Abstract
The disease resistance (R) genes play an important role in protecting plants from infection by diverse pathogens in the environment. The nucleotide-binding site (NBS)-leucine-rich repeat (LRR) class of genes is one of the largest R gene families. Chinese chestnut (Castanea mollissima) is resistant to Chestnut Blight Disease, but relatively little is known about the resistance mechanism. We identified 519 NBS-encoding genes, including 374 NBS-LRR genes and 145 NBS-only genes. The majority of Ka/Ks were less than 1, suggesting the purifying selection operated during the evolutionary history of NBS-encoding genes. A minority (4/34) of Ka/Ks in non-TIR gene families were greater than 1, showing that some genes were under positive selection pressure. Furthermore, Ks peaked at a range of 0.4 to 0.5, indicating that ancient duplications arose during the evolution. The relationship between Ka/Ks and Ks indicated greater selective pressure on the newer and older genes with the critical value of Ks = 0.4–0.5. Notably, species-specific duplications were detected in NBS-encoding genes. In addition, the group of RPW8-NBS-encoding genes clustered together as an independent clade located at a relatively basal position in the phylogenetic tree. Many cis-acting elements related to plant defense responses were detected in promoters of NBS-encoding genes.
Similar content being viewed by others

Introduction
Plants have a large number of disease RESISTANCE (R) genes for defense against numerous and various pathogens, including bacteria, fungi, oomycetes, viruses and nematodes1,2,3. R genes encode proteins that allow plants to systematically recognize and respond to pathogen infection4,5,6. NBS-LRR genes are one of the largest gene families in plant genomes and the largest class of known disease resistance genes. Among the over 140 R genes characterized from different flowering plants, approximately 80% belong to the NBS-LRR (nucleotide-binding site and leucine-rich repeat) gene families, members of which can directly or indirectly identify the pathogen2,7,8.
The NBS domain is thought to be required in binding and hydrolyzing ATP and GTP1. The LRR motif in NBS-encoding genes functions is to regulate direct or indirect interactions with pathogens9. Based on the presence or absence of an N-terminal Toll/Interleukin-1 receptor (TIR) domain, NBS-encoding genes are divided into TIR-NBS-LRR/TIR-NBS (TNL/TN) genes and non-TIR-NBS-LRR/non-TIR-NBS (non-TNL/non-TN) genes10. The latter usually has a coiled-coil (CC) or other domains at the N-terminus, therefore this category can be further divided into CC-NBS-LRR/CC-NBS genes (CNL/CN) and X-NBS-LRR/X-NBS genes (XNL/XN)1. CC and LRR domains co-regulate the signaling capacity of the NBS domain in a recognition-specific manner11. Further, some NBS-encoding genes have another domain RPW8, which confers resistance to powdery mildew with a transmembrane region before the CC structure at N-terminus12. The RPW8-NBS-LRR (RNL) group is generally regarded as a small but special subclass of non-TNL13,14. However, recent studies indicate that the RNL group is an individual sister group to the non-TNL group15,16,17.
Gene duplications have contributed to the high numbers and proportions of NBS-LRR genes in plant families18. The 174, 519, 416 NBS-LRR genes have been identified in Arabidopsis, rice and poplar, the model systems for dicots, monocots and woody plants, respectively2,19,20,21. Chestnut acts as a model species for the Fagaceae that dominate the hardwood forests of the northern hemisphere22 and have significant economic and ecological value. Chestnut can be infected with pathogens that cause diseases such as chestnut blight23,24, ink disease25 or bark disease26,27. Furthermore, many disease resistance genes belonging to the NBS-LRR gene families have been identified9,28,29,30,31,32,33,34,35.
Chestnuts (Castanea spp.) are important nuts and forest trees and play important roles in ecosystem and generate nuts for wildlife and specialty nuts for human consumption. Chinese chestnut is resistant to chestnut blight, caused by fungal pathogen Cryphonectria parasitica (formerly Endothia parasitica), which, when it was accidentally introduced to North America around 1904 from Japan with nursery stocks, has almost wiped out the American chestnut tree (Castanea dentata) in the early 1900’s, once plentiful tree in the eastern United States. However, the resistance mechanism to chestnut blight in Chinese chestnut is not clear. The whole genome sequence recently released22,36 provides the opportunity to undertake a whole-genome analysis of NBS-encoding R genes in Chinese chestnut to obtain insight into the evolutionary development of this gene family. We used poplar (Populus tricocarpa)21,37 as a reference, which was the most closely related to chestnut among all the sequenced species, to identify chestnut-specific duplications after the divergence between Chinese chestnut and poplar. Results of this genome-wide analysis suggests that ancient and species-specific duplications have contributed to the expansion of NBS-encoding genes in Chinese chestnut. This research lays a foundation for further characterizing these R genes and helps identifying R genes that may be involved in resistance to chestnut blight.
Results
Total number of NBS-encoding genes in Chinese chestnut
A total of 519 NBS-encoding genes were identified in the C. mollissima genome, including 374 NBS-LRR genes and 145 NBS-only genes (Table 1). The NBS-encoding genes comprised 1.36% of expressed genes in Chinese chestnut compared with 0.91% in poplar. Accordingly, the proportion of NBS-LRR genes in Chinese chestnut (0.98%) was also higher than that in poplar (0.72%). Among NBS-LRR and NBS-only genes, two types of genes could be subclassified based on their N-terminal structures: TIRs and non-TIRs. Among the NBS-encoding genes, 27 TIR genes (22 TNLs and 5 TNs) and 492 non-TIR genes (352 non-TNLs and 140 non-TNs) were found. These results demonstrated that the number of non-TIR genes was greater than that of TIR genes, which was similar to the numbers of the two types of NBS-encoding genes in poplar. Furthermore, the proportions of non-TNLs were greater than these of TNLs in both Chinese chestnut and poplar genomes. In the Chinese chestnut genome, 0.06% and 0.92% of genes detected were TNLs and non-TNLs, respectively, compared to proportions of 0.17% and 0.55% for those gene categories in poplar. However, although the number of NBS-encoding genes in Chinese chestnut was greater than that in poplar, the number of TIR genes in Chinese chestnut was smaller than that in poplar. Moreover, the non-TIR genes could also be further divided into 32 CNLs, 96 CNs, 320 XNLs and 44 XNs (Table 1).
The average number of exons identified in NBS-LRR genes from Chinese chestnut was 4.37, which was greater than the average of 2.35 exons for NBS-LRR genes in poplar. In addition, the average number of exon in TNL genes was less than that in non-TNL genes in Chinese chestnut, which was different from the average exon numbers in these genes in grape and poplar21. The average exon number in TNL, non-TNL, CNL, NBS-encoding and NBS-LRR genes was 2.50, 2.93, 3.32, 2.66 and 2.90, respectively, which were all less than for all genes predicted in the Chinese chestnut genome.
Duplication of NBS-encoding genes in the Chinese chestnut genome
According to criteria for both coverage ≥70% and identity ≥70%, 273 genes were detected in 64 NBS gene families in Chinese chestnut. Therefore, 246 of these genes were singletons in the Chinese chestnut genome (Table 2). The percentage of multiple NBS-encoding genes in Chinese chestnut (52.60%) was much lower than that in poplar (78.13%). The average number of members per family was 4.27 in chestnut and 5.33 in poplar and the maximal number members within a family of these genes in Chinese chestnut (19) was less than that in poplar (23), which indicated fewer NBS gene duplications and multi-gene families in Chinese chestnut than in poplar.
If coverage and identity criteria were changed to 80%, the proportion of multiple genes among all NBS-encoding genes decreased in both Chinese chestnut and poplar. The proportions of multiple NBS-encoding genes were 41.43% and 74.52% between Chinese chestnut and poplar, respectively, which could allow the inference that more relatively recent duplications have occurred in the poplar genome. When coverage and identity criteria were increased to 90%, 19.46% of the multiple genes were still identified in Chinese chestnut, indicating that recent duplication events partly contributed to the expansion of NBS-encoding genes.
The multi-gene families and number of multiple genes encoding TNLs and non-TNLs were diverse in Chinese chestnut. The numbers of both multi-gene families and multiple genes in non-TNLs were greater than those in TNLs, indicating that duplication of NBS-encoding genes occurred primarily among non-TNL genes (Table 2).
Duplication time of NBS-LRR genes in chestnut
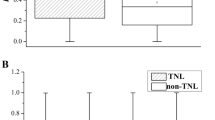
Ks is the time indicator for duplication events and the frequency distributions of individual Ks values reflect the relative time of genome duplications21. Firstly, we calculated the rate of synonymous substitutions and obtained the frequency distribution of Ks values in non-TIR-NBS-encoding genes (Fig. 1), which had a significant Ks peak in the range from 0.4 to 0.5. However the tendency decreased when Ks values ranged from 0.5 to 1. Ks values in the range from 0.1 to 0.2 accounted for 12.19% of the values in the range of Ks values between 0 and 1.0, which demonstrated relatively recent duplications in Chinese chestnut. However, the Ks range from 0.4 to 1.0 indicates that 35.41% of the duplication events occurred during chronologically relatively distant periods. Secondly, the curve between Ka/Ks and Ks for non-TIR-NBS-encoding genes in Chinese chestnut was fitted to detect any relationship between evolutionary pressure and duplication time for NBS paralogs. It was clear that the younger and older genes had larger Ka/Ks values, indicating that they were under greater selective pressures within the critical range of Ks values between 0.4 and 0.5.
However, among TIR-NBS-encoding genes, only two Ks values (0.4781 and 0.1624) were calculated from two gene families including four members, with Ka/Ks values of 0.3606 and 0.4554, respectively, which were lower than the average Ka/Ks value for non-TIR gene families, indicating greater functional constraints in TIR gene families.
Selective pressure on NBS-encoding genes in Chinese chestnut
Plant disease resistance genes have been shown to be subject to positive selection38. To better understand the evolutionary fate of NBS-encoding duplicates in Chinese chestnut, we used site and branch models in PAML4 to detect positive selection patterns. Because this analysis requires comparison of at least three genes, TIR-NBS-encoding genes were not evaluated and 34 non-TIR genes families with greater than three members each were analyzed.
We calculated the ratios of nonsynonymous to synonymous nucleotide substitutions (ω), a molecular evolutionary measure of selection pressure39, to detect positive selection in NBS-encoding genes. A value of ω greater than 1 indicates that a gene is evolving with more constraint on nonsynonymous substitutions than on synonymous substitutions, which is evidence of positive selection40. In contrast, value of ω less than 1 indicate purifying selection. A value for ω of 1 means that a gene is under neutral selection. As Fig. 2 shows, most of these gene families (30/34) have undergone purifying selection. Meanwhile, four gene families had values for ω of greater than 1, which demonstrated that some NBS-encoding gene families were under positive selection.
Selective pressure on non-TIR-NBS-encoding genes in Chinese chestnut.
Numbers represent the dN/dS ratio for each gene family using the branch model; 2∆ln represents the result of the LR test for the site model; * and ** represent, respectively, significant (2∆ln > 5.991, p < 0.05) and highly significant (2∆ln > 9.210, p < 0.01) tests for positive selection between model M7 and M8.
Subsequently, the result of the LR test, 2Δln detected positive selection of significant differences between the M7 and M8. It was important to note that 85.29% (29/34) (Fig. 2) of non-TIR gene families had some sites under highly significant positive selection pressure, showing positive selection on gene families played a certain role in the evolution of non-TIR-NBS-encoding genes. Specifically, the analysis revealed that two gene families (Family 30 and 33) (Fig. 2) were driven by significant positive selection. Further, Bayes Empirical Bayes (BEB) analysis was performed to detect amino acid sites that have been under positive selection41. The sites inferred to be under positive selection at the 95% (*) and 99% (**) confidence intervals are shown in Table S1 (Additional file 1), which suggested that these positively selected sites possessed relatively high substitutions compared with others among NBS-LRR genes.
Taken together, 530 amino acid sites from 22 families were revealed to be under positive selection in non-TIR gene families, which might have driven the evolution of function in NBS-encoding genes in Chinese chestnut (Additional file 1: Table S1).
Phylogenetic analysis of NBS-encoding genes
To confirm the phylogenetic relationships among the NBS-encoding genes, the NBS domains of 519 NBS-encoding genes were analyzed and compared with those in poplar and a phylogenetic tree was constructed. A few genes had longer branch lengths but the majority had relatively shorter branches (Additional file 2: Figure S1), which indicated two different evolutionary patterns occurred in NBS-encoding genes.
In general, the majority of the NBS-encoding genes were clustered according to species, which suggested species-specific duplication during the evolution of Chinese chestnut. In the phylogenetic tree, when the clade was defined by bootstrap values of greater than 50%, the clades resulting from species-specific duplication were counted. Specifically, 81 clades, including 401 NBS-encoding genes were identified with 4.95 R paralogs per clade. Additionally, the proportion of species-specific duplicated genes in Chinese chestnut was 77.26%. This result clearly indicated that NBS-encoding genes expanded into multiple gene family members after the divergence of Chinese chestnut and poplar.
The N-terminal RPW8 domain (RESISTANCE TO POWDERY MILDEW8) in the NBS-encoding genes, medicates broad-spectrum resistance in Arabidopsis12. The analysis of RPW8 genes helps explain the origin and relationships of RPW8 genes to other genes. In the present study, a total of 15 Chinese chestnut RPW8-NBS-encoding genes are marked with solid circle in phylogenetic tree (Additional file 2: Figure S1). For comparison, five NBS-encoding genes from poplar containing the RPW8 domain are marked with triangle in poplar (Additional file 2: Figure S1). Interestingly, the genes carrying the RPW8 domain were clustered together (Additional file 2: Figure S1).
Additionally, the 15 RPW8 genes from Chinese chestnut formed a relatively independent and monophyletic group, that was not phylogenetically embedded within other clades. Similar results were obtained for RNL genes from M. truncatula, potato, soybean, common bean and pigeon pea genomes15,16,17. Notably, the position of RPW8 genes of Chinese chestnut were located at a relatively basal, but not the most basal position on the phylogenetic tree.
The cis-element analysis of NBS-encoding genes promoter sequences
Plant defense is controlled by cis-regulatory elements corresponding to key genes involved in defense and pathogen-specific responses42. Therefore, the investigation and identification of cis-acting elements in the promoters of NBS-encoding genes will help us understand the function in plant defense responses. In this analysis, we performed the cis-elemtne analysis of all NBS-encoding genes in the PLACE. It is noted that many cis-regulatory elements associate with plants responding to pathogens, including DOFCOREZM, EECCRCAH1, GT1GAMSCAM4, GT1CONSENSUS and AGCBOXNPGLB43,44 in promoter regions of NBS-encoding genes (Table 3 and Additional file 3), which might demonstrate that NBS-encoding genes involve in response to pathogen infections. Moreover, the results (Additional file 3) point a way to identify candidate genes that might be used for conferring disease resistance.
Discussion
A small number of TIR-NBS-encoding genes in Chinese chestnut
Surveys for NBS-encoding genes in the sequenced genomes of many species, including Arabidopsis, rice, grapevine and poplar have found variable numbers of NBS-encoding genes2,19,20,21. Based on the structure of the N-terminal domain, NBS-encoding genes have been categorized into two classes, TIR type and non-TIR type. TNLs were absent in monocots, such as rice19,20, the dicot Aquilegia coerulea14 and the dicotyledonous order Lamiales, but were found in most dicots10,45,46. However, only 27 TIR-type genes including 22 TNL genes and five TN genes, were identified in Chinese chestnut. The proportion of TNLs in entire genome of Chinese chestnut was only 0.0577%, which was much lower than that in Arabidopsis (0.348%), grapevine (0.319%), or poplar (0.171%)2,21. Furthermore, the percentage of TNLs among the NBS-LRR genes of poplar (23.6%) was four times to that in Chinese chestnut (5.88%), which also demonstrated that the number of TNLs was lower in Chinese chestnut. In Chinese chestnut, the average number of exons estimated for TNLs (2.50) was lower than that in whole genome genes and non-TNLs, a result that differed from those of previous studies in Arabidopsis, poplar, grapevine, apple, pear and peach2,21,47.
Furthermore, because of the small number of TIR-type NBS-encoding genes, only two Ks values (0.4781 and 0.1623) were estimated for the two TIR gene families. The average Ka/Ks ratio for TIR-type genes, with Ks ranging from 0 to 1, was lower than that for non-TIR-type genes (0.4080 compared with 0.5871). This result was consistent with those of previous studies of TNLs and non-TNLs in Arabidopsis, grapevine, poplar, apple, pear and peach2,21,47. The lower Ka/Ks values for TIR gene families than for non-TIR gene families indicates greater functional constraints during the evolution of TIR gene subfamilies48 and stronger diversifying selection in non-TIR gene families, which could provide variation that would allow plants to adapt to different pathogens in their environment47.
Duplication events of NBS-encoding genes in Chinese chestnut
Gene duplication plays a critical role in the generation of new R genes, increasing the number of these genes and dispersing them in the genome7. The number of NBS-encoding genes in Chinese chestnut was 3- and 1.2-fold that in Arabidopsis and poplar, respectively. To identify duplicated gene pairs, we defined a gene family according to three criteria. Using 70% criterion of both cutoff of coverage and sequence identity of not less than 70%, 64 gene families were detected, including 273 NBS-encoding genes in Chinese chestnut (53.60%) (Table 2). The percentage of NBS-encoding genes occurring in multi-gene families in Chinese chestnut was nearly the same as that in the rice genome (53.7%) and lower than that in the poplar genome (78.1%). Using our 80% criterion of both cutoff of coverage and sequence identity of not less than 80%, 41.43% of NBS genes were found to belong to multi-gene families in Chinese chestnut, while 74.52% of NBS genes occurred in multi-gene families in the poplar genome. However, the proportion of genes in multi-gene families sharply declined to 19.46% when the 90% criterion was used, which showed that relatively recent duplications resulted in a small portion of the multi-gene families in Chinese chestnut.
The number of synonymous substitutions per synonymous site and the frequency distribution of Ks values could be used to infer the age of genome duplications21. The peak and distribution of Ks values was distinct in different species. Ks peaks varied greatly, occurring at Ks values from 0 to 0.1 in grape and poplar and at Ks values from 0.1 to 0.2 in Arabidopsis, apple, pear, peach and Prunus mume, but at Ks values from 0.3 to 0.4 in rice and strawberry21,47. In Chinese chestnut, Ks peaked in the range of values from 0.4 to 0.5 (Fig. 1), a much higher Ks value than that in poplar (Ks = 0–0.1) or in other species, indicating that the gene expansions of Chinese chestnut were more ancient. Furthermore, the Ks values for Chinese chestnut were mainly distributed in a range from 0.2 to 0.5 and were rarely greater than 0.5 (Fig. 1). The fact that 63.94% of multiple NBS-encoding genes were present in the range of Ks values from 0.2 to 0.5, demonstrating that relatively ancient duplications played an important role in the expansion of NBS-encoding genes in Chinese chestnut.
Species-specific duplication driving in the expansion of NBS-encoding genes in Chinese chestnut
Gene duplication has supplied raw genetic material for evolution49 and has been a major force for generating biological novelties that can lead to adaptation to environments50. To elucidate the evolutionary pattern of NBS-encoding genes in Chinese chestnut, a phylogenetic tree of conserved NBS domain from Chinese chestnut and poplar genomes was constructed (Fig. 3). The most distinct characteristic was species-specific duplication events, that might be responsible for the evolution of recognition of species-specific pathogens and that responsed to selective pressure imposed by species-specific pathogens21.
The 401 species-specific NBS-encoding genes detected in Chinese chestnut were classified into 81 clades with bootstrap values were greater than 50%. The proportion of the species-specific duplicates reached 77.26%, suggesting large-scale genes expansion after the divergence of Chinese chestnut and poplar. A similar result has also been reported for four gramineous plants (Zea mays, Sorghum bicolor, Brachypodium distachyon and Oryza sativa)51 and five species in the Rosaceae (Fragaria vesca, Malus × domestica, Pyrus bretschneideri, Prunus persica and Prunus mume)47.
The independent and relatively basal NBS group containing the RPW8 domain
The RPW8 domain (PF05659.6) mediated broad-spectrum pathogen resistance and was originally identified in the protein encoded by the polymorphic powdery mildew resistance locus RPW812. In previous studies, proteins containing the RPW8 domain have been categorized into the non-TIR-NBS-encoding genes subgroup13,14. However, 15 RPW8 domain-encoding genes (1 XNs, 2 TNLs, 3 CNLs and 9 XNLs) clustered into an independent lineage in phylogenetic relationships (Additional file 2: Figure S1). Also, five NBS-encoding-genes from poplar encoding the RPW8 domain clustered together with 15 RPW8-NBS-encoding genes from Chinese chestnut. The clustering of RPW8-NBS-encoding genes observed in Chinese chestnut also occurred with RPW8-NBS-LRR genes from M. truncatula15, potato16, soybean, common bean and pigeon pea17, which illustrated that RPW8-NBS-encoding genes did not comprise a special group of non-TIR genes but formed a separate group independent from the non-TIR genes. Moreover, the group of RPW8-NBS-encoding genes located at a relatively basal but not the most basal position in this phylogenetic tree of Chinese chestnut (Additional file 2: Figure S1). This result is similar to the study in Arabidopsis that a monophyletic group (the CNL-A group) consisted of RNL genes2,17. Thus, the RPW8-NBS-encoding genes were phylogenetically separate from the non-TIR genes.
Materials and Methods
Identification of NBS-encoding genes in Chinese chestnut and classification of gene family members
The entire genome sequence and annotation V1.0 of Chinese chestnut (C. mollissima) were downloaded from the hardwood genomics project (http://www.hardwoodgenomics.org/chinese-chestnut-genome). To identify NBS-encoding genes in Chinese chestnut, we used the amino acid sequence of the NB-ARC domain (PF00931) as blastp query against all known protein sequences with the threshold expectation value set to 1.0. All hits were further submitted to Pfam analysis (http://pfam.xfam.org/) to verify the presence of the NBS (NB-ARC) domain. Furthermore, the identified NBS-encoding genes were examined to detect whether they encode the LRR or TIR domain using the merged results from Pfam analysis and SMART protein motif analysis (http://smart.embl-heidelberg.de/). Finally, all identified genes were examined to detect the presence of CC domain using COILS (http://embnet.vital-it.ch/software/COILS_form.html) databases with a threshold of 0.952 in Chinese chestnut and RPW8 domain using Pfam analysis in Chinese chestnut and poplar.
Genes were grouped into gene families according to two criteria, including the cutoff of coverage (aligned sequence lengths/gene lengths) and the sequence identity of not less than 70%. Likewise, the stricter criteria for both coverage and sequence identity of not less than 80% and 90%, respectively, were used to detect the relatively recent duplications among NBS-encoding genes.
Sequence alignment and phylogenetic analyses
The NBS domain sequences of all identified NBS-encoding genes were aligned in MEGA 5.0 using the MUSCLE program53. A neighbor-joining (NJ) method was then applied to construct a phylogeny of NBS-encoding genes using ClustalW 2.0 with default options and 1000 bootstrap replications54. Data for NBS-encoding genes from poplar (Populus trichocarpa) were obtained from a previous study21.
The ratio of nonsynonymous substitutions to synonymous substitutions
To detect the mode of selection, we evaluated the ratio of nonsynonymous substitutions to synonymous substitutions. Firstly, based on the protein sequences, the CDSs (nucleotide coding sequences) of NBS-encoding genes in each gene family were aligned using ClustalW 2.054. Subsequently, nonsynonymous substitutions (Ka), synonymous substitutions (Ks) and the ratio between them (Ka/Ks) were calculated in each gene family using MEGA 5.053.
Detection of positive selection
The Phylogenetic Analysis by Maximum Likelihood 4 (PAML4) package55 was used for the site model and branch model test to determine selective pressure on NBS-LRR genes in Chinese chestnut. A single dN/dS ratio (model = 0), in addition to models M7 (beta) and M8 (beta-ω) (NS site = 7 8) were used for the site model in all gene families with at least three members. Subsequently, the LR test between model M7 and M8 was carried out with critical criteria of chi-square 5.991 (p < 0.05, df = 2) and 9.210 (p < 0.01, df = 2), respectively. For the branch model, a single dN/dS ratio (model = 0) and model 0 (NS site = 0) were applied in the codeml program.
Promoter regions analysis
To characterize the cis-acting element(s) of NBS-encoding genes, we isolated an approximately 1000 bp promoter sequence of each NBS-encoding gene. Analysis of promoter sequences was conducted using SIGNALSCAN program available in Plant cis-acting regulatory DNA Elements (PLACE) (http://www.dna.affrc.go.jp/PLACE/), a database containing mainly plant motifs extracted from the published reports56.
Conclusions
NBS-LRR genes, as one of the largest families of R genes, were analyzed in Chinese chestnut (Castanea mollissima), a model species for the Fagaceae, to determine their pattern of evolution. In the present study, several TNLs were identified in Chinese chestnut that were absent from monocot genomes. In addition, we found a relatively ancient duplication in Chinese chestnut compared with poplar. The expansion of NBS-encoding genes could be attributed to such species-specific duplications during the evolution of Chinese chestnut. The values for Ka/Ks in all TIR and most non-TIR gene families were less than 1, indicating purifying selection as a leading force in the evolution of NBS-encoding genes. However, the Ka/Ks values for four non-TIR gene families were greater than 1, demonstrating that their evolution was driven by positive selection. Furthermore, the relationship between Ka/Ks and Ks illustrated higher selective pressure on the newer and older genes compared with genes in the critical range of Ks from 0.4 to 0.5. Interestingly, RPW8-NBS-encoding genes clustered into an independent clade at a relatively basal, but not most basal, position in this phylogenetic analysis. Finally, many cis-elements in NBS-encoding genes promoter were related to disease resistance, which demonstrated the function in responsing pathogens and laid the foundation of identifying candidate R genes.
Additional Information
How to cite this article: Zhong, Y. et al. Species-specific duplications of NBS-encoding genes in Chinese chestnut (Castanea mollissima). Sci. Rep. 5, 16638; doi: 10.1038/srep16638 (2015).
References
Dangl, J. L. & Jones, J. D. Plant pathogens and integrated defence responses to infection. Nature 411, 826–833 (2001).
Meyers, B. C., Kozik, A., Griego, A., Kuang, H. & Michelmore, R. W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. The Plant cell 15, 809–834 (2003).
McHale, L., Tan, X., Koehl, P. & Michelmore, R. W. Plant NBS-LRR proteins: adaptable guards. Genome biology 7, 212 (2006).
Jones, J. D. G. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Mcdowell, J. M. & Simon, S. A. Recent insights into R gene evolution. Mol Plant Pathol 7, 437–448 (2006).
Bent, A. F. & Mackey, D. Elicitors, effectors and R genes: the new paradigm and a lifetime supply of questions. Annual review of phytopathology 45, 399–436 (2007).
Liu, J., Liu, X., Dai, L. & Wang, G. Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. Journal of genetics and genomics = Yi chuan xue bao 34, 765–776 (2007).
Yang, S. et al. Rapidly evolving R genes in diverse grass species confer resistance to rice blast disease. P Natl Acad Sci USA 110, 18572–18577 (2013).
Ellis, J. G., Lawrence, G. J., Luck, J. E. & Dodds, P. N. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. The Plant cell 11, 495–506 (1999).
Meyers, B. C. et al. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. The Plant journal: for cell and molecular biology 20, 317–332 (1999).
Rairdan, G. J. et al. The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. The Plant cell 20, 739–751 (2008).
Xiao, S. Y. et al. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291, 118–120 (2001).
Bonardi, V. et al. Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. P Natl Acad Sci USA 108, 16463–16468 (2011).
Collier, S. M., Hamel, L. P. & Moffett, P. Cell Death Mediated by the N-Terminal Domains of a Unique and Highly Conserved Class of NB-LRR Protein. Mol Plant Microbe In 24, 918–931 (2011).
Ameline-Torregrosa, C. et al. Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant physiology 146, 5–21 (2008).
Jupe, F. et al. Identification and localisation of the NB-LRR gene family within the potato genome. BMC genomics 13 (2012). 10.1186/1471-2164-13-75.
Shao, Z. Q. et al. Long-Term Evolution of Nucleotide-Binding Site-Leucine-Rich Repeat Genes: Understanding Gained from and beyond the Legume Family. Plant physiology 166, 217–234 (2014).
Leister, D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends in genetics: TIG 20, 116–122 (2004).
Zhou, T. et al. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol Genet Genomics 271, 402–415 (2004).
Yang, S. H. et al. Genome-wide investigation on the genetic variations of rice disease resistance genes. Plant Mol Biol 62, 181–193 (2006).
Yang, S. H., Zhang, X. H., Yue, J. X., Tian, D. C. & Chen, J. Q. Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Genet Genomics 280, 187–198 (2008).
Fang, G. C. et al. A physical map of the Chinese chestnut (Castanea mollissima) genome and its integration with the genetic map. Tree Genet Genomes 9, 525–537 (2013).
Kuhlman, E. G. Chestnut blight. Science 210, 1199–1200 (1980).
Anagnostakis, S. L. Biological control of chestnut blight. Science 215, 466–471 (1982).
Choupina, A. B., Estevinho, L. & Martins, I. M. Scientifically advanced solutions for chestnut ink disease. Appl Microbiol Biot 98, 3905–3909 (2014).
Williams, I. C. The New Chestnut Bark Disease. Science 34, 397–400 (1911).
Graves, A. H. Resistance In the American Chestnut To the Bark Disease. Science 48, 652–653 (1918).
Gassmann, W., Hinsch, M. E. & Staskawicz, B. J. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. The Plant journal: for cell and molecular biology 20, 265–277 (1999).
Ballvora, A. et al. The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. The Plant journal: for cell and molecular biology 30, 361–371 (2002).
Ernst, K. et al. The broad-spectrum potato cyst nematode resistance gene (Hero) from tomato is the only member of a large gene family of NBS-LRR genes with an unusual amino acid repeat in the LRR region. The Plant journal: for cell and molecular biology 31, 127–136 (2002).
Lokossou, A. A. et al. Exploiting Knowledge of R/Avr Genes to Rapidly Clone a New LZ-NBS-LRR Family of Late Blight Resistance Genes from Potato Linkage Group IV. Mol Plant Microbe In 22, 630–641 (2009).
Hayashi, N. et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. The Plant journal: for cell and molecular biology 64, 498–510 (2010).
Hayes, R. J. et al. The inheritance of resistance to bacterial leaf spot of lettuce caused by Xanthomonas campestris pv. vitians in three lettuce cultivars. Horticulture Research 1 (2014). 10.1038/hortres.2014.66.
Weng, K. et al. Transcriptome of Erysiphe necator-infected Vitis pseudoreticulata leaves provides insight into grapevine resistance to powdery mildew. Horticulture Research 1 (2014). 10.1038/hortres.2014.49.
Qiu, W., Feechan, A. & Dry, I. Current understanding of grapevine defense mechanisms against the biotrophic fungus (Erysiphe necator), the causal agent of powdery mildew disease. Horticulture Research 2 (2015). 10.1038/hortres.2015.20.
Kubisiak, T. L. et al. A transcriptome-based genetic map of Chinese chestnut (Castanea mollissima) and identification of regions of segmental homology with peach (Prunus persica). Tree Genet Genomes 9, 557–571 (2013).
Tuskan, G. A. et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 (2006).
Mondragon-Palomino, M., Meyers, B. C., Michelmore, R. W. & Gaut, B. S. Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res 12, 1305–1315 (2002).
Kimura, M. & Ohta, T. On some principles governing molecular evolution. P Natl Acad Sci USA 71, 2848–2852 (1974).
Hughes, A. L. & Nei, M. Pattern Of Nucleotide Substitution at Major Histocompatibility Complex Class-I Loci Reveals Overdominant Selection. Nature 335, 167–170 (1988).
Yang, Z. H., Wong, W. S. W. & Nielsen, R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol 22, 1107–1118 (2005).
Mazarei, M., Teplova, I., Hajimorad, M. R. & Stewart, C. N. Pathogen phytosensing: Plants to report plant pathogens. Sensors-Basel 8, 2628–2641 (2008).
Park, H. C. et al. Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant physiology 135, 2150–2161 (2004).
Shi, L. et al. Identification of promoter motifs regulating ZmeIF4E expression level involved in maize rough dwarf disease resistance in maize (Zea Mays L.). Mol Genet Genomics 288, 89–99 (2013).
Pan, Q. L. et al. Comparative genetics of nucleotide binding site-leucine rich repeat resistance gene homologues in the genomes of two dicotyledons: Tomato and Arabidopsis. Genetics 155, 309–322 (2000).
Goff, S. A. et al. A draft sequence of the rice genome (Oryza sativa L. ssp japonica). Science 296, 92–100 (2002).
Zhong, Y., Yin, H., Sargent, D. J., Malnoy, M. & Cheng, Z. M. Species-specific duplications driving the recent expansion of NBS-LRR genes in five Rosaceae species. BMC genomics 16 (2015). 10.1186/s12864-015-1291-0.
Cannon, S. B. et al. Diversity, distribution and ancient taxonomic relationships within the TIR and non-TIR NBS-LRR resistance gene subfamilies. J Mol Evol 54, 548–562 (2002).
Zhang, J. Z. Evolution by gene duplication: an update. Trends Ecol Evol 18, 292–298 (2003).
Kanazawa, A., Liu, B. H., Kong, F. J., Arase, S. & Abe, J. Adaptive Evolution Involving Gene Duplication and Insertion of a Novel Ty1/copia-Like Retrotransposon in Soybean. J Mol Evol 69, 164–175 (2009).
Li, J. et al. Unique evolutionary pattern of numbers of gramineous NBS-LRR genes. Mol Genet Genomics 283, 427–438 (2010).
Lupas, A., Van Dyke, M. & Stock, J. Predicting coiled coils from protein sequences. Science 252, 1162–1164 (1991).
Tamura, K. et al. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance and Maximum Parsimony Methods. Mol Biol Evol 28, 2731–2739 (2011).
Larkin, M. A. et al. Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Yang, Z. H. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol 24, 1586–1591 (2007).
Higo, K., Ugawa, Y., Iwamoto, M. & Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic acids research 27, 297–300 (1999).
Acknowledgements
This project was funded by National Natural Science Foundation of China (31501737), the National Agriculture Ministry “948 Project” (#2011-G21), China and by the Priority Academic Program Development of Modern Horticultural Science in Jiangsu Province, China.
Author information
Authors and Affiliations
Contributions
Y.Z. and Y.J.L. conceived and designed this work; Y.J.L. and K.H.H. carried out the bioinformatic analyses; Y.J.L. wrote the paper; Y.Z. and Z.-M.C. modified the paper. All authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhong, Y., Li, Y., Huang, K. et al. Species-specific duplications of NBS-encoding genes in Chinese chestnut (Castanea mollissima). Sci Rep 5, 16638 (2015). https://doi.org/10.1038/srep16638
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep16638
- Springer Nature Limited
This article is cited by
-
Genome-wide analysis of NBS-encoding resistance genes in the Mediterranean olive tree (Olea europaea subsp. europaea var. europaea): insights into their molecular diversity, evolution and function
Tree Genetics & Genomes (2020)
-
Lineage-specific duplications of NBS-LRR genes occurring before the divergence of six Fragaria species
BMC Genomics (2018)
-
Genome-scale examination of NBS-encoding genes in blueberry
Scientific Reports (2018)
-
UGT74S1 is the key player in controlling secoisolariciresinol diglucoside (SDG) formation in flax
BMC Plant Biology (2017)
-
Deep RNA-Seq profile reveals biodiversity, plant–microbe interactions and a large family of NBS-LRR resistance genes in walnut (Juglans regia) tissues
AMB Express (2016)