio SY et al (2019) The prevalence and clinical characteristics of TECTA-associated autosomal dominant hearing loss. Genes 10:744
Yokota Y, Moteki H, Nishio SY et al (2019) Frequency and clinical features of hearing loss caused by STRC deletions. Sci Rep 9:4408
Yuan Y, Li Q, Su Y et al (2020) Comprehensive genetic testing of Chinese SNHL patients and variants interpretation using ACMG guidelines and ethnically matched normal controls. Eur J Hum Genet 28:231–243
Acknowledgements
We would like to thank the following 102 collaborative hospitals for participating in the Deafness Gene Study Consortium; Asahikawa Medical University, Shinoro ENT Clinic, Hokkaido University, Sapporo Medical University, Hakodate Central General Hospital, Hirosaki University, Iwate Medical University, Tohoku Rosai Hospital, Tohoku University, Akita University, Omagari Kousei Medical Center, Yamagata University, Fukushima Medical University, Fukushima Medical University Aizu Medical Center, Hoshi General Hospital, Hitachinaka General Hospital, University of Tsukuba, Dokkyo Medical University, Jichi Medical University, Gunma University, Takasaki Ear Nose & Throat Clinic, Kawagoe Otology Institute, Jichi Medical University Saitama Medical Center, National Rehabilitation Center for Persons with Disabilities, National Defense Medical College, Showa General Hospital, Nippon Medical School, Juntendo University, Tokyo Medical and Dental University, Kamio Memorial Hospital, Keio University, Tokyo Medical University, Tokyo Metropolitan Children's Medical Center, Akasaka Toranomon Clinic, Toranomon Hospital, The Jikei University School of Medicine, International University of Health and Welfare, Abe ENT Clinic, Kitasato University, Yokohama City University Medical Center, Kanagawa Children's Medical Center, Tokai University, Yokohama City University, Niigata University, Kanazawa University, University of Fukui, University of Yamanashi, Miyagawa ENT Clinic, Shinshu University, Gifu University, Nagara Medical Center, Kizawa Memorial Hospital, Kyoai Clinic, Japanese Red Cross Shizuoka Hospital, Hamamatsu University School of Medicine, Nagoya University, Chubu Rosai Hospital, Aichi Children's Health and Medical Center, National Mie Hospital, Mie University, Shiga Medical Center for Children, Shiga University School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto University, Kyoto Teishin Hospital, Osaka Medical College, Osaka University, Kansai Medical University, Kitano Hospital, National Hospital Organization Osaka National Hospital, Japanese Red Cross Osaka Hospital, Osaka Women's and Children's Hospital, Izumi Clinic, Hyogo Prefectural Amagasaki General Medical Center, Hyogo Medical University, Kobe University, Kobe City Medical Center General Hospital, Nara Medical University, Wakayama Medical University, Tottori University, Okayama University, Hiroshima City Hiroshima Citizens Hospital, Hiroshima University, Hiroshima Prefectural Hospital, Yamaguchi University, Tokushima University, Ehime University, Kochi University, Fukuoka Children's hospital, Kyushu University, Fukuoka University, Kurume University, Saga University, Kanda ENT Clinic, Nagasaki University, Oita University, University of Miyazaki, Kagoshima City Hospital, Kagoshima University, and University of the Ryukyus. In addition, we thank BML, Inc. for a collaborative study on social health insurance-based genetic testing. We also thank Fumiko Tomioka and Sachiko Matsuda for their technical assistance with this research, and Dr. Yuichi Isaka for his discussions regarding the preparation and writing of the manuscript.
Funding
This research was funded by the Health and Labor Sciences Research Grant for Research on Rare and Intractable Diseases and Comprehensive Research on Disability Health and Welfare from the Ministry of Health, Labor and Welfare of Japan (S.U.: H29-Nanchitou(Nan)-Ippan-031, 20FC1048), Grants-in-Aid from the Japan Agency for Medical Research and Development (AMED) (S.U.: 16kk0205010h0001, 17kk0205010h0002, 18kk0205010h0003, 17ek0109114h0003, 18ek0109363h0001, 19ek0109363h0002, 20ek0109363h0003) and a Grant-in-Aid for Scientific Research (A) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (S.U.: 15H02565).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Usami, Si., Nishio, Sy. The genetic etiology of hearing loss in Japan revealed by the social health insurance-based genetic testing of 10K patients. Hum Genet 141, 665–681 (2022). https://doi.org/10.1007/s00439-021-02371-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-021-02371-3