Abstract
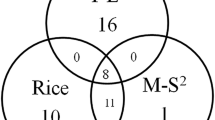
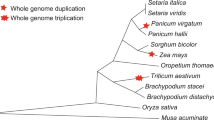
Nucleotide binding site (NBS)–leucine-rich repeat (LRR) genes belong to the largest class of disease-resistance gene super groups in plants, and their intra- or interspecies nucleotide variations have been studied extensively to understand their evolution and function. However, little is known about the evolutionary patterns of their copy numbers in related species. Here, 129, 245, 239 and 508 NBSs were identified in maize, sorghum, brachypodium and rice, respectively, suggesting considerable variations of these genes. Based on phylogenetic relationships from a total of 496 ancestral branches of grass NBS families, three gene number variation patterns were categorized: conserved, sharing two or more species, and species-specific. Notably, the species-specific NBS branches are dominant (71.6%), while there is only a small percentage (3.83%) of conserved families. In contrast, the conserved families are dominant in 51 randomly selected house-keeping genes (96.1%). The opposite patterns between NBS and the other gene groups suggest that natural selection is responsible for the drastic number variation of NBS genes. The rapid expansion and/or contraction may be a fundamentally important strategy for a species to adapt to the quickly changing species-specific pathogen spectrum. In addition, the small proportion of conserved NBSs suggests that the loss of NBSs may be a general tendency in grass species.





Similar content being viewed by others
References
Ameline-Torregrosa C, Wang BB, O’Bleness MS, Deshpande S, Zhu H, Roe B, Young ND, Cannon SB (2008) Identification and characterization of NBS–LRR genes in the model plant Medicago truncatula. Plant Physiol 146:5–21
Bailey TL, Elkan C (2005) The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol 3:21–29
Bakker EG, Toomajian C, Kreitman M, Bergelson J (2006) A genome-wide survey of R gene polymorphisms in Arabidopsis. Plan Cell 18:1803–1818
Brunner S, Fengler K, Morgante M, Tingey S, Rafalski A (2005) Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17:343–360
Chen Q, Han Z, Jiang H, Tian D, Yang S (2010) Strong positive selection drives rapid diversification of R-genes in Arabidopsis relatives. J Mol Evol 70:137–148
Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826–833
Ellis JG, Lawrence GJ, Luck JE, Dodds PN (1999) Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11:495–506
Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M (2008) Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320:1629–1631
Gaut BS (2002) Evolutionary dynamics of grass genomes. New Phytol 154:15–28
Hammond-Kosack KE, Jones JD (1997) Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol 48:575–607
Hulbert SH, Webb CA, Smith SM, Sun Q (2001) Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol 39:285–312
Huo N, Lazo GR, Vogel JP, You FM, Ma Y, Hayden DM, Coleman-Derr D, Hill TA, Dvorak J, Anderson OD, Luo MC, Gu YQ (2008) The nuclear genome of Brachypodium distachyon: analysis of BAC end sequences. Funct Integr Genom 8:135–147
Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329
Kohany O, Gentles AJ, Hankus L, Jurka J (2006) Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinform 7:474
Kuang H, Woo SS, Meyers BC, Nevo E, Michelmor RW (2004) Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell 16:2870–2894
Long M, Betrán E, Thornton K, Wang W (2003) The origin of new genes: glimpses from the young and old. Nat Rev Genet 4:865–875
Lupas A, Van Dyke M, Stock J (1991) Predicting coiled coils from protein sequences. Science 252:1162–1164
Lynch M, Crease TJ (1990) The analysis of population survey data on DNA sequence variation. Mol Biol Evol 7:377–394
Matsuoka Y, Vigouroux Y, Goodman MM, Sanche GJ, Buckler E, Doebley J (2002) A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci USA 99:6080–6084
McDowell JM, Simon SA (2008) Molecular diversity at the plant–pathogen interface. Dev Comp Immunol 32:736–744
McHale L, Tan X, Koehl P, Michelmore RW (2006) Plant NBS–LRR proteins: adaptable guards. Genome Biol 7:212
Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS–LRR-encoding genes in Arabidopsis. Plant Cell 15:809–834
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Paterson AH, Bowers JE, Bruggmann R, Dubchak I et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Porter BW, Paidi M, Ming R, Alam M, Nishijima WT, Zhu YJ (2009) Genome-wide analysis of Carica papaya reveals a small NBS resistance gene family. Mol Genet Genom 281:266–609
Sacristán S, Vigouroux M, Pedersen C, Skamnioti P, Thordal-Christensen H, Micali C, Brown JK, Ridout CJ (2009) Coevolution between a family of parasite virulence effectors and a class of LINE-1 retrotransposons. PLOS One 4:e7463
Scheideler M, Schlaich NL, Fellenberg K, Beissbarth T, Hauser NC, Vingron M, Slusarenko AJ, Hoheisel JD (2002) Monitoring the switch from housekeeping to pathogen defense metabolism in Arabidopsis thaliana using cDNA arrays. J Biol Chem 277:10555–10561
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Sun X, Zhang Y, Yang S, Chen JQ, Hohn B, Tian D (2008) Insertion DNA promotes ectopic recombination during meiosis in Arabidopsis. Mol Biol Evol 25:2079–2083
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J (2003) Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423:74–77
Wang QH, Dooner H (2006) Remarkable variation in maize ge- nome structure inferred from haplotype diversity at the bz locus. Proc Natl Acad Sci USA 103:17644–17649
Wang W, Zheng H, Fan C, Li J, Shi J et al (2006) High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell 18:1791–1802
Webb CA, Richter TE, Collins NC, Nicolas M, Trick HN, Pryor T, Hulbert SH (2002) Genetic and molecular characterization of the maize rp3 rust resistance locus. Genetics 162:381–394
Yang S, Feng Z, Zhang X, Jiang K, Jin X, Hang Y, Chen JQ, Tian D (2006) Genome-wide investigation on the genetic variations of rice disease resistance genes. Plant Mol Biol 62:181–193
Yang S, Zhang X, Yue J, Tian D, Chen JQ (2008) Recent duplication dominates the expansion of the NBS-encoding genes in two woody species. Mol Genet Genom 280:187–198
Zhang Y, Wang J, Zhang X, Chen JQ, Tian D, Yang S (2009) Genetic signature of rice domestication shown by a variety of genes. J Mol Evol 68:393–402
Zhou T, Wang Y, Chen JQ, Araki H, Jing Z, Jiang K, Shen J, Tian D (2004) Genome-wide identification of NBS genes in rice reveals significant expansion of divergent non-TIR NBS Genes. Mol Genet Genom 271:402–415
Acknowledgments
This work was supported by the National Natural Science Foundation of China (30970198 and 30870176), the Key Project of Chinese Ministry of Education (109071), and the Natural Science Foundation of Jiangsu Province (BK2009235) to S. Y. and J.-Q.C.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by Y. Van de Peer.
J. Li and J. Ding contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, J., Ding, J., Zhang, W. et al. Unique evolutionary pattern of numbers of gramineous NBS–LRR genes. Mol Genet Genomics 283, 427–438 (2010). https://doi.org/10.1007/s00438-010-0527-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-010-0527-6