Abstract
In this study, genome-wide identification, phylogenetic relationships, duplication time and selective pressure of the NBS-LRR genes, an important group of plant disease-resistance genes (R genes), were performed to uncover their genetic evolutionary patterns in the six Prunus species. A total of 1946 NBS-LRR genes were identified; specifically, 589, 361, 284, 281, 318, and 113 were identified in Prunus yedoensis, P. domestica, P. avium, P. dulcis, P. persica and P. yedoensis var. nudiflora, respectively. Two NBS-LRR gene subclasses, TIR-NBS-LRR (TNL) and non-TIR-NBS-LRR (non-TNL), were also discovered. In total, 435 TNL and 1511 non-TNL genes were identified and could be classified into 30/55/75 and 103/158/191 multi-gene families, respectively, according to three different criteria. Higher Ks and Ka/Ks values were detected in TNL gene families than in non-TNL gene families. These results indicated that the TNL genes had more members involved in relatively ancient duplications and were affected by stronger selection pressure than the non-TNL genes. In general, the NBS-LRR genes were shaped by species-specific duplications, and lineage-specific duplications occurred at recent and relatively ancient periods among the six Prunus species. Therefore, different duplicated copies of NBS-LRRs can resist specific pathogens and will provide an R-gene library for resistance breeding in Prunus species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prunus species are widely cultivated around the world for their economic and ornamental value. Such species include the cherry plum (P. cerasifera Ehrh.), sour cherry (P. cerasus L.), plums (P. domestica L. and P. insititia L.), invasive black cherry (P. serotina Ehrh.), and almond (P. communis Archang.) (Cici and Van Acker 2010). However, these Prunus trees can be infected by various pathogens. Thus, it is necessary to conduct a genetic investigation of the NBS-LRR genes in Prunus species. Based on the release of the whole-genome sequences of six Prunus species, P. yedoensis, P. domestica, P. avium, P. dulcis, P. persica and P. yedoensis var. nudiflora, a genome-wide identification and genetic evolutionary analysis of the NBS-LRR genes in these species was performed in this study.
Plants are constantly subjected to varying environmental conditions and stresses during their growth and development stages. In particular, plants are frequently attacked by a series of bacteria, nematodes, fungi, and insects (Dangl and Jones 2001). As important constituents of the plant innate immune system, disease-resistance genes (R genes) encode specific receptors that recognize pathogenic avirulence (Avr) genes (Keen 1990; Dangl and Jones 2001; Jones and Dangl 2006). A large proportion of plant R genes are called NBS-LRR genes and encode a conserved nucleotide-binding site (NBS) domain and leucine-rich repeat (LRR) motifs (McHale et al. 2006). The N-terminal structures of NBS-LRR proteins contribute to pathogen recognition and downstream signal transduction (Maekawa et al. 2011). According to the characteristics of the N-terminal domain, NBS-LRR proteins are divided into two subgroups. Proteins in one subgroup have a Toll/interleukin-1 receptor (TIR) domain at the N-terminal region (TIR-NBS-LRR proteins), and proteins of the other subgroup contain a coiled-coil (CC) domain, resistance to powdery mildew8 (RPW8) domain, or another type of domain (X) (non-TIR-NBS-LRR proteins) (Xiao et al. 2001; Meyers et al. 2003; Collier et al. 2011).
Plant NBS-LRR proteins are abundant in quantity and ancient in origin. The main function of NBS-LRR genes is resistance to disease by way of pathogen recognition (Dangl and Jones 2001; Meyers et al. 2003). NBS-LRR genes exist widely and have been detected in many different plant species, such as Arabidopsis thaliana, rice (Oryza sativa), cassava (Manihot esculenta), Medicago species, and potato (Solanum tuberosum) (Dangl and Jones 2001; Monosi et al. 2004; Ameline-Torregrosa et al. 2008; Lozano et al. 2012, 2015). However, variations in the copy numbers of NBS-LRR genes have been commonly found among different species. For example, there are only 50 NBS-LRR genes in papaya but 1015 NBS-LRRs in the apple genome (Porter et al. 2009; Arya et al. 2014). A previous study assumed that all plant R genes originate from a common ancestor and that gene duplications have led to variations in the copy numbers of NBS-LRR genes among different species (Meyers et al. 2005). Therefore, the expansion of NBS-LRR genes in specific species may help the plant adapt to rapid changes in the genome of the specific pathogens that harm it (Li et al. 2010). For example, the NBS-LRR genes were shaped by gene amplification in kiwifruit (Actinidia chinensis), in which some of them exhibited a positive response to bacterial canker disease infection (Wang et al. 2020); the Pi2/Pi9 genes encoding NBS-LRR proteins, located in a duplicated cluster of rice, showed broad-spectrum resistance against rice blast (Zhou et al. 2006; Zhu et al. 2012).
Prunus contains approximately 250 plant species and consists of five subgenera (Mace 1940; Robertson et al. 2004; Baek et al. 2018). This study was supported by relatively good whole-genome sequencing data of the six Prunus species. In P. yedoensis, an artificial cross from P. spachiana and P. speciosa (or closely related species), the total sequences with 192-fold genome coverage were assembled into 690.1 Mb and identified 95,076 high-confidence annotated genes (Shirasawa et al. 2019). For P. yedoensis var. nudiflora, also named Cerasus x nudiflora, a natural hybrid from P. pendula f. ascendens and P. jamasakura, which is a distinguishable species of P. yedoensis (Jung and Oh 2005; Roh et al. 2007; Baek et al. 2018). In the heterozygous genome of P. yedoensis var. nudiflora, 323.8 Mb assembled draft genome sequences with annotation of 41,294 protein-encoding genes were obtained based on 73-fold coverage reads capturing 126% of the estimated haploid genome (Baek et al. 2018). In P. avium (sweet cherry), 272.4 Mb of assembly sequences occupied 77.8% of the estimated genome (352.9 Mb) (Shirasawa et al. 2017). In addition, the total read coverage of P. domestica was 210-fold, in which 130,866 genes were annotated as predicted (Genome Database for Rosaceae, GDR, https://www.rosaceae.org/). The assembly completion rate of P. dulcis exceeds 96% according to the 285-fold coverage reads (GDR, https://www.rosaceae.org/). Finally, for the genome sequences of P. persica, higher accuracy and more mapped sequences were performed compared with the high-quality first assembly version (Verde et al. 2017). In total, 1946 NBS-LRR genes were identified among the six Prunus species. Subsequently, the duplication time, selection pressure, and phylogenetic relationships of the NBS-LRR genes were examined here. It was found that both species-specific duplications and lineage-specific duplications jointly contributed to the expansion of NBS-LRR genes in the six Prunus species. In addition, the TNL and non-TNL genes showed distinct evolutionary patterns.
Materials and methods
Identification of NBS-LRR genes
To detect the NBS-LRR genes in the six Prunus species, the whole-genome sequences of P. yedoensis, P. domestica, P. avium, P. dulcis, P. persica and P. yedoensis var. nudiflora were downloaded from the site (GDR, https://www.rosaceae.org/). The NB-ARC domain was searched in the annotated proteins of the whole genomes of all six species using local InterProScan with default settings. Genes encoding NB-ARC domains were considered NBS-encoding candidate genes for further LRR detection. SMART (a Simple Modular Architecture Research Tool; http://smart.embl-heidelberg.de/) was used to examine whether the NBS-encoding genes encoded the LRR motifs. NLR-parser analysis (https://github.com/steuernb/NLR-Parser) was also performed to enhance the accuracy of LRR motif annotation. Subsequently, Pfam analysis was used to determine whether these NBS-LRR genes encoded TIR, RPW8, or other domains. The CC domain was confirmed using the COILS databases (http://embnet.vital-it.ch/software/COILS_form.html).
Classification of the NBS-LRR gene families
The nucleotide coding sequences (CDSs) of the TNL and non-TNL genes in the six Prunus species were processed using all-versus-all BLASTN searches with an E-value of 1.0 (Zhong et al. 2018). Based on the BLAST results, two parameters, the coverage and identity between sequences, both larger than 70%, were used to classify the TNL and non-TNL genes into gene families. Subsequently, to analyze the relatively recent duplications of NBS-LRR genes in the six Prunus species, both the coverage and identity criteria were increased to stricter standards of > 80% and > 90% (Yang et al. 2008; Zhong et al. 2015).
All of the CDSs of the NBS-LRR genes were aligned with the guidance of amino acid sequence alignments using Clustalw2.0 with default settings (Larkin et al. 2007). Subsequently, MEGA X (Kumar et al. 2018) was employed to calculate synonymous substitutions (Ks), nonsynonymous substitutions (Ka), and the ratios of nonsynonymous to synonymous substitutions (Ka/Ks) in each gene family of TNLs and non-TNLs. The mutation rate of peach was 9.48 × 10–9 point mutations per site per generation (Xie et al. 2016), experiencing less than 3 years per generation (Dennis 2009). The duplication time of the NBS-LRR genes was calculated based on the formula T = Ks/9.48 × 10–9/3.
In addition, the nucleotide diversity (Pi value) of each TNL and non-TNL gene family was determined by MEGA X (Kumar et al. 2018), and sequence exchange events were conducted by GENECONV 1.81 (http://www.math.wustl.edu/sawyer/geneconv/) using the default setting with 10,000 permutations (P value < 0.05) (Chen et al. 2010).
Phylogenetic tree of NBS-LRR genes
The nucleotide sequences of the NB-ARC domains of the obtained TNL and non-TNL genes were aligned using the MUSCLE (Multiple Sequence Comparison by Log-Expectation) program with MEGA X (Kumar et al. 2018). Subsequently, the alignments were used to construct two maximum likelihood (ML) phylogenetic trees of TNLs and non-TNLs with MEGA X (Kumar et al. 2018) using the Tamura-Nei model with 1000 replicates. Species-specific duplications were defined as NBS-LRR duplications that appeared in only one Prunus species, and lineage-specific duplications occurred in two or more Prunus species.
Differentially expressed NBS-LRR genes in peach after aphid infection
The RNA-seq data of two P. persica lines, resistant individual R36 and susceptible individual S38, were detected after infection with the green peach aphid (GPA) after 0, 3, 6, 9, 12, 24, 48 and 72 h (Niu et al. 2018a). NBS-LRRs belonging to differentially expressed genes (DEGs), with the criteria of |logFC|≥ 2 (P < 0.05) and FDR < 0.05, and their FPKM values (fragments per kilobase of transcript per million mapped fragments) were screened out to generate heatmaps in R36 and S38 by R script.
Results
NBS-LRR genes in six Prunus species
Using BLAST searches, a total of 1946 NBS-LRR genes were detected in the six Prunus genomes. Of the six species, P. yedoensis possessed the largest number of NBS-LRR genes at 589. For the other five species, P. domestica, P. avium, P. dulcis, P. persica and P. yedoensis var. nudiflora, 361, 284, 281, 318, and 113 NBS-LRR genes were identified, respectively (Table 1). Because the assembly genome of P. yedoensis var. nudiflora was based on homozygous reads possessing half the size of P. yedoensis, leading to a distinct difference in NBS-LRR gene numbers between the two species (Baek et al. 2018). Among the NBS-LRR genes, the two subgroups of genes, TNLs and non-TNLs, could be classified based on their N-terminal domains (Zhong et al. 2015). In total, 435 TNL genes and 1511 non-TNL genes were identified in the six species. Specifically, 162, 3, 76, 87, 105, and 2 TNL genes and 427, 358, 208, 194, 213, and 111 non-TNL genes were identified in P. yedoensis, P. domestica, P. avium, P. dulcis, P. persica and P. yedoensis var. nudiflora, respectively. The numbers of non-TNL genes were significantly greater than the numbers of TNL genes among the six species (t test, P < 0.05).
According to the existence of different N-terminal protein domains, the non-TNL genes of the six Prunus species were classified into two subtypes: 837 CC-NBS-LRR (CNL) genes and 674 X-NBS-LRR (XNL) genes. The CNL subtype contained 827 CC-NBS-LRR’ (CNL’) and 10 RPW8-CC-NBS-LRR (RPW8-CNL) genes. The XNL subtype incorporated 662 X-NBS-LRR’ (XNL’) and 12 RPW8-X-NBS-LRR (RPW8-XNL) genes (Table 1). The RPW8-CNL and RPW8-XNL genes, which contained an N-terminal RPW8 domain, were found in all six of the surveyed species.
NBS-LRR multi-genes in six Prunus species
To detect the multi-gene families of the six Prunus species, gene families were defined based on two criteria: (1) a coverage greater than 70% and (2) an identity between sequences larger than 70%. A total of 409 TNL multi-genes and 1357 non-TNL multi-genes were detected in 30 TNL gene families and 103 non-TNL gene families, respectively (Table 2). The proportion of TNL genes in multi-gene families (94.02%) was greater than that of non-TNL genes in multi-gene families (89.81%). Overall, 90.75% (1766/1946) of all the NBS-LRR genes were contained in multi-gene families (Table 2). Although different numbers of multi-genes were detected in each species, distinctly similar proportions of multi-genes were found in both TNL and non-TNL gene families.
For the TNL gene families, the proportions of multi-genes in four of the species were approximately 90%; specifically, 97.53% in P. yedoensis, 92.11% in P. avium, 89.66% in P. dulcis, and 94.29% in P. persica, except P. domestica had the highest proportion of TNL multi-genes (100%), and P. yedoensis var. nudiflora had the lowest proportion of 50% (Table 2). Similarly, for non-TNL genes, approximately 90% of the genes could be clustered into multi-gene families in four of the species: 92.74% in P. yedoensis, 91.62% in P. domestica, 92.96% in P. persica, and 89.19% in P. yedoensis var. nudiflora, except P. avium (81.73%) and P. dulcis (85.57%), possessed relatively lower proportions of non-TNL multi-genes. In addition, the proportion of TNL multi-genes was greater than that of non-TNL multi-genes in five of the Prunus species. The exception was P. yedoensis var. nudiflora, with 50% of TNL genes in multi-gene families and 89.19% of non-TNL genes in families (Table 2). Furthermore, the average identities within the TNL multi-gene families and the non-TNL multi-gene families were 87.44% and 88.27%, respectively (Table 2). The identity values significantly differed between TNL and non-TNL gene families, which indicated that the non-TNL gene families possessed significantly greater identity values than the TNL gene families (t test, P < 0.01; Table S1).
Subsequently, stricter criteria for coverage and identity (> 80% and > 90%) were used to further elucidate the relatively recent duplications of the NBS-LRR genes among the six species. When the coverage and identity thresholds were changed to > 80%, the proportions of TNL and non-TNL multi-genes decreased among the six Prunus genomes. The proportion of TNL multi-genes was still greater than 80% in the five species but was only 50% in P. yedoensis var. nudiflora (Table 2). The proportion of non-TNL multi-genes exceeded 80% in five Prunus species, but P. avium displayed the lowest proportion of non-TNL multi-genes at 69.23% (Table 2). However, the proportions of multi-genes were significantly lower with the stricter criteria (> 80%) than with the criteria of > 70% in the six species (t test, P < 0.05).
When the cutoff values for coverage and identity were increased to the most stringent criteria (> 90%), the proportions of TNL multi-genes in the six species all exceeded 50%. For the TNL genes in P. domestica under the three criteria (> 70%, > 80% and > 90%), the three TNL genes always belonged to the multi-genes, indicating that they might undergo recent and species-specific duplications. The proportion of non-TNL multi-genes still exceeded 50% in five species but was only 40.38% in P. avium (Table 2). The proportions of multi-genes were significantly lower in all six species using the strictest thresholds (> 90%) than when using standards of > 80% (t test, P < 0.01). Taken together, these results indicated that recent duplication events contributed to the expansion of NBS-LRR genes in the six Prunus species.
NBS-LRR gene expansions in six Prunus species
Ks commonly represents the time elapsed since gene duplication events (Peterson and Masel 2009) and thus could be used to explore the duplication ages of the NBS-LRR genes in the six Prunus genomes. The Ks values were calculated for each TNL and non-TNL gene family according to the criteria of identity and coverage values of > 70%. Because of the influence of nucleotide substitution saturations, only Ks values below 1 were kept for further analysis in this study.
TNL genes had higher median, first and third quartile and mean Ks values than non-TNL genes (Fig. 1A). A t test analysis also showed that the TNL genes had significantly larger Ks values than the non-TNL genes (P < 0.01). Moreover, there were two obvious peaks within the Ks distribution for the TNL gene paralogs, the frequencies of which were relatively similar. The higher peak was from 0.1 to 0.3, and the relatively lower peak was from 0.5 to 0.8. These two peaks represented two distinct expansion periods at a relatively ancient stage (0.5–0.8) and a relatively recent time (0.1–0.3) during the ongoing duplication events of the TNL genes. Within the Ks distribution for non-TNL gene paralogs, there was a more prominent peak, which ranged from 0.1 to 0.2. This peak was indicative of relatively young duplications that occurred in the non-TNL genes. In addition, a less conspicuous peak was located from 0.5 to 0.6 with a lower frequency. This peak suggested that a smaller-scale expansion occurred in a relatively ancient period (Fig. S1).
Ks values of NBS-LRR genes in six Prunus species. A The Ks values of TNLs and non-TNLs. B Ks values of paralogs and orthologs in TNLs and non-TNLs. The top and bottom bars are maximum and minimum values of the Ks ratios; the top and bottom of the box borders show third and first quartiles of the Ks ratios; the bar and small square in the box mean average and median values
These results indicated that there were persistent occurrences of duplication events during the evolution of TNL and non-TNL genes among the six species. Recent duplications can play an important role in the expansion of both TNL and non-TNL genes. However, a higher proportion of TNL genes was generated by ancient duplications than by non-TNL genes. Furthermore, for both TNL and non-TNL genes, the Ks values of the orthologs were significantly higher than those of the paralogs (t test, P < 0.01; Fig. 1B). These results demonstrated that, in general, the species differentiation of the six Prunus species emerged earlier than the duplications of the NBS-LRR genes.
Selective pressure on NBS-LRR genes in six Prunus species
The ratio of nonsynonymous to synonymous nucleotide substitutions (Ka/Ks) is an important parameter for detecting selective constraints on target genes (Zhong et al. 2018). A Ka/Ks ratio larger than 1 indicates positive selection on genes; if the ratio is equal to 1, this demonstrates neutral selection; and a Ka/Ks ratio less than 1 indicates purifying selection.
A fraction of the NBS-LRR genes had Ka/Ks values larger than 1, including 6.07% of the TNL gene pairs (512/8438) and 1.10% of the non-TNL gene pairs (314/28454). This indicated that positive selection was working on these genes in response to the rapid changes in the genomes of different pathogens. In contrast, the remaining TNL and non-TNL gene pairs had Ka/Ks ratios less than 1, illustrating that their evolution was driven by purifying selection. In addition, Ka/Ks values were approximately equal to 1 in seven TNL and 11 non-TNL gene pairs; these genes might face pseudogenization or no functionalization under neutral mutations.
The TNL genes had significantly larger Ka/Ks values than the non-TNL genes (t test, P < 0.01). This was supported by higher third and first quartile, median and average Ka/Ks ratio values of TNL genes than of non-TNL genes (Fig. 2A). This result indicated that the TNL genes might be under stronger selective pressure and experience more rapid evolutionary courses than the non-TNL genes. Moreover, there were significant differences between the Ka/Ks values of the paralogs and orthologs in both the TNL and non-TNL gene families (t test, P < 0.01; Fig. 2B). These differences demonstrated that NBS-LRR gene paralogs were subjected to weaker selection pressure in the six species.
A scatter matrix analysis was adopted to draw confidence ellipses of Ka and Ka/Ks values using the default confidence level of 95%. The relationships between the Ka and Ka/Ks values of TNLs and non-TNLs showed a broader distribution of Ka values for the non-TNLs than for the TNLs over the same Ka/Ks ratio ranges (Fig. 3C and F). Similarly, at the same Ka/Ks values, the Ka values of orthologs spread over a wider scope than those of paralogs in non-TNL gene families (Fig. 3D and E). For the relationships between Ka and Ka/Ks values of TNL genes, higher Ka values were observed for orthologs than for paralogs at the same Ka/Ks values (Fig. 3A and B), which illustrated that the paralogs possessed more functional conservation than the orthologs.
Distribution of Ka and Ka/Ks values of NBS-LRR genes in six Prunus species. The orange triangles represent the paralogs in TNL (A) and non-TNL multi-genes (D), and the green triangles represent the orthologs in TNL (B) and non-TNL multi-genes (E). The blue and purple triangles represent all TNL (C) and non-TNL multi-genes (F), respectively
Variation in NBS-LRR genes among six Prunus species
The nucleotide diversity (Pi value) represents the divergence distance between two genes in each gene family.
In terms of the first and third quartiles, media and average values of the Pi values, the TNL genes were always higher than the non-TNL genes (Fig. 4A). In addition, t test analysis was carried out and revealed that the Pi values of TNL genes were significantly higher than those of non-TNL genes (P < 0.01). In addition, the orthologs had slightly higher Pi values than paralogs in TNL genes. Moreover, a t test analysis showed that the Pi values of orthologs were significantly greater than those of paralogs in non-TNL gene families (P < 0.01; Fig. 4B).
A total of 1604 sequence exchange events were identified in NBS-LRR gene families, among which more exchange events were detected in non-TNLs (1244) than in TNLs (360). Moreover, orthologs (247) had higher sequence exchange events than paralogs (113) in TNL genes. The sequence exchange events of orthologs (849) were significantly greater than those of paralogs (395) in non-TNLs (t test, P < 0.05; Table S2), speculating that the frequent sequence exchange between the orthologs might contribute to the diversification of non-TNL genes.
Phylogenetic analysis of NBS-LRR genes in six Prunus species
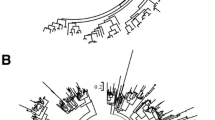
Phylogenetic trees were constructed based on the nucleotide sequences of the NBS domains of the TNL (Fig. 5) and non-TNL (Fig. 6) genes. Within the two phylogenetic ML trees, two types of clades could be defined using bootstrap values larger than 50, including species-specific and lineage-specific duplicated clades. The species-specific duplicated clades, which are represented by the vertical blue lines, include NBS-LRR genes for which duplications only appeared in one Prunus species (Fig. S2). The lineage-specific duplicated clades, represented with vertical red lines, encompass gene duplications that occurred in two or more Prunus species (Fig. S3).
The TNL phylogenetic tree displayed 44 species-specific duplicated clades, including 99 NBS-LRR genes involved in species-specific duplications (Fig. S2). Interestingly, the highest number of TNL genes related to species-specific duplications was 51 in P. yedoensis. Two of the three TNL genes in P. domestica clustered together in a species-specific duplicated clade, which might indicate gene contraction of the TNLs in this Prunus species. However, no TNL genes were found in species-specific duplicated clades of P. yedoensis var. nudiflora. The results showed that 22.76% (99/435) of the TNL genes were generated by species-specific duplications, demonstrating that species-specific duplications were partially responsible for the expansions of TNL genes among five of the Prunus species (all species except P. yedoensis var. nudiflora). In addition, there were 17 lineage-specific duplicated clades in the TNL tree, consisting of 100 genes, showing that 22.99% (100/435) of the TNL genes were produced by lineage-specific duplications. This indicated that lineage-specific duplications also played a partial role in TNL gene expansions.
Compared with the TNL tree, the non-TNL tree contained more species-specific duplicated clades (181) and lineage-specific duplicated clades (29; Fig. S3). There were 432 non-TNL genes in species-specific duplicated clades, illustrating that 28.59% (432/1511) of the non-TNL genes were generated from species-specific duplications among the six Prunus species. Only 13.83% of the non-TNL genes (209/1511) were discovered in lineage-specific duplicated clades. Moreover, 22 RPW8-NBS-LRR (RNL) genes were located in the relatively basal clade of the non-TNL tree (Fig. 6). Among the species-specific duplicated non-TNL genes, nine RNL genes were discovered in four species-specific duplicated clades (Fig. S3). Therefore, both species-specific duplications and lineage-specific duplications partially contributed to the expansions of the NBS-LRR genes among the six Prunus species (Fig. 5 and 6).
DEGs of NBS-LRRs in peaches R36 and S38 after aphid infestation
To verify the disease-resistance function of the NBS-LRR genes, we performed differential expression analysis of the NBS-LRR genes in P. persica infected by aphids. According to the RNA-seq data of two peach genotypes infected by aphids (Niu et al. 2018a), 48 NBS-LRR genes were found to exhibit differential expression among all NBS-LRR genes of P. persica among R36 and S38.
There were different expression patterns of the NBS-LRR genes in R36 and S38 based on hierarchical clustering analysis (Fig. 7). For the resistance genotype R36, the time points of 0, 3, 6, 9, and 12 h clustered in a close group, and the later periods of infection (24, 48, and 72 h) were in another group. That is, the expression levels presented a general increasing trend with the infection time, and the differentially expressed NBS-LRR genes reached their peak from 24 to 72 h after infestation in R36 leaves. For example, the expression levels of Pp5G025500, Pp7G062600 and Pp7G160100 slowly increased from 0 to 36 h but decreased slightly at 72 h. Interestingly, the Pp1G389500 gene had a relatively high expression level at 0 h, was basically steadily upregulated from 0 to 72 h, and reached its expression peak at 72 h. However, the differences in expression mode in susceptible genotype S38 were that the infection time points of 0, 24 and 72 h clustered together, and the relatively early periods of 3, 6, 9, 12, and 48 h clustered in another class. These results indicated a general waved regulation tendency of NBS-LRR expression; in other words, obvious upregulation and downregulation appeared alternately during the whole infection process in S38. The maximum levels of many differentially expressed NBS-LRRs were observed at 6 h or 12 h (Fig. 7B). Pp1G389500, Pp2G027200 and Pp2G037800 are representative examples of the waved expression pattern in S38. They first upregulated from 0 to 6 h, after a slight decrease at 9 h, and then peaked at 12 h; subsequently, they underwent downregulation, upregulation and downregulation from 12 to 72 h. In addition, another two genes, Pp2G055200 and Pp8G027300, also possessed waved expression levels, but their highest expression quantities were found at 6 h.
Expression heatmaps of DEGs of NBS-LRRs in P. persica R36 and S38 treated with aphid infection. A and B represent the resistant genotype R36 and the susceptible genotype S38, respectively. The h means hour after the infection treatment. The eight NBS-LRR genes were marked as representative DEGs in response to aphid infection
Therefore, the different expression patterns between R36 and S38 resulted in relatively lower average expression levels at the early time points of 0 to 12 h in R36 than in S38. However, the average expression levels at the later time points from 24 to 72 h were larger in the resistant genotype R36 than in the susceptible genotype S38. One notable exception was the gene Pp1G389500, whose expression quantities were significantly larger in R36 than in S38 (t test, P < 0.05). Taken together, the results demonstrated that the resistant strain R36 had a longer defense duration than the susceptible strain S38.
According to the expression level analysis, eight representative DEGs of NBS-LRR genes were screened out, including 2 TNLs and 6 non-TNLs. After locating their positions on the two phylogenetic trees and conforming their Ka/Ks values with other family members, one TNL gene (p2G055200) was found to possess a Ka/Ks value larger than 1 and was located with a paralog in a clade with relatively short branch lengths in the phylogenetic tree (Fig. S4). Taken together, it could be inferred that this gene might undergo a relatively fast evolutionary rate and might be a candidate gene for disease-resistance breeding in peaches.
Discussion
In this study, 1946 NBS-LRR genes were detected in the six Prunus plant genomes. Based on the multi-gene classification and Ks distributions, the NBS-LRR genes revealed that large-scale expansions occurred at recent and relatively ancient stages. Interestingly, these NBS-LRR gene expansions were collectively driven by species-specific and lineage-specific duplications among the six Prunus species. In addition, the TNL and non-TNL genes experienced different duplication times and selective pressures, demonstrating that the two subtype NBS-LRR genes might respond to distinct Avr genes with different variation rates. These results supported that the NBS-LRR genes of the six Prunus species were shaped by different genetic events in conflict with specific pathogens in the environments, which could be considered a resistance gene pool for Prunus plant breeding.
Expansions of NBS-LRR genes occurred at different periods
Gene duplication plays an active and crucial role in the enhancement of genetic diversity and the emergence of novel functions in organisms (van der Hoorn and Kamoun 2008). In plants, large numbers of duplicated genes are produced by ancient or recent gene duplications and are then retained for environmental adaptation and growth and development requirements (Panchy et al. 2016). For example, the disease-resistance NBS-LRR genes experience duplications on different scales and at different time points across plant species (van der Hoorn and Kamoun 2008). In the model plant A. thaliana, a total of 149 NBS-LRR genes have been created by widespread duplications and some small-scale genetic events (Meyers et al. 2003). For Rosaceae species, approximately half of the NBS-LRR genes have been derived from recent duplications in apple and pear genomes (Zhong et al. 2015). In this study, although the six Prunus species were derived from a common ancestor, the absolute numbers of NBS-LRR genes differed. The Yoshino cherry (P. yedoensis) genome had a high number of NBS-LRR genes, which was approximately 5.21-, 2.07-, and 1.85-times greater than the number of NBS-LRR genes in P. yedoensis var. nudiflora, P. avium and P. persica, respectively. The distinct variations in NBS-LRR gene number might be due to more excessively large predicted genes in the whole-genome sequences of P. yedoensis than in those of the other species.
The classification of multi-gene families reflected the periods of NBS-LRR gene duplications. The results indicated that duplications mainly emerged in more recent times (recent duplications or young duplications). Previous studies have stated that recent duplications have led to wide arrays of NBS-LRR- and NBS-encoding multi-genes in the genomes of grapevine, poplar, kiwi fruit (Actinidia chinensis), apple, pear, and mei (Yang et al. 2008; Zhong et al. 2015; Li et al. 2016). In the present study, for the strictest standards of coverage and identity values (> 90%), there were significantly lower percentages of TNL and non-TNL multi-genes than when using the undemanding standards of > 70% and > 80% (t test, P < 0.01). Nevertheless, more than 50% of the NBS-LRR genes were incorporated into multi-gene families in the investigated species under the criteria of > 90%. An exception to this was that only 40.38% of non-TNL genes were classified into families in P. avium (Table 2). These results clearly showed that approximately half of the NBS-LRR genes were generated via recent duplications in the six Prunus species.
In addition to the multi-gene classification, the distribution of the Ks values of paralogs highlighted periods of continuous duplication of the NBS-LRR genes across the six species. During the uninterrupted duplications of the NBS-LRR genes, two expansion peaks with similar Ks ranges were detected in the TNL (0.1–0.3) and non-TNL gene families (0.1–0.2; Fig. S1). The smaller Ks values may demonstrate that recent duplications contributed to the expansions of these related genes after the speciation of the six Prunus plants. As ligneous plants with long life cycles, Prunus species are likely to be challenged by various pathogens during their growth, development, and aging stages. Similarly, the concentrations at smaller Ks ratios in other ligneous plants, such as poplar, have previously been reported to manifest the importance of recent duplication events in the evolution of the NBS-LRR genes (Yang et al. 2008). The genetic mechanism underlying the influence of recent duplications on the NBS-LRR genes might be that the duplications provide a large material library for heritable variations, and the specific host plants can use them to resist species-specific pathogens (Parniske et al. 1997; Yang et al. 2008; Zhong et al. 2015).
Nevertheless, relatively ancient duplications were also discovered underlying the NBS-LRR genes of the six species. These ancient duplications were indicated by the other two peaks at relatively large values within the Ks distributions for TNL gene paralogs (0.5–0.8) and non-TNL gene paralogs (0.5–0.6; Fig. S1). It could be inferred that these NBS-LRR gene expansions emerged before the speciation of the six plants. After a variety of genetic events, the offspring copies after a variety of genetic events were reserved to provide defense against lineage-specific bacterial and viral pathogens or insects.
Species-specific and lineage-specific duplications jointly contributed to NBS-LRR gene expansions
According to the gene duplications emerging in one or more species, species-specific duplications and lineage-specific duplications of the NBS-LRR genes could be elucidated in the six Prunus species. Species-specific duplications represent gene duplications that occur in one species in response to selective pressure exerted by certain species-specific pathogens (Yang et al. 2008). Lineage-specific duplications represent gene duplications that occur in two or more species with close genetic relationships (Cannon et al. 2002). In this study, according to the TNL gene phylogenetic tree, 22.76% and 22.99% of the TNL genes were associated with species-specific and lineage-specific duplications, respectively. However, for the non-TNL genes, more species-specific duplicated genes (28.59%) were found than lineage-specific duplicated genes (13.83%). Therefore, both species-specific and lineage-specific duplications participated in the evolutionary processes of NBS-LRR genes. Species-specific duplications contributed more strongly to non-TNL gene expansions than lineage-specific duplications in the six Prunus species. Combining the results of the phylogenetic tree and Ks value analyses, it could be deduced that the species-specific duplications may correspond to the recent duplications (Ks peaks at 0.1–0.2) that occurred after the speciation of the six Prunus species. Furthermore, the lineage-specific duplications showed a great correlation with the relatively ancient duplications (Ks peaks at 0.5–0.8 in TNL and 0.5–0.6 in non-TNL) that arose before Prunus diversification.
The inference could be examined (not shown in Results) and further consolidated on the basis of the previously estimated mutation rate for peach, which was 9.48 × 10–9 point mutations per site per generation (Xie et al. 2016). Considering that the species studied were from the same genus, this same mutation rate could be used in combination with the two paralog Ks peaks to estimate the expansion times of the NBS-LRR genes. Recent species-specific duplications were roughly predicted 31.65–63.29 million years ago (MYA), and relatively ancient lineage-specific duplications began over 158.23 MYA. The recent duplications thus coincided with the Prunus speciation that occurred at approximately 36–44 MYA (Baek et al. 2018) and represented species-specific duplications. Relatively ancient duplications occurred before the appearance of the Prunus genus approximately 61–88 MYA (Chin et al. 2014; Baek et al. 2018) and represented lineage-specific duplications. Therefore, both the species-specific duplications and the lineage-specific duplications, by means of NBS-LRR gene expansions, were constantly in conflict with specific pathogens driven by various environmental factors in the six Prunus species.
The two subclasses of NBS-LRR genes evolved differently among the six Prunus species
The two subclasses of NBS-LRR genes, TNLs and non-TNLs, can be traced back to the ancient period of green algae. Since then, TNLs and non-TNLs have exhibited different copy number variations and evolutionary rates and have been subjected to different selective pressures (Shao et al. 2016, 2019). In this study, significantly fewer TNL gene members were found than non-TNL gene members (t test, P < 0.05) across the plant genomes. This could be explained by the fact that TNL genes were exposed to an extended period of gene contraction, and CNLs, the main essential component of non-TNLs, experienced gradual gene expansions during the stages after angiosperm divergence at approximately 100–225 MYA (Shao et al. 2016). In addition to copy number differences, distinct evolutionary speeds between the two subclasses have also frequently been reported in plants. For example, TNL genes were found to have significantly larger Ks values than non-TNL genes in Arabidopsis, Fragaria and soybean species (Chen et al. 2010; Zhang et al. 2011; Zhong et al. 2015, 2018). In the present study, as expected, the TNL genes possessed significantly greater Ks values than the non-TNL genes (t test, P < 0.01). Broadly speaking, the TNL genes duplicated earlier than the non-TNL genes in the six Prunus species.
Through further analysis of the Ks ranges, it could be deduced that a larger percentage of TNL genes were anciently duplicated than non-TNL genes, which would explain the Ks differences between the two subclasses. These results illustrated that more TNL genes were manufactured by relatively ancient duplications than non-TNL genes, and TNL genes were shaped more rapidly than non-TNL genes for environmental adaptation. Furthermore, it has been found that stronger selection pressures work on TNL genes than on non-TNL genes in the investigated genomes, Rosaceae plants and soybean plants (Zhang et al. 2011; Zhong et al. 2015, 2018). This could be deduced by the significantly greater Ka/Ks values for TNL genes than for non-TNL genes. The arms race between plants and pathogens leads to long-term coevolutionary histories between plant R genes and pathogenic Avr genes (Petit-Houdenot and Fudal 2017). Consequently, the diverse evolutionary profiles of the TNLs and non-TNLs might be due to the coevolution of corresponding Avr genes along with faster or slower rates of genetic variation.
Moreover, the Pi values of TNLs were greater than those of non-TNL gene families, revealing higher sequence divergence between TNL genes than non-TNLs. These results also demonstrated the different evolution patterns between TNL and non-TNL genes, in which the TNL genes evolved faster than non-TNLs (Yang et al. 2008; Chen et al. 2010). The sequence exchange events of non-TNLs were greater than those of TNLs, which might contribute to accumulating more variations for creating novel R genes in six Prunus species (Chen et al. 2010).
Within the non-TNL subclass, the RNL genes (which include RPW8-CNL and RPW8-XNL here) have been regarded as an independent subgroup of NBS-LRR genes for the past few years. The RNL genes also have an ancient origin; the rapid emergence and divergence of TNLs, CNLs, and RNLs can be traced back to the stage prior to green algae divergence (Shao et al. 2019). Accordingly, the RNL genes always cluster into an independent clade distributed in the basal nodes of the phylogenetic tree and can be easily distinguished from the TNLs and CNLs. This has been shown in potato and legume genomes (Jupe et al. 2012; Shao et al. 2014, 2016). This was also the case for the RNL genes of the genomes investigated in the present study (Fig. 6), which further verifies their ancient origin and their sister relationship with CNL genes in the six Prunus species.
NBS-LRR genes have different expression patterns in response to aphid infection between resistant and susceptible P. persica
Previous studies have shown that DEGs mainly play a role in perception, signal transduction, secondary metabolism, transcriptional regulation and plant–pathogen interactions (Gervasi et al. 2018; Ma et al. 2020; Wan et al. 2021). Among the two genotypes of flax (Linum usitatissimium L.), the upregulated genes in the resistant cultivar to Fusarium oxysporum were significantly higher than those in the susceptible cultivar (Dmitriev et al. 2017). Similarly, after bananas (Musa spp.) were infected with F. oxysporum, more DEGs had higher expression levels detected in resistant cultivars ‘Yueyoukang 1’ than in susceptible cultivars ‘Baxijiao’ (Niu et al. 2018b). These results were consistent with those in P. sogdiana, in which the expression level of the R-gene PsoRPM2 in resistant plants was significantly higher than that in susceptible plants after infection with root-knot nematodes (RKNs) (Zhu et al. 2017). In the present study, the differentially expressed NBS-LRR genes from the two peach lines R36 and S38 had distinct responses to aphid infection, showing that the NBS-LRR genes from the resistant genotype had relatively longer defense durations than those from the susceptible genotype. Surprisingly, a similar phenomenon was also previously reported in two peach cultivars, and more DEGs were involved in the response to Xanthomonas arboricola pv. pruni in the susceptible cultivar ‘JH Hale’ at the early infection period, while the resistant cultivar ‘Redkist’ had a greater number of DEGs during the later period (Gervasi et al. 2018).
References
Ameline-Torregrosa C, Wang BB, O’Bleness MS, Deshpande S, Zhu H, Roe B, Young ND, Cannon SB (2008) Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol 146:5–21
Arya P, Kumar G, Acharya V, Singh AK (2014) Genome-wide identification and expression analysis of NBS-encoding genes in Malus x domestica and expansion of NBS genes family in Rosaceae. PLoS ONE 9:e107987
Baek S, Choi K, Kim GB, Yu HJ, Cho A, Jang H, Kim C, Kim HJ, Chang KS, Kim JH, Mun JH (2018) Draft genome sequence of wild Prunus yedoensis reveals massive inter-specific hybridization between sympatric flowering cherries. Genome Biol 19:127
Cannon SB, Zhu HY, Baumgarten AM, Spangler R, May G, Cook DR, Young ND (2002) Diversity, distribution, and ancient taxonomic relationships within the TIR and non-TIR NBS-LRR resistance gene subfamilies. J Mol Evol 54:548–562
Chen QH, Han ZX, Jiang HY, Tian DC, Yang SH (2010) Strong positive selection drives rapid diversification of R-genes in arabidopsis relatives. J Mol Evol 70:137–148
Chin SW, Shaw J, Haberle R, Wen J, Potter D (2014) Diversification of almonds, peaches, plums and cherries—molecular systematics and biogeographic history of Prunus (Rosaceae). Mol Phylogenet Evol 76:34–48
Cici SZ, Van Acker RC (2010) Gene flow in Prunus species in the context of novel trait risk assessment. Environ Biosaf Res 9:75–85
Collier SM, Hamel LP, Moffett P (2011) Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant Microbe Interact 24:918–931
Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826–833
Dennis R Jr (2009) The Peach: botany, production and uses. Choice: Curr Rev Acad Libr 46:1726–1726
Dmitriev AA, Krasnov GS, Rozhmina TA, Novakovskiy RO, Snezhkina AV, Fedorova MS, Yurkevich OY, Muravenko OV, Bolsheva NL, Kudryavtseva AV, Melnikova NV (2017) Differential gene expression in response to Fusarium oxysporum infection in resistant and susceptible genotypes of flax (Linum usitatissimum L). BMC Plant Biol 17:253
Gervasi F, Ferrante P, Dettori MT, Scortichini M, Verde I (2018) Transcriptome reprogramming of resistant and susceptible peach genotypes during Xanthomonas arboricola pv pruni early leaf infection. PLoS ONE 13:e0196590
Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329
Jung YH, Oh MY (2005) Phylogenetic relationships of Prunus (Rosaceae) in Korea and Japan inferred from chloroplast DNA sequences. Korean J Genet 27:279–288
Jupe F, Pritchard L, Etherington GJ, MacKenzie K, Cock PJA, Wright F, Sharma SK, Bolser D, Bryan GJ, Jones JDG, Hein I (2012) Identification and localisation of the NB-LRR gene family within the potato genome. BMC Genom 13:75
Keen NT (1990) Gene-for-gene complementarity in plant-pathogen interactions. Annu Rev Genet 24:447–463
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and clustal X version 2.0. Bioinformatics 23:2947–2948
Li J, Ding J, Zhang W, Zhang Y, Tang P, Chen JQ, Tian D, Yang S (2010) Unique evolutionary pattern of numbers of gramineous NBS-LRR genes. Mol Genet Genom 283:427–438
Li Y, Zhong Y, Huang K, Cheng ZM (2016) Genomewide analysis of NBS-encoding genes in kiwi fruit (Actinidia chinensis). J Genet 95:997–1001
Lozano R, Ponce O, Ramirez M, Mostajo N, Orjeda G (2012) Genome-wide identification and mapping of NBS-encoding resistance genes in Solanum tuberosum group phureja. PLoS ONE 7:e34775
Lozano R, Hamblin MT, Prochnik S, Jannink JL (2015) Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genom 16:360
Ma L, Wang Q, Mu J, Fu A, Wen C, Zhao X, Gao L, Li J, Shi K, Wang Y, Zhang X, Zhang X, Fei Z, Grierson D, Zuo J (2020) The genome and transcriptome analysis of snake gourd provide insights into its evolution and fruit development and ripening. Hortic Res 7:199
Mace MT (1940) A manual of graduate training in surgery published by the american college of surgeons. Can Med Assoc J 43:273–274
Maekawa T, Cheng W, Spiridon LN, Toller A, Lukasik E, Saijo Y, Liu P, Shen QH, Micluta MA, Somssich IE, Takken FLW, Petrescu AJ, Chai J, Schulze-Lefert P (2011) Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9:187–199
McHale L, Tan X, Koehl P, Michelmore RW (2006) Plant NBS-LRR proteins: adaptable guards. Genome Biol 7:212
Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15:809–834
Meyers BC, Kaushik S, Nandety RS (2005) Evolving disease resistance genes. Curr Opin Plant Biol 8:129–134
Monosi B, Wisser RJ, Pennill L, Hulbert SH (2004) Full-genome analysis of resistance gene homologues in rice. Theor Appl Genet 109:1434–1447
Niu L, Pan L, Zeng WF, Lu ZH, Cui GC, Fan ML, Xu Q, Wang ZQ, Li GH (2018a) Dynamic transcriptomes of resistant and susceptible peach lines after infestation by green peach aphids (Myzus persicae Sulzer) reveal defence responses controlled by the Rm3 locus. BMC Genom 19:846
Niu YQ, Hu B, Li XQ, Chen HB, Takac T, Samaj J, Xu CX (2018b) Comparative digital gene expression analysis of tissue-cultured plantlets of highly resistant and susceptible banana cultivars in response to Fusarium oxysporum. Int J Mol Sci 19:350
Panchy N, Lehti-Shiu M, Shiu SH (2016) Evolution of gene duplication in plants. Plant Physiol 171:2294–2316
Parniske M, Hammond-Kosack KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BB, Jones JD (1997) Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91:821–832
Peterson GI, Masel J (2009) Quantitative prediction of molecular clock and K-a/K-s at short timescales. Mol Biol Evol 26:2595–2603
Petit-Houdenot Y, Fudal I (2017) Complex interactions between fungal avirulence genes and their corresponding plant resistance genes and consequences for disease resistance management. Front Plant Sci 8:1072
Porter BW, Paidi M, Ming R, Alam M, Nishijima WT, Zhu YJ (2009) Genome-wide analysis of Carica papaya reveals a small NBS resistance gene family. Mol Genet Genom 281:609–626
Robertson A, Newton AC, Ennos RA (2004) Multiple hybrid origins, genetic diversity and population genetic structure of two endemic Sorbus taxa on the Isle of Arran, Scotland. Mol Ecol 13:123–134
Roh MS, Cheong EJ, Choi IY, Joung YH (2007) Characterization of wild Prunus yedoensis analyzed by inter-simple sequence repeat and chloroplast DNA. Sci Hortic 114:121–128
Shao ZQ, Zhang YM, Hang YY, Xue JY, Zhou GC, Wu P, Wu XY, Wu XZ, Wang Q, Wang B, Chen JQ (2014) Long-term evolution of nucleotide-binding site-leucine-rich repeat genes: understanding gained from and beyond the legume family. Plant Physiol 166:217–234
Shao ZQ, Xue JY, Wu P, Zhang YM, Wu Y, Hang YY, Wang B, Chen JQ (2016) Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol 170:2095–2109
Shao ZQ, Xue JY, Wang Q, Wang B, Chen JQ (2019) Revisiting the origin of plant NBS-LRR genes. Trends Plant Sci 24:9–12
Shirasawa K, Isuzugawa K, Ikenaga M, Saito Y, Yamamoto T, Hirakawa H, Isobe S (2017) The genome sequence of sweet cherry (Prunus avium) for use in genomics-assisted breeding. DNA Res 24:499–508
Shirasawa K, Esumi T, Hirakawa H, Tanaka H, Itai A, Ghelfi A, Nagasaki H, Isobe S (2019) Phased genome sequence of an interspecific hybrid flowering cherry, ‘Somei-Yoshino’ (Cerasus x yedoensis). DNA Res 26:379–389
van der Hoorn RAL, Kamoun S (2008) From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20:2009–2017
Verde I, Jenkins J, Dondini L, Micali S, Pagliarani G, Vendramin E, Paris R, Aramini V, Gazza L, Rossini L, Bassi D, Troggio M, Shu SQ, Grimwood J, Tartarini S, Dettori MT, Schmutz J (2017) The Peach v2.0 release: high-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genom 18:225
Wan R, Guo C, Hou X, Zhu Y, Gao M, Hu X, Zhang S, Jiao C, Guo R, Li Z, Wang X (2021) Comparative transcriptomic analysis highlights contrasting levels of resistance of Vitis vinifera and Vitis amurensis to Botrytis cinerea. Hortic Res 8:103
Wang T, Jia ZH, Zhang JY, Liu M, Guo ZR, Wang G (2020) Identification and analysis of NBS-LRR Genes in Actinidia chinensis genome. Plants-Basel 9:1350
Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M, Turner JG (2001) Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291:118–120
Xie ZQ, Wang L, Wang LR, Wang ZQ, Lu ZH, Tian DC, Yang SH, Hurst LD (2016) Mutation rate analysis via parent-progeny sequencing of the perennial peach. I. A low rate in woody perennials and a higher mutagenicity in hybrids. Proc Royal Soc B-Biol Sci 283:20161016
Yang SH, Zhang XH, Yue JX, Tian DC, Chen JQ (2008) Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Genet Genom 280:187–198
Zhang X, Feng Y, Cheng H, Tian D, Yang S, Chen JQ (2011) Relative evolutionary rates of NBS-encoding genes revealed by soybean segmental duplication. Mol Genet Genom 285:79–90
Zhong Y, Yin H, Sargent DJ, Malnoy M, Cheng ZM (2015) Species-specific duplications driving the recent expansion of NBS-LRR genes in five Rosaceae species. BMC Genom 16:77
Zhong Y, Zhang X, Cheng ZM (2018) Lineage-specific duplications of NBS-LRR genes occurring before the divergence of six Fragaria species. BMC Genom 19:128
Zhou B, Qu SH, Liu GF, Dolan M, Sakai H, Lu GD, Bellizzi M, Wang GL (2006) The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol Plant Microbe Interact 19:1216–1228
Zhu XY, Chen S, Yang JY, Zhou SC, Zeng LX, Han JL, Su J, Wang L, Pan QH (2012) The identification of Pi50(t), a new member of the rice blast resistance Pi2/Pi9 multi-gene family. Theor Appl Genet 124:1295–1304
Zhu X, Xiao K, Cui HY, Hu JF (2017) Overexpression of the Prunus sogdiana NBS-LRR subgroup gene PsoRPM2 promotes resistance to the root-knot nematode meloidogyne incognita in tobacco. Front Microbiol 8:2113
Acknowledgements
This study was supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions, China, and the high-performance computing platform of Bioinformatics Center, Nanjing Agricultural University.
Author information
Authors and Affiliations
Contributions
YZ designed and initiated this study. YZ and ZC carried out the bioinformatics analyses and wrote the manuscript. YZ and ZMC critically revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Research involved with human or animals participants
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Bing Yang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file2 Fig. S2. Phylogenetic tree of TNL genes among six Prunus species. (TIF 26823 KB)
438_2021_1849_MOESM4_ESM.tif
Supplementary file4 Fig. S4. The clade of the TNL phylogenetic tree containing the p2G055200 gene. The gene p2G055200 is marked by red highlight. (TIF 522 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhong, Y., Chen, Z. & Cheng, ZM. Different scales of gene duplications occurring at different times have jointly shaped the NBS-LRR genes in Prunus species. Mol Genet Genomics 297, 263–276 (2022). https://doi.org/10.1007/s00438-021-01849-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-021-01849-z